Triple-Site Antigen Capture ELISA for Human Myoglobin Can Be More Effective Than Double-Site Assay
V. A. Nikulina1, E. A. Kizim2, Yu. S. Massino2, O. L. Segal2, M. B. Smirnova2, V. V. Avilov3, D. B. Saprygin3, S. P. Smotrov3, G. I. Kolyaskina1, and A. D. Dmitriev2*
1Mental Health Center, Russian Academy of Medical Sciences, Zagorodnoe Shosse 2/2, Moscow, 113152 Russia; fax: (7-095) 952-89402Institute of Higher Nervous Activity and Neurophysiology, Russian Academy of Sciences, ul. Butlerova 5a, Moscow, 117865 Russia; fax: (7-095) 952-8940; E-mail: dmitr@rcmh.msk.ru
3Association of Medical Laboratory Diagnostics, ul. Barrikadnaya 2, Moscow, 117312 Russia
* To whom correspondence should be addressed.
Received February 4, 1999; Revision received April 8, 1999
Using a panel of monoclonal antibodies against human myoglobin (Mb), we have shown that the sensitivity of antigen-capture ELISA can be significantly increased by simultaneous immobilization of two cooperating capture monoclonal antibodies on a solid phase. This method ("triple-site ELISA") uses three monoclonal antibodies to different epitopes of the same antigen (two capture/one tracer) unlike the traditional double-site assay using one capture and one tracer monoclonal antibody. We developed double- and triple-site ELISA for Mb by varying the capture and tracer monoclonal antibodies. Triple-site assays showed 4-6-fold increase in sensitivity compared to the double-site assays. A model for this effect is suggested; according to the model, in triple-site ELISA, high-affinity cyclic configurations can be formed by an antigen, two capture monoclonal antibodies, and the surface of the solid phase.
KEY WORDS: antigen capture ELISA, triple-site ELISA, monoclonal antibodies, synergistic effect, human myoglobin
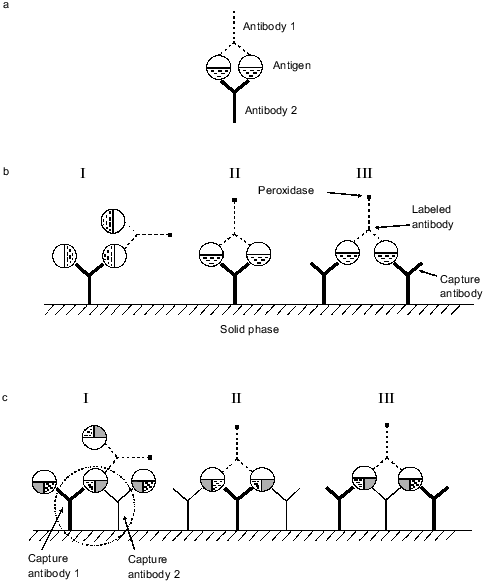
Standard solid-phase ELISA with monoclonal antibodies involves the formation of a complex between antibody adsorbed on a solid phase (capture antibody), antigen, and antibody to another epitope of the antigen conjugated with an enzyme (tracer antibody). Thus, a sandwich is formed: solid phase--capture antibody--antigen--tracer antibody. In the reaction catalyzed by the sandwich, the activity of the antibody-conjugated enzyme is proportional to the antigen concentration in the incubation medium. The standard sandwich method is also called double-site because capture and tracer antibodies bind to different epitopes of the antigen. The affinities of capture and tracer antibodies is the main factor determining the sensitivity of the sandwich method. The sensitivity and resolution of competition analysis in solution (single-phase system) can be significantly enhanced using two monoclonal antibodies to different epitopes of the antigen [1]. We have studied the influence of adsorption on a solid phase of two monoclonal antibodies to different epitopes of myoglobin (with tracer antibody to a third epitope) on sensitivity and resolution of the sandwich method. This modification of the sandwich method is called the triple-site method. It significantly increases sensitivity and resolution versus double-site analysis. Models explaining this phenomenon are discussed.
MATERIALS AND METHODS
1. Purification of human myoglobin. Myoglobin was purified from heart as described previously [2]. The protocol includes ammonium sulfate fractionation and Sephadex G-75 gel-filtration. High performance liquid chromatography is used as the last purification stage. The purified myoglobin migrated as a single band in SDS electrophoresis in 15% polyacrylamide gel in the system of Laemmli [3].
2. Protein assay. Myoglobin concentration was assayed by the Lowry method using bovine serum albumin as the standard. The standard was prepared assuming A1 cm280 nm = 7.6 for a 1% albumin solution [4]. The concentration of purified myoglobin solution and purified antibodies was determined by optical methods. For 1% myoglobin solution, A1 cm280 nm = 17.2 [4] and A1 cm220 nm = 132 [4]. The three methods of myoglobin concentration assay (Lowry and optical density at 280 and 220 nm) gave similar results, the deviation not exceeding 10%. To assay the concentration of purified antibodies, A1 cm280 nm = 14 was used for 1% solution [4].
3. Hybridoma preparation. Hybridomas were prepared by the method of Milstein and Cuello [5] with certain modifications as described previously [6, 7].
4. Assay of anti-myoglobin antibodies. Anti-myoglobin antibodies were determined by solid-phase ELISA. Wells of ELISA plates were saturated with myoglobin overnight at room temperature (5 µg/ml in 0.05 M carbonate buffer, pH 9.5; 100 µl/well). After 4-5 washes with distilled water, the plates were incubated for 1 h at 37°C with culture supernatants diluted 100-1000-fold and with horseradish peroxidase conjugated anti-myoglobin antibody (1:3000 dilution); 100 µl of both components were added per well. Culture supernatants and the conjugate were diluted in 0.025 M sodium phosphate buffer (pH 7.4) containing 0.15 M NaCl (PBS), 0.2% BSA, and 0.05% Tween-20 (ELISA buffer). Color was generated by the reaction with o-phenylenediamine (1 mg/ml in 0.1 M citrate-phosphate buffer, pH 5, containing 0.03% H2O2) at room temperature for 30 min. The reaction was terminated by addition of 50 µl of 0.5 M H2SO4, and absorption at 472 nm was determined using a Titertek Multiscan MC microplate scanner.
5. Purification of monoclonal antibodies. Antibodies were purified by two protocols. To purify antibodies by ion-exchange chromatography, 5-10 ml of ascitic fluid were centrifuged (2000g, 20 min, 0°C) and the supernatant was diluted 5-fold with PBS; antibodies were precipitated with ammonium sulfate (0.4 g/ml). The antibodies were pelleted by centrifugation (2000g, 20 min, 0°C) and dissolved in 0.025 M Tris-HCl buffer, pH 9. The solution was dialyzed against the same buffer overnight and loaded onto a DEAE-Sepharose 4B column (2 × 20 cm) equilibrated with 0.025 M Tris-HCl buffer (pH 9). The column was eluted with a 0-0.4 M NaCl gradient in the same buffer; the gradient volume was 240 ml. Peak fractions were pooled and anti-myoglobin activity was determined as described in section 4.
Affinity purification of anti-myoglobin antibodies was described in detail previously [8].
6. Conjugation of monoclonal antibodies with horseradish peroxidase and determination of efficiency of the conjugates. Antibodies (8 mg/ml in 0.05 M sodium carbonate buffer, pH 9.5) were conjugated with 4 mg of horseradish peroxidase (Rz ~ 3.3) by the periodate method [9]. The conjugate was stored in PBS at -70°C.
To determine relative efficiency of the anti-myoglobin antibody conjugates with horseradish peroxidase, a series of dilutions of the conjugate were prepared from 1:103 to 3:106. Anti-myoglobin activity of the diluted conjugates was determined as described in section 4. The conjugates of all anti-myoglobin antibodies were compared (table). The conjugates significantly differed (by over 10-fold) in color reaction at high dilution (3:106). The conjugate of 14D6 antibody was the most efficient (color developed at dilution exceeding 3:106); conjugates of other antibodies developed color at dilution lower by 10-15-fold. Thus, the 14D6 antibody was used as tracer in this study.
8. Selection of pairs of monoclonal antibodies forming a sandwich with myoglobin (grouping test). Wells of ELISA plates were saturated with antibodies overnight at room temperature (10 µg/ml in 0.05 M carbonate buffer, pH 9.5; 100 µl/well). The plates were washed 4-5 times with distilled water and 50 µl of myoglobin solution was added per well (200 ng/ml) followed by 50 µl of anti-myoglobin antibody conjugate with horseradish peroxidase (dilution from 1:500 to 1:5000; both solutions were prepared in ELISA buffer). Plates were incubated for 2.5 h at 37°C, and peroxidase activity was determined as described above. Detection of peroxidase activity indicates that immobilized (capture) and labeled antibodies bind to different epitopes of myoglobin and form a sandwich: immobilized antibody--myoglobin--labeled antibody. Lack of peroxidase activity indicates that capture and tracer antibodies bind to the same epitope of myoglobin (or adjacent epitopes) and a sandwich is not formed.
9. Sandwich assay of myoglobin. Antibodies were dissolved in 0.05 M carbonate buffer (pH 9.5). Wells of ELISA plates were saturated with antibody of the same clone (10 µg/ml) or with the mixture of antibodies of two clones (5 µg/ml of antibody of each clone) overnight at room temperature (100 µl solution per well). Plates were washed 4-5 times under a stream of distilled water. Incubation medium (final volume 100 µl) contained 25 µl of myoglobin standard (0, 2.5, 5, 10, ..., 320 ng/ml) and 75 µl of anti-myoglobin antibody conjugated with horseradish peroxidase at a working dilution. Plates were incubated for 2.5-3 h at 37°C and, after a series of washes, reacted with o-phenylenediamine as described in section 4.
To compare the sensitivity of the triple-site and double-site sandwich methods, the methods were used for testing of myoglobin in serum. Plates were saturated with individual antibodies or with the mixture of 13G6 and 18H2 antibodies as described above but the final saturation volume was 200 µl. The incubation mixture (final volume 200 µl) included 25 µl of myoglobin standard diluted with myoglobin-free serum [8] and 175 µl of antibody 14D6 conjugated with horseradish peroxidase (final dilution in the incubation medium 1:10,000). Color was developed by addition of 200 µl of o-phenylenediamine solution per well; reaction was terminated by addition of 25 µl of 1.5 M H2SO4 per well.
RESULTS
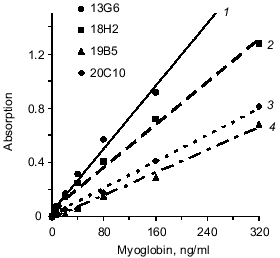
Fusion of splenocytes of immune mice with X63Ag8.653 myeloma resulted in a number of hybridomas; anti-myoglobin activity of produced antibodies was determined in the culture supernatants. Five clones were selected for the study because their anti-myoglobin activity was detected by solid phase ELISA (see section 4 of "Materials and Methods") when the culture supernatants were diluted 100-1000-fold. Antibodies of these clones (after two reclonings) were purified from ascitic fluid by ion-exchange and affinity chromatography. Antibodies of each clone were conjugated with horseradish peroxidase. The sandwich-forming clones were selected as described in section 8 of "Materials and Methods". Antibodies of the sandwich-forming clones can simultaneously bind to the myoglobin molecule without interfering with each other. The data of the table indicate that the selected antibodies bind to three different epitopes of myoglobin. Antibodies 18H2, 19B5, and 20C10 bind to the same epitope (or adjacent epitopes); antibodies 13G6 and 14D6 bind to two other epitopes. Epitopes are designated by Roman numbers in the table. Calibration curves were constructed (as described in section 9 of "Materials and Methods") with all pairs of antibodies which were efficient in the sandwich test (single stage incubation). For example, two calibration curves were constructed with the pair of antibodies 18H2 and 13G6: capture antibody adsorbed on the well 18H2, tracer antibody 13G6; and capture antibody 13G6, tracer antibody 18H2. All variants of calibration curves were constructed in a similar way. The data indicated that antibody 14D6 is the most efficient. The conjugate of this antibody with horseradish peroxidase was used in the sandwich method at a final dilution 1:10000, whereas conjugates of other antibodies were efficient at significantly lower dilution (1:500-1:2000). Calibration curves with labeled antibody 14D6 are shown in Fig. 1. The best characteristic (slope) of the calibration curve was found with capture antibody 13G6; the slope of the calibration curve was slightly less with capture antibody 18H2.
Selection of pairs of monoclonal antibodies forming a sandwich with
myoglobin in solid-phase ELISA
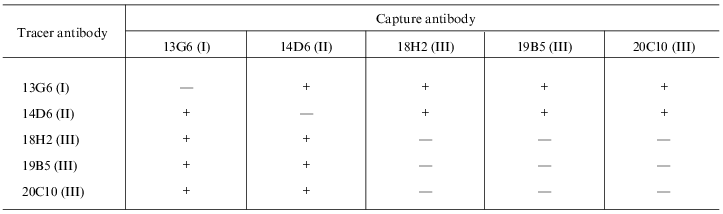
Note: ELISA plates were saturated with antibodies and incubated with myoglobin in the presence of antibodies conjugated with horseradish peroxidase. The plus sign corresponds to the presence of peroxidase activity; the minus sign corresponds to the absence of peroxidase activity. Other conditions are described in section 8 of "Materials and Methods".
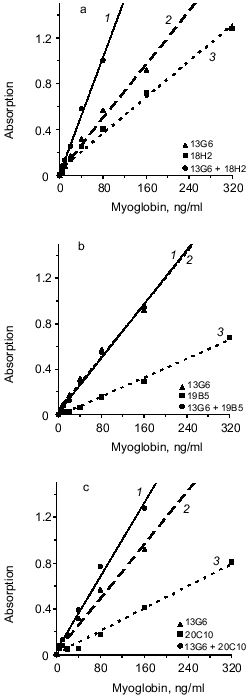
Next, the influence of the mixture of two capture antibodies to different epitopes of myoglobin on sensitivity and resolution of the method was investigated. This method is called triple-site ELISA because three antibodies (two capture and one tracer) raised against different epitopes of the antigen are used. The panel of antibodies enables three variants of the mixture of two capture antibodies to different epitopes: 13G6 + 18H2, 13G6 + 19B5, and 13G6 + 20C10 if tracer antibody 14D6 is used. Calibration curves constructed for plates saturated with antibody of each clone (double-site analysis) were compared versus their mixture (triple-site method). Antibodies were used at similar saturating concentrations; for individual antibodies the concentration was 10 µg/ml and when the mixture of two antibodies was used for saturation, each of them was added at 5 µg/ml. The data of Fig. 2 indicate that when a mixture of two antibodies is adsorbed on the solid phase, the slope of the calibration curve is enhanced versus the double-site variants of the analysis, i.e., when one capture antibody is used. Enhanced calibration curve slope is found in the case of the mixture of antibodies 13G6 + 20C10 (Fig. 2c), but the effect is more pronounced when the mixture of antibodies 13G6 + 18H2 is used (Fig. 2a). For example, at 80 ng/ml myoglobin and the mixture 13G6 + 18H2 as capture antibodies, the color developed corresponds to optical density 1. Under the same conditions, capture antibody 13G6 gives optical density 0.57 and capture antibody 18H2 gives optical density 0.4. In Fig. 2, calibration curves drawn with dotted lines (double-site analysis) should be compared to the calibration curves drawn with solid lines (triple-site method). However, the mixture of capture antibodies 13G6 + 19B5 did not enhance resolution and sensitivity of the method versus the variant when only 13G6 antibody was used as capture (curves 1 and 2 coincide; see Fig. 2b).Fig. 1. Standard calibration curves of solid-phase ELISA of myoglobin constructed by saturation of ELISA plates with various antibodies. Capture antibody: 1) 13G6 antibody; 2) 18H2 antibody; 3) 20C10 antibody; 4) 19B5 antibody; antibody 14D6 was used as the tracer.
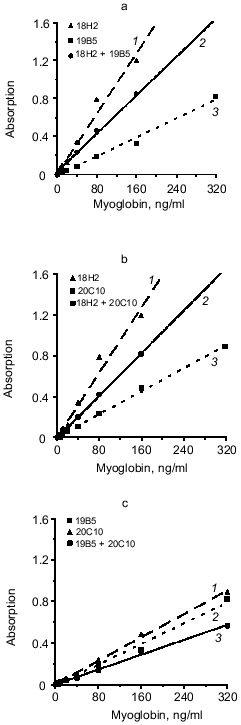
The effect of mixture of two different antibodies with similar epitope specificity adsorbed on the solid phase on the sensitivity of the sandwich method was tested. The hybridoma panel enables analysis of three combinations of the mixture of two capture antibodies to the same or adjacent epitopes of myoglobin: 18H2 + 19B5, 18H2 + 20C10, and 19B5 + 20C10 if the 14D6 antibody is used as the tracer. The data of Fig. 3 (a, b, c) indicate that in all cases the combination of different antibodies to the same epitope does not enhance the slope of the calibration curves (calibration curves with the mixture of capture antibodies are drawn with solid lines and the curves with individual capture antibodies are drawn with dotted lines).Fig. 2. Standard calibration curves of solid-phase sandwich ELISA of myoglobin using ELISA plates saturated with individual antibodies (dotted lines) and the mixtures of two antibodies to different epitopes of the antigen molecule (solid lines). Capture antibody: a) mixture of antibodies 13G6 + 18H2 (1); antibody 13G6 (2); antibody 18H2 (3); b) mixture of antibodies 13G6 + 19B5 (1); antibody 13G6 (2); antibody 19B5 (3); c) mixture of antibodies 13G6 + 20C10 (1); antibody 13G6 (2); antibody 20C10 (3). ELISA plates were saturated with individual antibodies (10 µg/ml) or with the mixture of two antibodies (each at 5 µg/ml). The labeled antibody was 14D6.
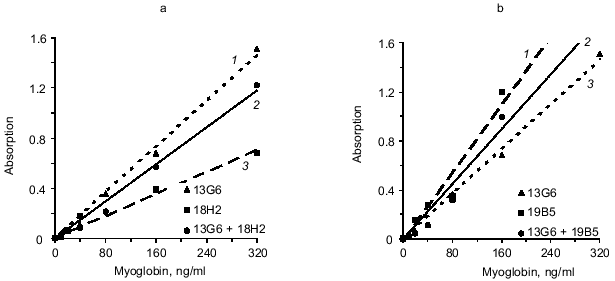
We analyzed another variant of the triple-site sandwich method in which one antibody was used for capture and the tracer was composed of a mixture of two conjugates of antibodies with peroxidase (conjugated antibodies to different epitopes of myoglobin). Antibody 14D6 was adsorbed on the ELISA plate wells and the mixtures of conjugated antibodies (13G6 + 18H2 and 13G6 + 19B5) were used as tracer. Calibration curves constructed with individual labeled antibodies (double-site analysis) were compared to the curves corresponding to the mixture of two labeled antibodies to different epitopes (triple-site method). Labeled antibodies were used at the same effective concentrations. For example, conjugated antibody 13G6 was used at final dilution 1:1000 in the double-site variant and the conjugate of 18H2 antibody was used at dilution 1:500; the mixture included 13G6 at final dilution 1:2000 and 18H2 at final dilution 1:1000. The data of Fig. 4 indicate that the triple-site method using the mixture of two labeled antibodies to different epitopes is not superior to the double-site variant using individual labeled antibodies. The calibration curves constructed using the mixture of labeled antibodies are drawn with the solid lines, and the curves constructed using individual labeled antibodies are drawn with the dotted lines.Fig. 3. Standard calibration curves of solid-phase sandwich ELISA of myoglobin using ELISA plates saturated with individual antibodies (dotted lines) and the mixtures of two antibodies to the same epitope of the antigen molecule (solid lines). Capture antibody: a) antibody 18H2 (1); mixture of antibodies 18H2 + 19B5 (2); antibody 19B5 (3); b) antibody 18H2 (1); mixture of antibodies 18H2 + 20C10 (2); antibody 20C10 (3); c) antibody 20C10 (1); antibody 19B5 (2); mixture of antibodies 19B5 + 20C10 (3). ELISA plates were saturated with individual antibodies (10 µg/ml) or with the mixture of two antibodies (each at 5 µg/ml). The labeled antibody was 14D6.
To evaluate the sensitivity of the triple-site method as compared to the double-site method, the assays were used to determine myoglobin in serum (see section 9 of "Materials and Methods"). Calibration curve standards were diluted with myoglobin-free serum [8]; each standard was run in six parallels and the final volume of the incubation mixture was 200 µl. The variant analyzed included adsorption of the mixture of antibodies 13G6 + 18H2 on the solid phase and detection with 14D6 antibody. The minimal significantly determined myoglobin concentration (mean absorption for zero standard + 3 standard deviations) in the triple-site variant was 6 ng/ml. When antibody 13G6 was used as capture, the sensitivity of analysis was 27 ng/ml. If antibody 18H2 was used as capture, the sensitivity was even lower (40 ng/ml). Thus, in case of the triple-site analysis, the sensitivity was increased by 4.5-fold versus the best variant of the double-site analysis.Fig. 4. Standard calibration curves of solid-phase sandwich ELISA of myoglobin constructed using individual labeled antibodies (dotted lines) and the mixtures of two antibodies to different epitopes of the antigen molecule (solid lines). Tracer antibody: a) antibody 13G6 (1); mixture of antibodies 13G6 + 18H2 (2); antibody 18H2 (3); b) antibody 19B5 (1); mixture of antibodies 13G6 + 19B5 (2); antibody 13G6 (3). The ELISA plates were saturated with 14D6 antibody.
DISCUSSION
Immunization with myoglobin (Mr ~ 18,600) results in the generation of antibodies to five or six regions of the protein (epitopes) [10, 11]. Up to four antibody molecules can bind to the myoglobin molecule in solution [10]. Thus, myoglobin is a suitable model for the study of the sandwich method variant when more than two antibody molecules are bound to the antigen.
In the sandwich method of myoglobin assay, adsorption of two antibodies to different epitopes on a solid phase and use of an antibody to the third epitope (triple-site sandwich) significantly enhances the sensitivity. When the mixture of antibodies 13G6 + 18H2 is adsorbed on a solid phase (labeled antibody 14D6), the sensitivity is enhanced 4.5-fold versus the variant of the method when antibody 13G6 alone is used as capture (from 27 to 6 ng/ml). This enhancement is even more pronounced in the triple-site method when antibody 18H2 is used as capture. The sensitivity is enhanced by 6.5-fold (from 40 to 6 ng/ml). Interestingly, none of the variants including adsorption of the mixture of two different antibodies against the same epitope enhanced the slope of the calibration curve (Fig. 3).
To explain the enhancement of the sensitivity in the triple-site sandwich method, an effect demonstrated in 1981-1983 should be considered. It was shown that if an antigen and the mixture of two antibodies to different epitopes of the antigen are present in solution, stable cyclic complexes are formed similar to those shown in Fig. 5a. The complex includes two antigen and two antibody molecules (reviewed in [1]). The cyclic complex is characterized by significantly higher stability than the simple antigen**--antibody complex because to destroy the cycle simultaneous dissociation of two bonds between the antigen and antibody is required. The mixture of two monoclonal antibodies to different epitopes sometimes increased the sensitivity of competitive radioimmune assay by over an order of magnitude [1, 12]. It was shown that cycles can be formed when antibody to the same epitope of the antigen are bound to a solid phase and antibody to another epitope is soluble [13]. One can suggest that in the double-site variant of the sandwich method employing a pair of monoclonal antibodies to different epitopes, cycles shown in Fig. 5b, II and III, can be formed along with sandwiches shown in Fig. 5b, I. The cyclic complexes include one or two molecules of adsorbed antibody, two antigen molecules, and a molecule of conjugated antibody. Even if a relatively small fraction of the molecules forms the cyclic complexes in the sandwich, this would significantly improve the characteristics of the analysis. Adsorption of the mixture of two monoclonal antibodies to different epitopes of myoglobin on a solid phase can form not only the cyclic complexes characteristic for the double-site variant of the analysis, but also the complexes shown in Fig. 5c. In these complexes, some of the myoglobin molecules simultaneously binds to two antibodies adsorbed on the solid phase forming the cycle: antibody 1--myoglobin--antibody 2 that is linked to the solid phase (outlined by dotted line in Fig. 5c, I). This cycle is characterized by higher stability than the standard sandwich between the adsorbed antibody and antigen because dissociation of two antibody--antigen bonds is required for myoglobin dissociation from the adsorbed antibody. This cycle is rather stable and its presence can explain the enhanced sensitivity of the method when two antibodies to different epitopes are adsorbed on a solid phase. Moreover, apart from the complex with labeled antibody formed in the double-site variant of analysis (Fig. 5b, II and III), complexes shown in Fig. 5c (II and III) can be formed in the triple-site sandwich. This can also enhance the sensitivity of the analysis.
Three variants of the triple-site method were analyzed (same tracer antibody and three combinations of mixtures of two capture antibodies). In two cases, when the mixtures of antibodies 13G6 + 18H2 and 13G6 + 20C10 were used as capture, the sensitivity of the analysis was improved. In a single case (with mixture of antibodies 13G6 + 19B5), the slope of the calibration curve was not improved. It should be noted that sometimes when a mixture of two monoclonal antibodies to different epitopes of an antigen was used in competitive analysis in solution, the sensitivity was not enhanced. Thus, cyclic complexes as shown in Fig. 5a are not always formed [14]. A similar effect was observed when the mixture of antibodies 13G6 + 19B5 is adsorbed on the surface of ELISA plates. These antibodies bind to different epitopes of myoglobin, but the slope of the calibration curve was not enhanced. Apparently, the epitopes of the myoglobin molecule that bind to these antibodies interfere with simultaneous interaction of the antibodies adsorbed on the solid phase.Fig. 5. Complexes of adsorbed antibodies, antigen, and labeled antibody which can be formed in solid-phase ELISA. a) Cyclic complexes formed in solution by two antigen molecules and two molecules of antibodies against different epitopes of the antigen; b) complexes that can be formed by adsorbed antibodies, antigen, and labeled antibodies in the double-site variant of the sandwich method; c) complexes that can be formed by adsorbed antibodies, antigen, and labeled antibodies in the triple-site variant of the sandwich method.
The development of sandwich methods for assay of proteins usually requires a panel of monoclonal antibodies to different epitopes. Adsorption on a solid phase of the mixture of antibodies to two different epitopes and use of tracer antibody to another epitope can enhance the sensitivity of the method.
REFERENCES
1.Thompson, R. G., and Jackson, A. P. (1984)
Trends Biochem. Sci., 9, 1-4.
2.Strausser, H. R., Rothfeld, E. L., and Bucsi, R. A.
(1966) Proc. Soc. Exp. Biol. (N. Y.), 122, 661.
3.Laemmli, U. K. (1970) Nature, 277,
680-685.
4.Fasman, G. D. (ed.) (1976) Handbook of
Biochemistry and Molecular Biology, Vol. II, 3rd ed., CRC Press, p.
383.
5.Milstein, C., and Cuallo, A. C. (1984) Immunol.
Today, 5, 299-304.
6.Massino, Y. S., Kizim, E. A., Dergunova, N. N.,
Vostrikov, V. M., and Dmitriev, A. D. (1992) Immunol. Lett.,
33, 217-222.
7.Massino, Y. S., Dergunova, N. N., Kizim, E. A.,
Smirnova, M. B., Tereshkina, E. B., Kolyaskina, G. I., and Dmitriev, A.
D. (1997) J. Immunol. Meth., 201, 57-66.
8.Nikulina, V. A., Kizim, E. A., Massino, Yu. S.,
Segal, O. L., Smirnova, M. B., Avilov, V. V., Saprygin, D. B., Smotrov,
S. P., Kolyaskina, G. I., and Dmitriev, A. D. (1999) Byul. Eksp.
Biol. Med., in press.
9.Nakane, P. K., and Kawaoi, A. (1974) J.
Histochem. Cytochem., 22, 1084.
10.Atassi, M. Z. (ed.) (1977) Immunochemistry of
Proteins, Plenum Press, New York, p. 1.
11.Juronen, E. I., Viikma, M. H., and Mikelsaar, A.
V. N. (1988) J. Immunol. Meth., 111, 109.
12.Moyle, W. R., Anderson, D. M., and Ehrlich, P. H.
(1983) J. Immunol., 131, 1900.
13.Ehrlich, P. H., Moyle, W. R., Moustafa, Z. A.,
and Canfield, R. E. (1982) J. Immunol., 128, 2709.
14.Moyle, W. R., Lin, C., Corson, R. L., and
Ehrlich, P. H. (1983) Mol. Immunol., 20, 439.