Primary Structure of a 21-kD Protein from Potato Tubers
T. A. Valueva1*, T. A. Revina1, G. V. Kladnitskaya1, V. V. Mosolov1, and P. Mentele2
1Bach Institute of Biochemistry, Russian Academy of Sciences, Leninskii pr. 33, Moscow, 117071 Russia; fax: (095) 954-2732; E-mail: inbio@glas.apc.org2Department of Clinical Chemistry and Clinical Biochemistry, Munich University, Nussbaumstrasse 20, 80336 Munich, Germany
* To whom correspondence should be addressed.
Received June 21, 1999; Revision received August 11, 1999
A 21-kD protein isolated earlier from potato tubers (Solanum tuberosum L.) has two isoforms, with pI 6.3 and 5.2, which were separated by fast protein ion-exchange chromatography on a Mono Q column. The primary structures of the two forms consisted of 187 and 186 amino acid residues. Both isoforms are composed of two polypeptide chains, designated A and B, linked by a single disulfide bond between Cys-146 of the A chain and Cys-7 of the B chain. The amino acid sequences of the A chains of the two forms, consisting of 150 residues each, differ in a single amino acid residue at position 52 (Val --> Ile), while the B chains, containing 37 and 36 residues, respectively, have substitutions at nine positions (Leu-8 --> Ser-8, Lys-25--Asp-26 --> Asn-25--Glu-26, Ile-31--Ser-32 --> Val-31--Leu-32, Lys-34--Gln-35--Val-36--Gln-37 --> Gln-34--Glu-35--Val-36). Both isoforms form stable inhibiting complexes with human leukocyte elastase and are less effective against chymotrypsin and trypsin.
KEY WORDS: primary structure, serine proteinase inhibitor, human leukocyte elastase, trypsin, chymotrypsin, potato tubers
Abbreviations: HLE) human leukocyte elastase; SBTI) Kunitz soybean trypsin inhibitor; FPLC) fast protein liquid chromatography; HPLC) high-performance liquid chromatography; PE) pyridylethyl; PTH) phenylthiohydantoin.
In our previous communications [1-3], we described a 21-kD protein isolated from potato
tubers (Solanum tuberosum L.) that acts as a potent inhibitor of
serine proteinases. It consists of two polypeptide chains (a large A
chain (16.5 kD) and a small B chain (4.5 kD) connected by a disulfide
bond) and contains two independent active sites. One of these is
responsible for interactions with human leukocyte elastase (HLE) or
chymotrypsin and has methionine in the P1 position, while in the other
reactive site, arginine is the P1 residue involved in binding to
trypsin [3]. We have shown that the 21-kD protein
can form a triple complex with one molecule of chymotrypsin (or HLE)
and one molecule of trypsin [3]; this shows the two
active sites do not interfere with each other. N-Terminal sequencing
(21 amino acid residues) allowed us to assign this protein to the
Kunitz soybean trypsin inhibitor superfamily (SBTI) [1, 2]. However, only one reactive
site involved in binding both trypsin and chymotrypsin is
characteristic of this superfamily [4]. An
inhibitor of cathepsin D and trypsin from potato tubers (PCDI) [5] and chymotrypsin inhibitor from the winged bean
Psophocarpus tetragonolobus (WCI) [6] are
the only exceptions. PCDI contains two active sites, one of which is
involved in interactions with an aspartic proteinase (cathepsin D),
while the other binds trypsin [5]. In common with
the 21-kD protein, the WCI molecule is composed of two polypeptide
chains linked by a disulfide bond, and it binds simultaneously two
molecules of chymotrypsin [7]. It should be noted
that the structure of the active sites in these inhibitors is still
unknown.
The primary structures of Kunitz soybean trypsin inhibitors are characterized by a high number of conserved amino acid residues evenly distributed along the polypeptide chain [8]. The cysteine residues and the active site amino acid residues are most conserved [9]. Complete sequencing of the 21-kD protein inhibitor of serine proteinases from potato tubers is necessary for its more precise classification and for the location and determination of the primary structure of the active sites.
In this work, we report the primary structure and specificity of two isoforms of the 21-kD protein from potato tubers, detected by isoelectrofocusing in polyacrylamide gels.
MATERIALS AND METHODS
The 21-kD protein was isolated from potato tubers by precipitation with ammonium sulfate, gel-filtration on Sephadex G-75, and ion-exchange chromatography on DEAE-cellulose, as described earlier [1].
Multiple forms of the 21-kD protein were separated by fast protein liquid chromatography (FPLC) on a Mono Q HR 5/5 column (Pharmacia, Sweden) equilibrated with 20 mM Tris-HCl buffer, pH 8.0. The proteins were eluted with a linear NaCl gradient (from 0 to 0.3 M) in the above-mentioned buffer. The proteins were rechromatographed under the same conditions; the protein fractions were desalted and freeze-dried.
The inhibitors were assayed with relevant enzymes (HLE, trypsin, and alpha-chymotrypsin). Proteinase activity was measured by the rate of hydrolysis of chromogenic substrates: N-Suc-L-Ala-L-Ala-L-Val-pNa (for HLE [10]), N,alpha-Bz-L-Arg-pNa (BAPA, for trypsin [11]), and N-Suc-L-Val-L-Pro-L-Phe-pNa (for chymotrypsin [12]).
The concentration of active enzymes was determined by titrating their active sites: trypsin, with p-nitrophenyl-p´-guanidine benzoate [13]; chymotrypsin, with N-trans-cinnamoylimidazole [14], HLE, with alpha1-antitrypsin, in which the concentration of the active inhibitor was determined by titration with trypsin with a known concentration of active sites. The concentration of inhibitors was determined by titration of each proteinase used, taking into account that the enzyme and the inhibitor form an equimolar complex [1, 3]. To calculate the apparent values of equilibrium dissociation constants (Ki) of the inhibitors with proteinases, nonlinear regression analysis using the Enzfitter program was used [15].
The A and B chains were separated after oxidizing the protein with performic acid [16] or reducing it with beta-mercaptoethanol in 6 M guanidine chloride and subsequent alkylation with beta-vinylpyridine [17].
Oxidized or S-pyridylethylated (S-PE) A chains were digested with trypsin treated with the chloromethyl ketone of N,alpha-tosyl-DL-phenylalanine in 25 mM Tris-HCl buffer, pH 8.0, for 2 h at 37°C. S-PE-A chains and the native protein were digested with pepsin in 5% formic acid for 2.5 h at room temperature. To modify Lys residues, S-PE-B chains were treated with phenyl isothiocyanate, and phenylthiohydantoin (PTH) derivatives were hydrolyzed with trypsin as described.
The A and B chains of the protein and all peptides were separated by reversed-phase high-performance liquid chromatography (HPLC) on a Superspher RP 8 column (125 × 2 mm, Merck, Germany). The A and B chains were separated in a linear acetonitrile gradient (from 0 to 60%) in 0.1% trifluoroacetic acid. Peptides were eluted with a linear gradient from 100% of solvent A to 40% of solvent A and 60% of solvent B (solvent A is 0.1% trifluoroacetic acid in water (v/v); solvent B is 0.1% trifluoroacetic acid in acetonitrile (v/v)). In all instances, the flow rate was 0.3 ml/min. Proteins and peptides were detected at 206, 254, and 280 nm using a HP 1040A spectrophotometer (Hewlett-Packard, USA). Separated oxidized or S-PE-chains of the 21-kD protein were desalted by RP-HPLC on an Aquapore RP 300 column (2.1 × 30 mm, Applied Biosystems, USA).
Amino acid sequences were determined by automatic Edman degradation [18] using a model 473 A gas-phase sequencer (Applied Biosystems). PTH-derivatives of amino acids were identified by RP-HPLC.
The molecular masses of proteins and peptides were measured by mass spectrometry on a API II mass spectrometer (Sciex, Thornhill, Canada). Average molecular masses were calculated from spectra containing multiple charged ions [19]. The instrument was calibrated with ions of ammonium and polypropylene glycol adduct.
Isoelectrofocusing in a pH gradient from 3 to 9 was conducted in polyacrylamide slabs (4 × 5 cm) on a PhastSystem apparatus (Pharmacia, Sweden) according to the manufacturers' protocols. Gels were stained with 0.4% aqueous AgNO3 [20].
The inhibitors studied in this work were assayed with the following enzymes: HLE (EC 3.4.21.36), trypsin (EC 3.4.21.4), and alpha-chymotrypsin (EC 3.4.21.1) containing 85, 51, and 81% of active sites, respectively (Sigma, USA); trypsin and pepsin of sequencing grade for protein digestion were purchased from Boehringer Mannheim (Germany). Other reagents were as follows: p-nitrophenyl-p´-guanidinobenzoate, N-trans-cinnamoylimidazole, and alpha1-antitrypsin containing 49.5% of active protein (Sigma); Suc-L-Ala-L-Ala-L-Val-pNa, BAPA, and N-Suc-L-Val-L-Pro-L-Phe-pNa (Bachem, Switzerland).
RESULTS
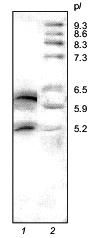
SDS-electrophoresis and N-terminal sequencing demonstrated the homogeneity of the 21-kD protein from potato tubers consisting of two polypeptide chains (A and B) linked by a disulfide bond [1]. However, isoelectrofocusing in a pH gradient from 3 to 9 showed that it consists of two major components with pI 6.3 and 5.2 (Fig. 1).
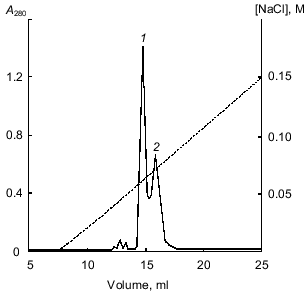
The 21-kD protein was subjected to ion-exchange chromatography (FPLC) on a Mono Q column at pH 8.0. The proteins retained on the column were eluted with a linear NaCl gradient (0-0.3 M) in 20 mM Tris-HCl buffer, pH 8.0. Figure 2 shows that two major fractions were eluted at NaCl concentrations of 0.06 and 0.07 M, respectively. These fractions were further purified by FPLC under the same conditions. Isoelectrofocusing in a pH gradient from 3 to 9 showed that each fraction contained only one protein band: the pI of the protein eluted at 0.06 M NaCl was 6.3, while that of the protein eluted at 0.07 M NaCl was 5.2. Mass spectrometry showed that the molecular masses of the proteins with pI 6.3 and 5.2 were 20.262 and 20.174 kD, respectively. Thus, the molecular masses of both proteins were similar and close to 21 ± 1 kD determined for the 21-kD protein by SDS-electrophoresis [1, 2] and gel chromatography [3]. Furthermore, these data indicated that the isoform with pI 6.3 had at least one more amino acid residue than the isoform with pI 5.2.Fig. 1. Isoelectrofocusing of the 21-kD protein from potato tubers in a polyacrylamide gel: 1) 21-kD protein (4 µg); 2) standard proteins (trypsinogen, lectin B, bovine myoglobin, horse myoglobin, human carbonic anhydrase B, conalbumin, beta-lactalbumin A; pI values are given on the right); pH gradient is from 3 to 9.
Oxidation with performic acid cleaved each protein into two fragments, which were separated by RP-HPLC (data not shown). This indicated that both forms of the 21-kD protein were composed of two polypeptide chains linked by a disulfide bond. The molecular masses of the separated A and B chains of the form with pI 6.3 were 16.082 and 4.182 kD and those of the form with pI 5.2 were 16.068 and 4.082 kD, respectively, as determined by mass spectrometry. These values were very close to those of the native proteins (see above).Fig. 2. FPLC separation of 21-kD protein isoforms with pI 6.3 (1) and 5.2 (2) on a Mono Q column.
S-PE-A and S-PE-B chains were obtained by reduction with beta-mercaptoethanol, alkylation with vinylpyridine, and subsequent RP-HPLC separation.
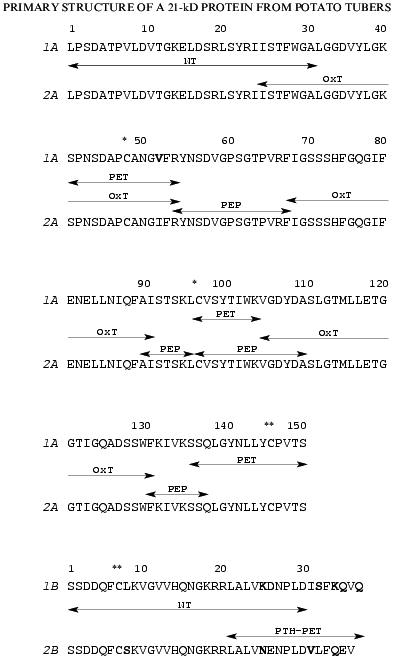
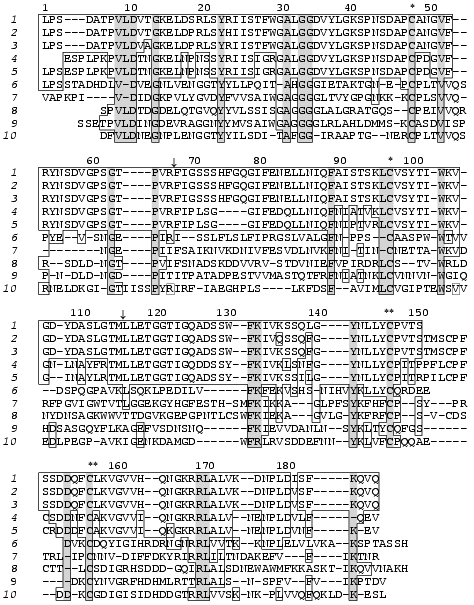
The strategy used for determination of amino acid sequences of the isolated chains of both proteins is shown in Fig. 3. It included N-terminal sequencing of S-PE-A and S-PE-B chains (NT) and sequencing of peptides obtained by enzymatic digestion of: a) S-PE-A chains, with trypsin (PET) or pepsin (PEP); b) A-chains obtained by protein oxidation with performic acid, with trypsin (OxT); and c) S-PE-B chains modified with phenyl isothiocyanate, with trypsin (PTH-PET).
N-Terminal sequencing allowed us to identify 32 amino acid residues in A and 30 residues in B chains of both isoforms. The N-terminal amino acid sequences (32 amino acid residues) of the A chains of both forms were identical, while the B chains differed at positions 8, 25, and 26 (Fig. 3).
The peptides obtained by enzymatic hydrolyses were separated by RP-HPLC; their amino acid composition and amino acid sequences were determined by automatic Edman degradation.Fig. 3. Amino acid sequences of two isoforms of the 21-kD protein from potato tubers: 1A and 1B are the A and B chains of the 21-kD protein with pI 6.3; 2A and 2B are the A and B chains of the 21-kD protein with pI 5.2. The fragments used for amino acid sequencing are marked with arrows: NT, N-terminal amino acid sequence; OxT, tryptic peptides generated from oxidized proteins; PET and PEP, peptides produced by tryptic and peptic hydrolyses of A chains of S-PE-proteins, respectively; PE-PTHT, tryptic peptides of PTH-derivatives of B chains of S-PE-proteins. * and **, disulfide bonds between Cys-48 and Cys-97 of the A chains and Cys-146 of A-chains and Cys-7 of B chains (divergent amino acid residues are given in bold).
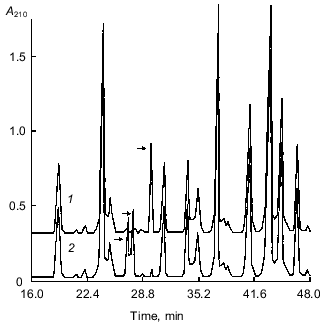
The peptide maps obtained by RP-HPLC separation of the tryptic peptides of S-PE-A chains of both isoforms of the 21-kD protein are shown in Fig. 4. As seen from the figure, the elution profiles of the peptides of both forms are similar and differ only in three fractions, two of which are present in the form with pI 6.3 and one in the form with pI 5.2. Sequencing of peptides marked with arrows in Fig. 4 showed that the unique fraction found in a tryptic hydrolyzate of the form with pI 5.2 (Fig. 4, curve 1) contained one peptide with the following sequence: SPNSDAPCANGIFR. However, both fractions specific to the isoform with pI 6.3 (Fig. 4, curve 2) contained a single 14-membered peptide, whose sequence was SPNSDAPCANGVFR; it differed from the above-mentioned sequence by a single substitution of Ile-12 for Val-12. These peptides correspond to position 41-54 in the 21-kD protein sequence (Fig. 3). Accordingly, the sequences of A chains of both forms differ only in the amino acid residue at position 52: Val in the peptide with pI 6.3 is replaced by Ile in the peptide with pI 5.2.
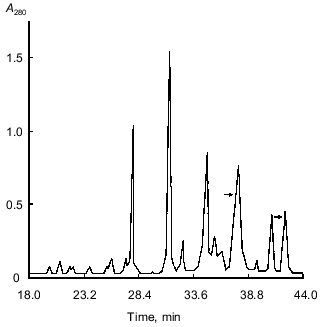
The peptide maps obtained by RP-HPLC separation of the tryptic fragments of S-PE-B chains of the two forms differed significantly and were not further analyzed. This indicated that the major differences in amino acid sequences of the two forms were concentrated in just this region.Fig. 4. Peptide maps obtained by RP-HPLC separation of tryptic peptides of S-PE-A chains of two isoforms of the 21-kD protein from potato tubers with pI 5.2 (1) and 6.3 (2). Divergent peptides are marked with arrows.
Amino acid analysis of the B chains of the two isoforms showed that they contained two Arg residues located at positions 19 and 20, as determined by N-terminal sequencing (Fig. 3). This allowed us to selectively digest the S-PE-B chains at Arg residues after modification of epsilon-amino groups of lysine with phenyl isothiocyanate. The peptides produced, designated PTH-PET in Fig. 3, allowed us to reconstruct the amino acid sequences of the B chains of both forms. The sequence of the isoform of B chain with pI 6.3 differed from that of the isoform with pI 5.2 in nine positions: Leu-8 --> Ser-8, Lys-25--Asp-26 --> Asn-25--Glu-26, Ile-31--Ser-32 --> Val-31--Leu-32, Lys-34--Gln-35--Val-36--Gln-37 --> Gln-34--Glu-35--Val-36.
Using the peptide sequences, we determined the complete amino acid sequences of both forms of the 21-kD protein, each of which consists of two polypeptide chains linked by a disulfide bond (Fig. 3). The forms with pI 6.3 and 5.2 contain 187 and 186 amino acid residues, respectively; the A chains of both forms have equal number of residues (150) while the B chains differ by one residue (37 and 36 amino acid residues, respectively).
To locate disulfide bonds, the native 21-kD protein with pI 6.3
was digested with pepsin, fragments formed were separated by RP-HPLC
(Fig. 5), and their N-terminal sequences were
determined. Only in two fragments (marked with arrows in Fig. 5), two different N-terminal amino acid sequences were
identified. Amino acid analysis showed that these fragments contained
two half-cystine residues. After oxidation with performic acid and
RP-HPLC rechromatography, each fragment generated two peptides, whose
sequences were determined by automatic Edman degradation (Fig. 5). The peptides eluted at 37 and 42 min (Fig. 5) had N-terminal sequences as follows:
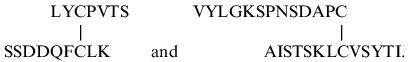
The position of these fragments in the 21-kD protein sequence (Fig. 3) indicates that the first disulfide bridge between Cys-48 and Cys-97 closes a large intrachain peptide loop in chain A, while the other interchain disulfide bridge is formed by Cys-146 of chain A and Cys-7 of chain B.
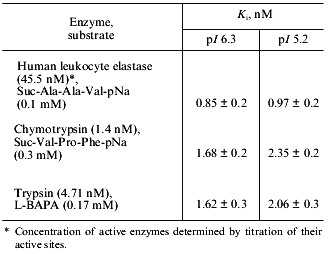
We have also studied the effect of both forms of the 21-kD protein on the activity of a number of proteolytic enzymes. We have shown that both forms are inactive against subtilisin, plant cysteine proteinases (papain, bromelain, and ficin), and aspartic proteinases (pepsin and cathepsin D), but inhibit HLE, chymotrypsin, and trypsin. Therefore, we examined the effect of increasing concentrations of both isoforms on the activity of these serine proteinases. Using the equation for slow formation of the stable enzyme--inhibitor complex [21], we calculated the inhibition constants (Ki) for each target enzyme (see the table). As it is seen from this table, both isoforms of the 21-kD protein are potent inhibitors of HLE and are less effective against chymotrypsin and trypsin; their interactions with all proteinases tested are characterized by similar Ki values.Fig. 5. RP-HPLC separation of the peptic peptides of the native 21-kD protein with pI 6.3 from potato tubers. Disulfide-containing peptides are marked with arrows.
Inhibition of target serine proteinases by two isoforms of the 21-kD
protein from potato tubers with pI 6.3 and 5.2
DISCUSSION
The 21-kD protein isolated from potato tubers consists of two isoforms having pI 6.3 and 5.2. Each isoform is composed of two polypeptide chains linked by a disulfide bond--the large A chain (150 amino acid residues) representing N-terminal region and the small B chain (37 and 36 amino acid residues) comprising C-terminal region. The A chains of both forms have identical N-terminal amino acid sequences, except for one substitution at position 52, while the B chains differ in nine amino acid residues (Fig. 3). One molecule of each form has two disulfide bonds. One of them is formed by homologous Cys residues (Cys-48--Cys-97) closing large peptide loops in the A chains; the second disulfide bond connects two polypeptide chains via Cys-146 of chain A and Cys-7 of chain B. Both isoforms of the 21-kD protein are potent inhibitors of HLE, chymotrypsin, and trypsin and have similar Ki values for these enzymes. Therefore, it seems reasonable to suggest that the isoforms of the 21-kD protein from potato tubers with pI 6.3 and 5.2, which share high sequence homology (95%) and display similar specificity towards target proteinases, are isoinhibitors encoded by two alleles at the same locus.
The sequence of the isoform with pI 6.3 was compared with the available sequences using the Gen Bank NCBI (National Center of Biotechnological Information, USA) database [22]. These data are shown in Fig. 6. The 21-kD protein reveals high sequence homology with several plant proteins of the SBTI (Kunitz soybean trypsin inhibitor) superfamily [5, 6, 23-29]. In the primary structure of the proteins presented in Fig. 6, several amino acid residues characteristic of this superfamily are conserved. However, the amino acid sequence of the 21-kD protein is closer to the proteins of potato tubers: a proteinase inhibitor (pKEN14-28) and two inhibitors of cathepsin D and trypsin (PIGENI and PCDI) [5, 23, 24]. The primary structures of the 21-kD protein with pI 6.3, pKEN14-28, PIGENI, and PCDI have 96 and 79% sequence homology, respectively. However, the 21-kD protein differs from these inhibitors in the existence of two chains and in its enzyme specificity. Thus, in contrast to pKEN14-28 acting as a specific trypsin inhibitor [23], the 21-kD protein is effective against HLE, chymotrypsin, and trypsin. In contrast to PIGENI and PCDI, the 21-kD protein does not inhibit aspartic proteinases [5, 24].
Thus, these findings confirm our previous suggestion [1, 2] that the 21-kD protein, whose molecule consists of two chains linked by a disulfide bond, belongs to the subfamily of potato Kunitz-type proteinase inhibitors (PKPIs) [23].Fig. 6. Comparison of amino acid sequences of the 21-kD protein with pI 6.3 (1) from potato tubers with other proteins of the Kunitz inhibitor family (SBTI): 2) pKEN14-28, a proteinase inhibitor from potato tubers [23]; 3, 4) PIGENI and PCDI, inhibitors of cathepsin D and trypsin from potato tubers [24, 5]; 5) the inhibitor of aspartic proteinases induced by mechanical injury of potato leaves or by their treatment with abscisic acid [25]; 6) WCI, a chymotrypsin inhibitor from winged bean Psophocarpus tetragonolobus (L.) DC [6]; 7) the inhibitor of serine proteinases from latex of melon tree Carica papaya L. [26]; 8) a 21-kD protein, the trypsin inhibitor from Theobroma cacao L. seeds [27]; 9) sporamin A, a trypsin protein inhibitor from Ipomoea batatas L. [28]; 10) SBTI [29]. Amino acid residues are numbered according to the sequence of the 21-kD protein from potato tubers; identical amino acid residues are boxed; conserved residues are hatched; * and **, disulfide bonds Cys-48--Cys-97 and Cys-146--Cys-157 (P1 residues of the reactive sites of the inhibitors are marked with arrows).
It is noteworthy that two-chained proteins have not been found previously among PKPIs. However, the Kunitz-type inhibitors in the leguminous plants of the subfamily Mimosoideae are composed of two polypeptide chains with molecular masses of 16 and 5 kD held together by a disulfide bond, which is located in the C-terminal region between residues 125 and 155 [6, 30, 31]. The chymotrypsin inhibitor WCI from winged bean belongs to this group [6]; it shows high sequence homology (up to 30%) with the 21-kD protein (Fig. 6).
In our previous works [2, 32], we showed that the 21-kD protein inhibited hyphal growth and development of zoospores in a phytopathogenic microorganism Phytophthora infestans. We also demonstrated that this protein was induced in potato tubers upon infection with Ph. infestans. It is, therefore, of particular interest that the primary structure of the 21-kD protein is very similar (up to 76% of sequence homology) to that of the inhibitor of aspartic proteinase induced in potato leaves after mechanical injury or treatment with abscisic acid [25]. The 21-kD protein is also highly homologous to stress proteins accumulated in willow and poplar leaves after mechanical wounding (up to 35 and 50% of sequence homology, respectively); the first protein is a trypsin inhibitor, while the second does not manifest any inhibitory activity [33, 34]. This may indicate that both forms of the 21-kD protein (or at least one of them) are involved in defense mechanisms in potato.
The Kunitz-type inhibitors are so-called canonical inhibitors of serine proteinases, which interact with enzymes by a substrate-type mechanism [35]. On the surface of such inhibitors, there is a specific structure, a binding loop, containing a peptide P1--P1´ bond, which forms the active site of the inhibitor [36].
As mentioned above, the molecule of the 21-kD protein contains two independent active sites, one of which is responsible for binding trypsin and has Arg as the P1 residue, while the other site binds HLE (or chymotrypsin) and has Met in this position [3]. Based on sequence homology of the 21-kD protein with the SBTI family (Fig. 6), we conclude that the peptide bond Arg-67--Phe-68 is located in the first active site of the inhibitor, which is encompassed in the peptide loop between Cys-48--Cys-97. It should be noted that the sequences around the active site peptide bond located between P3--P2´ residues of the active site are identical in the inhibitors from potato tubers shown in Fig. 6. Therefore, the occurrence of Thr-64--Pro-65 in P3--P2 positions seems significant because they are supposed to play an essential role in the formation of the specific structure of the binding loop in canonical inhibitors of serine proteinases [37].
A molecule of the 21-kD protein has a single methionine residue at position 115. This provides evidence that the peptide bond between Met-115 and Leu-116 is located in the second active site of the inhibitor, which is responsible for binding HLE (or chymotrypsin). In this region, the amino acid sequences between Thr-114 and Leu-117 (Fig. 6) in the inhibitors from potato tubers are identical. We assume that the disulfide bond between Cys-146 and Cys-157 connecting the A and B chains of the 21-kD protein forms a canonical binding loop involving the peptide bond Met-115--Leu-116, which forms the second active site of the inhibitor. The formation of such structure is supported by the ability of the 21-kD protein to form triple complexes with target proteinases, in which one molecule of the inhibitor binds simultaneously one molecule of trypsin and one molecule of chymotrypsin [3]. However, further investigations of formation mechanisms and structure of complexes between the 21-kD inhibitor and enzymes binding to the second reactive site are necessary.
This work was supported in part by the Russian Foundation for Basic Research (grant No. 98-04-48191).
REFERENCES
1.Valueva, T. A., Revina, T. A., and Mosolov, V. V.
(1997) Biochemistry (Moscow), 62, 1375-1384.
2.Valueva, T. A., Revina, T. A., Kladnitskaya, G. V.,
and Mosolov, V. V. (1998) FEBS Lett., 426, 131-134.
3.Valueva, T. A., Revina, T. A., and Mosolov, V. V.
(1999) Biochemistry (Moscow), 64, 1074-1078.
4.Carcia-Olmedo, F., Salcedo, G., Sanchez-Monge, R.,
Gomez, L., Royo, J., and Carbonero, P. (1987) Oxford Surv. Plant
Mol. Cell Biol., 4, 275-334.
5.Maganja, D. B., Strukelj, B., Pungercar, J.,
Gubensek, K., Turk, V., and Kregar, I. (1992) Plant Mol. Biol.,
20, 311-313.
6.Odani, S., Yokogawa, Y., Takeda, H., and Abe, S.
(1996) Eur. J. Biochem., 241, 77-82.
7.Shibata, H., Hara, S., and Ikenaka, T. (1988) J.
Biochem., 104, 537-543.
8.Valueva, T. A., and Mosolov, V. V. (1999)
Fiziol. Rast., 46, 307-321.
9.Mosolov, V. V., and Valueva, T. A. (1993) Plant
Protein Inhibitors of Proteolytic Enzymes [in Russian], Bach
Institute of Biochemistry, Russian Academy of Sciences, Moscow.
10.Wenzel, H. R., Engelbrecht, S., Reich, H.,
Mondry, W., and Tschesche, H. (1980) Hoppe-Seyler's Z. Physiol.
Chem., 361, 1413-1416.
11.Erlanger, B. F., Kokowsky, N., and Cohen, E.
(1961) Arch. Biochem. Biophys., 96, 271-278.
12.Powers, J. C., Tanaka, T., Haper, J. W.,
Minematsu, Y., Barker, L., Lincoln, D., Grumley, K. V., Fraki, J. E.,
Schecter, N. M., Lazarus, G. G., Nakajima, K., Nakashino, K., Neurath,
H., and Woodbury, R. G. (1985) Biochemistry, 24,
2048-2059.
13.Chase, T., and Shaw, E. (1970) Meth.
Enzymol., 19, 20-27.
14.Shonbaum, G. R., Lerner, B., and Bender, M. Z.
(1961) J. Biol. Chem., 236, 2930-2935.
15.Knight, C. G. (1986) Proteinase Inhibitors
(Barrett, A. J., and Salvesen, G., eds.) Elsevier, Amsterdam, pp.
23-51.
16.Moore, S. (1963) J. Biol. Chem.,
238, 235-237.
17.Friedman, M., Krull, L. H., and Cavins, J. F.
(1970) J. Biol. Chem., 245, 3868-3871.
18.Sollner, C., Mentele, R., Eckerskorn, C., Fritz,
H., and Sommerhoff, C. P. (1994) Eur. J. Biochem., 219,
937-943.
19.Mann, M., Meng, C. K., and Fenn, F. B. (1989)
Anal. Chem., 61, 1702-1708.
20.Heukeshoven, J., and Dernick, R. (1985)
Electrophoresis, 6, 103-112.
21.Morrison, J. F. (1982) Trends Biochem.
Sci., 7, 102-115.
22.Instructions to Authors (1997) Nucleic
Acids, 25, No. 1.
23.Ishikawa, A., Ohta, S., Matsuoka, K., Tsukaho,
H., and Nakamura, K. (1994) Plant Cell Physiol., 35,
303-312.
24.Krizaj, M., Drobnic-Kosorok, J., Brzin, J.,
Jerata, R., and Turk, V. (1993) FEBS Lett., 333,
15-20.
25.Hildmann, T., Ebneth, M., Pena-Cortes, H.,
Sanchez-Serrano, J. J., Willmitzer, L., and Prat, S. (1992) Plant
Cell, 4, 1157-1170.
26.Habu, Y., Peyachoknagul, S., Umemoto, K., Sakata,
Y., and Ohno, T. (1992) J. Biochem., 111, 249-258.
27.Tai, H., McHenry, L., Fritz, P. J., and Furtek,
D. B. (1991) Plant Mol. Biol., 16, 913-915.
28.Hattori, T., Yoshida, N., and Nakamura, K. (1989)
Plant Mol. Biol., 13, 563-572.
29.Hoffman, L. M., Sengupta-Gopalanm, C., and
Paaren, H. E. (1984) Plant Mol. Biol., 3, 111-117.
30.Odani, S., Ono, K., and Ikenaka, T. (1979) J.
Biochem., 86, 1795-1805.
31.Souza-Pinto, J. G., Oliva, M. L. V., Sampaio, C.
A. M., Damas, J., Auerswald, E. A., Limaos, E., Fritz, H., and Sampaio,
M. (1996) Immunopharmacology, 33, 330-332.
32.Valueva, T. A., Kladnitskaya, G. V., Il'inskaya,
L. I., Gerasimova, N. G., Ozeretskovskaya, O. L., and Mosolov, V. V.
(1998) Bioorg. Khim., 24, 346-349.
33.Saarikiski, P., Clapham, D., and von Arnold, S.
(1996) Plant Mol. Biol., 31, 465-478.
34.Bradshaw, H. D., Jr., Hollick, J. B., Parsons, T.
J., Clarke, H. R., and Gordon, M. P. (1990) Plant Mol. Biol.,
14, 51-59.
35.Laskowski, M., Jr., and Kato, I. (1980) Ann.
Rev. Biochem., 49, 593-626.
36.Bode, W., and Huber, R. (1992) Eur. J.
Biochem., 204, 433-451.
37.Apostoluk, W., and Otlewski, J. (1998)
Proteins, 32, 459-474.