Telomerase Is a True Reverse Transcriptase. A Review
T. R. Cech,1,2 T. M. Nakamura,1 and J. Lingner3
1Department of Chemistry and Biochemistry, Howard Hughes Medical Institute, University of Colorado, Boulder, Colorado 80309-0215, USA.2To whom correspondence should be addressed.
3Swiss Institute for Experimental Cancer Research, 1066 Epalinges/VD, Switzerland.
Submitted August 15, 1997.
Synthesis of telomeric repeats at chromosome ends requires telomerase, a ribonucleoprotein enzyme. The RNA subunit, which contains the template for DNA synthesis, has been identified in many organisms. Recently, the protein subunit that catalyzes telomeric DNA extension has also been identified in Euplotes aediculatus and Saccharomyces cerevisiae. It has sequence and functional characteristics of a reverse transcriptase related to retrotransposon and retroviral reverse transcriptases, so this new family of telomerase subunits has been named TRT (Telomerase Reverse Transcriptase). We find it remarkable that the same type of protein structure required for retroviral replication is now seen to be essential for normal chromosome telomere replication in diverse eukaryotes.
KEY WORDS: telomere, telomerase, chromosome, DNA replication, reverse transcriptase, Euplotes aediculatus, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Tetrahymena thermophila, Drosophila melanogaster.
It has long been recognized that conventional DNA replication machinery is unable to replicate the very ends of a linear DNA molecule [1, 2]. If the initial chromosome is blunt-ended, the problem becomes one of incomplete lagging strand (discontinuous strand) DNA replication. In contrast, if the starting chromosome has a 3´-terminal single-stranded overhang, as has been documented in ciliated protozoa, yeast, and mammals [3-8], the problem is one of incomplete replication of the leading strand (continuous DNA replication), as described by Lingner et al. [9].
How, then, is the chromosome end maintained? The molecular basis of telomere replication came to light in 1985 with the discovery by Greider and Blackburn of the enzyme telomere terminal transferase or telomerase in Tetrahymena thermophila [10]. They later showed that telomerase was a ribonucleoprotein enzyme, with essential RNA and protein components [11, 12]. By extending the chromosome 3´ end using deoxyribonucleotides as substrates and a sequence within the telomerase RNA subunit as a template, telomerase compensates for the limitations of conventional replication.
Telomerase RNA subunits were later found in other ciliated protozoa [13-16], budding yeast [17, 18], and mammals [19, 20]. The isolation of the genes for RNA subunits allowed the key genetic tests to be performed to show that the telomerase RNA was responsible for dictating the sequence of chromosomal telomeres in vivo [17, 21].
The first demonstrated catalytic subunit of telomerase came from Euplotes aediculatus through standard biochemical purification [22]. Three features of the purification provided considerable confidence that the final product was authentic. 1) The specific activity (incorporation of dTTP per fmol of RNA subunit) was constant from the crude nuclear extract through the purification, showing that the major activity was purified in active form. 2) The native gel electrophoretic mobility of telomerase was similar throughout the purification, suggesting that no major proteins that were associated with telomerase in nuclear extracts were lost (although there was a small increase in mobility at one stage, perhaps due to proteolysis). 3) The yield of the two proposed protein subunits (p123 and p43) relative to the yield of RNA subunit, as well as the estimated molecular mass of the complex, were consistent with a 1:1:1 complex of RNA--p123--p43.
When the gene for the p123 subunit was cloned and sequenced [23], it was found to have sequence similarity to a yeast protein, Est2p (Ever Shorter Telomeres). EST2 had been previously found in a screen for mutants defective in telomerase action [24]. Because p123 but not p43 appears to be a phylogenetically conserved telomerase subunit, the following discussion will focus on this protein and its yeast homolog.
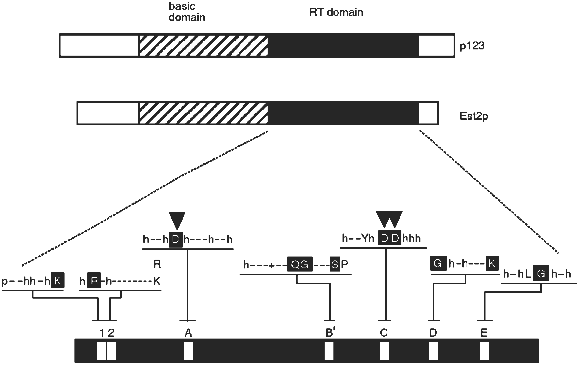
The derived amino acid sequences of the Euplotes p123 and yeast Est2p telomerase proteins revealed reverse transcriptase (RT) motifs. As shown in Fig. 1, these occur in the carboxy terminal half of each protein. Telomerase has for many years been called a "specialized reverse transcriptase" because it synthesizes DNA on an RNA template. The implication of the reverse transcriptase sequence motifs was that the protein structure responsible for telomerase activity is in fact directly related to that of other known reverse transcriptases. However, the sequence similarity between reverse transcriptases is not so extensive as to make a convincing argument by itself. A critical test of the functional importance of the reverse transcriptase sequence motifs was required. This test was performed with the yeast Est2 protein. As described by Lingner et al. [23], single amino acid changes in the aspartic acid residues predicted to be required for reverse transcriptase activity (arrowheads in Fig. 1) resulted in telomere replication defects equivalent to those attained by knocking out the entire EST2 gene. Knocking out the RNA subunit gives the same phenotypes [17]. More specifically, cells containing these mutant proteins underwent senescence (cell division stopped after about 75 generations), and their telomeres became progressively shorter.
Because such a genetic result could reflect either a direct or an indirect involvement of the protein in the replication process, in vitro assays were also performed on extracts from wild-type and mutant cells. Using an assay similar to that described by Cohn and Blackburn [25], it was shown that deletion of the EST2 gene or single amino acid mutations in the critical aspartic acid residues eliminated telomerase activity in extracts [23]. Of the four genes that are required for telomerase activity in vivo, EST1, EST2, EST3, and EST4 [24], only EST2 is required for catalysis of telomere DNA extension in an in vitro assay (J. Lingner, T. R. Cech, T. R. Hughes, and V. Lundblad, manuscript submitted; see also [25]). Presumably Est1p, Est3p, and Est4p associate with the telomere substrate or have some role in regulating or targeting telomerase, but do not form the active site of the enzyme. Alternatively, they might be involved in lagging strand synthesis, which is thought to occur after telomerase action [26].Fig. 1. Telomerase reverse transcriptase subunits from Euplotes (p123) and yeast (Est2p). Each bar represents the protein from N-terminus (left) to C-terminus (right). The reverse transcriptase (RT) domain is shaded and expanded below to indicate the amino acids conserved among reverse transcriptases in general: p, polar; h, hydrophobic; -, nonconserved; +, basic. Capital letter, conserved amino acid; white capital letter in black box, most conserved amino acid. Downward-pointing arrowheads show aspartic acid residues critical for telomerase activity in vivo and in vitro [23].
Because the TRT (Telomerase Reverse Transcriptase) protein subunit was found in two phylogenetically unrelated organisms, Euplotes (a protozoan) and Saccharomyces (a fungus), it appears to be widespread in evolution. In fact, it has recently been found in Schizosaccharomyces pombe and in human [27], and it may turn out to be universal among eukaryotes that have telomerase.
Many of the properties of the telomerase reverse transcriptases are not so different from those of other known reverse transcriptases. Use of a DNA primer for in vivo function has been documented for non-LTR retrotransposon and group II intron RTs [28, 29]. Although those of us who use retroviral reverse transcriptases as tools for copying RNA in the laboratory think of the enzyme as being only transiently associated with its RNA template, a number of reverse transcriptases are found as stable ribonucleoprotein complexes [29, 30]. Furthermore, even HIV reverse transcriptase can be converted into an enzyme that acts in a telomerase-like manner by the simple substitution of Mn2+ ion for the normal Mg2+ ion; under these conditions, the reverse transcriptase remains in a stable complex with its RNA template and repetitively copies a small portion of the template sequence [31].
On the other hand, the TRTs also have features which distinguish them from the previously known reverse transcriptases. They are much larger, have an amino-terminal basic domain, and show an unusually large spacing between motifs A and B´. Even within the RT domain, there are substitutions within amino acids that are highly conserved among all the other RTs. For example, in motif C there are two aspartic acid residues (DD) that serve to coordinate the active site Mg2+ ion [32]. They normally occur in the context (F/Y)hDDh (where h is a hydrophobic amino acid), whereas in the telomerase RTs the local sequence is hxDD(F/Y) (where x is a nonconserved amino acid). This motif occurs in a beta-strand--turn--beta-strand structural element [32], and the amino acid changes are in positions that may shift the critical DD residues in the active site. The functional consequences of this and other differences are yet to be explored.
An exception to chromosome end maintenance by telomerase has been found in Drosophila melanogaster, where telomeres consist of retrotransposable elements [33, 34]. Interestingly, the TART reverse transcriptase thought to be responsible for replication for these non-LTR retrotransposons is closely related to the Euplotes and yeast TRTs. This suggests that D. melanogaster telomere replication may occur by a mechanism that involves similar active sites as used in other eukaryotes which have telomerase.
The recent finding of telomerase reverse transcriptases leads to a model for the molecular constitution of this enzyme. Before presenting the model, it is important to recognize that it is speculative, but fortunately it is subject to experimental test. It is likely that it will need to be revised or refined in the future. Our model is that catalytic activity of telomerase requires only two components, an RNA subunit and the TRT protein subunit. A strong test of this model would be to reconstitute telomerase activity from purified, recombinant RNA and TRT subunits. In our model, other proteins are associated with the TRT--RNA complex in vivo and have important roles other than formation of the catalytic center. Some of these other subunits may be conserved in evolution. For example, two proteins (p80 and p95) are associated with the telomerase RNA of Tetrahymena [35], and p80 has homologs in human, mouse, and rat [36, 37]. In general, there are two ways to reconcile the TRT model with p80 and other proteins found to be associated with telomerase RNA: 1) p80-like proteins may interact in the same complex as TRT, or 2) the telomerase RNA may be alternatively in a complex with TRT and in a complex with a p80-like protein (in those organisms that have such a protein). The function of these other protein subunits is not established. Perhaps they are critical for regulating telomerase or targeting it to chromosome telomeres, and other functions can also be envisioned. On the other hand, it has been proposed that the Tetrahymena p80--p95--RNA complex is a catalytic unit [35], which provides an alternative to our model that TRT will always provide the catalytic active site. Thus, as is common in science, we have a few answers which lead to many more questions.
The work described herein could not have been done without a very fruitful collaboration with Tim Hughes and Victoria Lundblad (Baylor College of Medicine, Houston, Texas), and Andrej Shevchenko and Matthias Mann (European Molecular Biology Laboratory, Heidelberg). Our research on telomerase is supported by the Howard Hughes Medical Institute and a grant from the National Institutes of Health.
LITERATURE CITED
1.Olovnikov, A. M. (1971) Dokl. Akad. Nauk
SSSR, 201, 1496-1499.
2.Watson, J. D. (1972) Nature, 239,
197-201.
3.Klobutcher, L. A., Swanton, M. T., Donini, P., and
Prescott, D. M. (1981) Proc. Natl. Acad. Sci. USA, 78,
3015-3019.
4.Henderson, E. R., and Blackburn, E. H. (1989)
Mol. Cell. Biol., 9, 345-348.
5.Wellinger, R. J., Wolf, A. J., and Zakian, V. A.
(1993) Cell, 72, 51-60.
6.Wellinger, R. J., Ethier, K., Labrecque, P., and
Zakian, V. A. (1996) Cell, 85, 423-433.
7.Makarov, V. L., Hirose, Y., and Langmore, J. P.
(1997) Cell, 88, 657-666.
8.McElligott, R., and Wellinger, R. J. (1997) EMBO
J., 16, 3705-3714.
9.Lingner, J., Cooper, J. P., and Cech, T. R. (1995)
Science, 269, 1533-1534.
10.Greider, C. W., and Blackburn, E. H. (1985)
Cell, 43, 405-413.
11.Greider, C. W., and Blackburn, E. H. (1987)
Cell, 51, 887-898.
12.Greider, C. W., and Blackburn, E. H. (1989)
Nature, 337, 331-337.
13.Shippen-Lentz, D., and Blackburn, E. H. (1990)
Science, 247, 546-552.
14.Romero, D. P., and Blackburn, E. H. (1991)
Cell, 67, 343-353.
15.Lingner, J., Hendrick, L. L., and Cech, T. R.
(1994) Genes Dev., 8, 1984-1998.
16.McCormick-Graham, M., and Romero, D. P. (1995)
Nucleic Acids Res., 23, 1091-1097.
17.Singer, M. S., and Gottschling, D. E. (1994)
Science, 266, 404-409.
18.McEachern, M. J., and Blackburn, E. H. (1995)
Nature, 376, 403-409.
19.Feng, J., Funk, W. D., Wang, S.-S., Weinrich, S.
L., Avilion, A. A., Chiu, C.-P., Adams, R. R., Chang, E., Allsopp, R.
C., Yu, J., Le, S., West, M. D., Harley, C. B., Andrews, W. H.,
Greider, C. W., and Villeponteau, B. (1995) Science,
269, 1236-1241.
20.Blasco, M. A., Funk, W., Villeponteau, B., and
Greider, C. W. (1995) Science, 269, 1267-1270.
21.Yu, G.-L., Bradley, J. D., Attardi, L. D., and
Blackburn, E. H. (1990) Nature, 344, 126-132.
22.Lingner, J., and Cech, T. R. (1996) Proc.
Natl. Acad. Sci. USA, 93, 10712-10717.
23.Lingner, J., Hughes, T. R., Shevchenko, A., Mann,
M., Lundblad, V., and Cech, T. R. (1997) Science, 276,
561-567.
24.Lendvay, T. S., Morris, D. K., Sah, J.,
Balasubramanian, B., and Lundblad, V. (1996) Genetics,
144, 1399-1412.
25.Cohn, M., and Blackburn, E. H. (1995)
Science, 269, 396-400.
26.Cech, T. R., and Lingner, J. (1997) Proc. CIBA
Foundation Symp., No. 211, in press.
27.Nakamura, T. M., Morin, G. B., Chapman, K. B.,
Weinrich, S. L., Andrews, W. H., Lingner, J., Harley, C. B., and Cech,
T. R. (1997) Science, in press.
28.Luan, D. D., Korman, M. H., Jakubczak, J. L., and
Eickbush, T. H. (1993) Cell, 72, 595-605.
29.Zimmerly, S., Guo, H., Eskes, R., Yang, J.,
Perlman, P. S., and Lambowitz, A. M. (1995) Cell, 83,
529-538.
30.Kennell, J. C., Wang, H., and Lambowitz, A. M.
(1994) Mol. Cell. Biol., 14, 3094-3107.
31.Ricchetti, M., and Buc, H. (1996)
Biochemistry, 35, 14970-14983.
32.Kohlstaedt, L. A., Wang, J., Friedman, J. M.,
Rice, P. A., and Steitz, T. A. (1992) Science, 256,
1783-1790.
33.Biessmann, H., Mason, J. M., Ferry, K.,
d’Hulst, M., Vageirsdottir, K., Traverse, K. L., and Pardue, M.-L.
(1990) Cell, 61, 663-673.
34.Levis, R. W., Ganesan, R., Houtchens, K., Tolar,
L. A., and Sheen, F. (1993) Cell, 75, 1083-1093.
35.Collins, K., Kobayashi, R., and Greider, C. W.
(1995) Cell, 81, 677-686.
36.Harrington, L., McPhail, T., Mar, V., Zhou, W.,
Oulton, R., Bass, M. B., Arruda, I., and Robinson, M. O. (1997)
Science, 275, 973-977.
37.Nakayama, J., Saito, M., Nakamura, H., Matsuura,
A., and Ishikawa, F. (1997) Cell, 88, 875-884.