Mini-REVIEW: Novel Function of the Eukaryotic Polypeptide-Chain Releasing Factor 3 (eRF3/GSPT) in the mRNA Degradation Pathway
S. Hoshino*, N. Hosoda, Y. Araki, T. Kobayashi, N. Uchida, Y. Funakoshi, and T. Katada
Department of Physiological Chemistry, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo 113-0033, Japan; fax: + 81 3-5841-4751; E-mail: hoshino@mol.f.u-tokyo.ac.jp* To whom correspondence should be addressed.
Received June 3, 1999
The mammalian GTP-binding protein GSPT, whose carboxy-terminal sequence is homologous to the eukaryotic elongation factor EF1alpha, binds to the polypeptide chain releasing factor eRF1 to function as eRF3 in translation termination. However, the amino-terminal domain of GSPT, which contains a prion-like sequence, is not required for the binding. Instead, the amino-terminal domain is capable of binding to the carboxy-terminal domain of polyadenylate-binding protein (PABP), whose amino terminus is associating with the poly(A) tail of mRNAs, presumably for their stabilization. Interestingly, multimerization of PABP with poly(A), which is ascribed to the action of its carboxy-terminal domain, was completely inhibited by the interaction with the amino-terminal domain of GSPT. This may facilitate shortening of the poly(A) tail of mRNAs by an RNase. Thus, GSPT/eRF3 appears to function not only as a stimulator of eRF1 in the translation termination but also as an initiator of the mRNA degradation machinery. Further physiological and cell biological approaches will be necessary to show whether our current in vitro findings on GSPT/eRF3 indeed reflect its bifunctional properties in living cells.
KEY WORDS: translation termination, mRNA degradation, GSPT, eRF, poly(A)-binding protein
Abbreviations: GSPT) GTP-binding protein apparently essential for the G1-to-S phase transition of the cell cycle (initially termed GST); IF, EF, RF) initiation, elongation, and release factors involved in protein synthesis, respectively; PABP) polyadenylate-binding protein.
THE DISCOVERY OF GSPT AS A YEAST CELL-CYCLE REGULATOR
The yeast GSPT gene, whose product is a GTP-binding protein structurally related to the translation elongation factor EF1alpha, was first isolated based on its ability to complement a temperature-sensitive gst1 mutation of Saccharomyces cerevisiae [1]. Since DNA synthesis in this mutant was substantially arrested at the non-permissive temperature, the GSPT gene product appears to play an essential role at the G1 to S phase transition in the yeast cell cycle. On the other hand, SUP35 was cloned by another group investigating omnipotent suppressor mutant of S. cerevisiae, and the gene turned out to be identical to GSPT [2].
Omnipotent suppressor is a class of nonsense suppressors that is recessive and effective towards three types of nonsense codons [3]. Mutations in the GSPT/SUP35 gene were shown to increase the level of translational ambiguity [4, 5], suggesting that this gene product might also function as a positive regulator of translational accuracy in yeast. However, the relationship between the yeast cell-cycle control and translational regulation by the GSPT/SUP35 gene product remained to be determined.
We previously cloned a human homologue of the yeast GSPT gene and two mouse GSPT genes, the counterpart of human GSPT1 and a novel member of the GSPT gene family, GSPT2 [6, 7]. The mammalian GSPT1 and GSPT2 proteins could associate with a eukaryotic polypeptide-chain releasing factor, eRF1, to function as eRF3, though the two GSPTs were distinct from each other in terms of their amino-terminal sequences, the expression during cell-cycle progression, and tissue distribution [7]. It thus appears that there is functional conservation of this family from yeast to mammals in terms of translation termination.
However, we recently found that GSPTs may have other function by interacting with PABP that binds to the 3´-poly(A) tail of eukaryotic mRNAs [8]. In this brief report, we will discuss the possibility that GSPT can function as an initiator of the mRNA degradation machinery in addition to its involvement in translation termination as eRF3 [9].
STRUCTURE OF EUKARYOTIC GSPT/eRF3
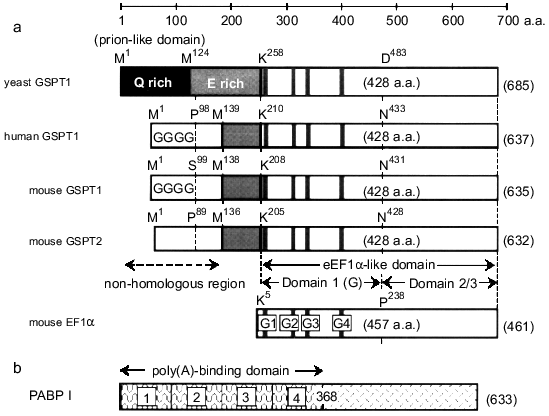
The cDNAs of eukaryotic polypeptide-chain releasing factors (GSPT/eRF3) have been cloned from yeast to human species [1, 2, 6, 7, 9, 10], and their primary sequences reveal that this family may structurally belong to an EF1alpha-type GTP-binding protein. As shown in Fig. 1, GSPT/eRF3 consists of at least two regions characterized as an amino-terminal region (204-257 amino acids), which is unique in this family and specifies the subtypes (at least 1 or 2), and a conserved EF1alpha-like domain (428 amino acids). There are prion-like and E-rich domains in the amino-terminal region of this family, and human and mouse GSPT1s have a G-repeat structure (GGGG) in their amino-terminal regions. Since EF1alpha is expected to be structurally related to prokaryotic elongation factor Tu [11], the EF1alpha-like domain of GSPT/eRF3 may be further divided into three subdomains--domain 1 (or G domain) involved in GTP binding (G1-G5) [12] and carboxy-terminal domains 2 and 3.
In addition to the differences in amino acid sequences, mouse proteins GSPT1 and GSPT2 were clearly distinct from each other in terms of expression during cell-cycle progression and tissue distribution [7]. In contrast to GSPT1, the expression of GSPT2 gene was constant during the cell-cycle progression of 3T3 cells, and was relatively abundant in mouse brain. Although the existence of two forms of GSPT/eRF3 has been reported only in mouse at the present time, our previous study on the chromosome mapping of GSPT1 gene suggested the existence of a homologous gene on the X chromosome [13], and this may represent the locus of human GSPT2 gene.Fig. 1. Schematic representation of the primary structure of the GSPT family and PABP I. a) The GSPT family consists of at least two regions characterized as an amino-terminal non-homologous region (204-257 amino acids) and a conserved EF1alpha-like domain (428 amino acids). The EF1alpha-like domain is further divided into domain 1 (G domain) involved in GTP binding (G1-G5) and C-terminal domains 2 and 3. There are prion-like and Glu (E)-rich domains in the amino-terminal region of this family, and human and mouse GSPT1s have a G-repeat structure (GGGG) in their amino-terminal regions. b) PABP I consists of two regions--four RNA-binding domains (1-4) in tandem repeat at the amino-terminal side and the carboxy-terminal portion.
IDENTIFICATION OF GSPT AS eRF3
In eukaryotic protein synthesis, all three types of termination codons are directly recognized by a polypeptide chain releasing factor, eRF1, to release a synthesized polypeptide chain from the ribosome [14]. Based on the analogy of prokaryotic translation termination system, an additional GTP-binding releasing factor (eRF3) was expected to be present in the ribosomal binding of eRF1. The first demonstration that GSPT functions as eRF3 came from studies in S. cerevisiae [15] and Xenopus laevis [9]. The product of the GSPT1/SUP35 gene appeared to biochemically form a binary complex with SUP45p, which was another member of omnipotent suppressors to function as eRF1. These findings allowed us to postulate that mammalian GSPT may also function as eRF3.
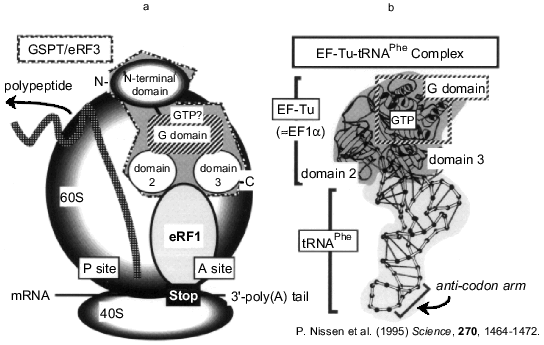
In previous papers [7, 8], we reported that mammalian proteins GSPTs are capable of binding to eRF1 in yeast two-hybrid and biochemical in vitro binding assays. The carboxy-terminal region of GSPT/eRF3 was essentially required for the interaction; GSPT2 deleted with the carboxy-terminal domains 2 and 3 failed to bind eRF1. Moreover, GSPT/eRF3 lacking the unique amino-terminal region could bind to eRF1, indicating that the carboxy-terminal site of the EF1alpha-like domain (presumably domains 2 and 3) constitutes an eRF1-binding region. This is consistent with the finding that there was no significant difference between the two subtypes of GSPT in binding to eRF1 [7]. The same results were reported by other groups [16, 17], and functional interactions were demonstrated by Frolova et al. [18]. Although further structural analyses are required for the detailed picture of the two termination factors, one can speculate an interaction model (Fig. 2) that is comparable with the elongation system; eRF1 may structurally mimic the stem of an aminoacyl-tRNA, whereas GSPT/eRF3 may mimic the function of EF1alpha or the prokaryotic elongation factor Tu, which carries the aminoacyl-tRNA in its GTP-bound forms to the A site of the ribosome. The carboxy-terminal domains 2 and 3 (possibly consisting of two beta-barrel structures) of GSPT/eRF3 protein might be responsible for the GTP-dependent interaction with eRF1, as observed in the association between Tu and aminoacyl-tRNA [19-21].
Fig. 2. A structural model for GSPT/eRF3 and the crystal structure of the EF-Tu--tRNA complex. A possible structure of GSPT/eRF3 (a) is illustrated based on the crystal structure of EF-Tu--tRNA complex (b) (see [19]). The EF1alpha-like domain of GSPT family has an amino acid sequence homologous to EF-Tu. The domains 2 and 3 may constitute an eRF1-binding region.
IDENTIFICATION OF GSPT/eRF3 AS A PABP-BINDING PROTEIN
The existence of the unique amino-terminal region of GSPT/eRF3, that was not involved in the association with eRF1, allowed us to speculate that this family may have another function. To investigate molecules interacting with the GSPT family, we screened a human T cell lymphoma cDNA library in a yeast two-hybrid assay system [8]. The screening resulted in the identification of polyadenylate-binding protein (PABP) I as a binding protein to the amino terminus of GSPT/eRF3. As shown in Fig. 1b, PABP I contains four RNA-binding domains in tandem repeat at its amino-terminal site; these domains bind to the 3´-poly(A) tail of mRNAs, probably for their stabilization and/or translocation from nuclei to cytoplasm (for review see [22, 23]). The carboxy-terminal domain of PABP is suggested to contribute to multiple, regularly spaced organization of the RNA-binding protein on the poly(A) tail [24].
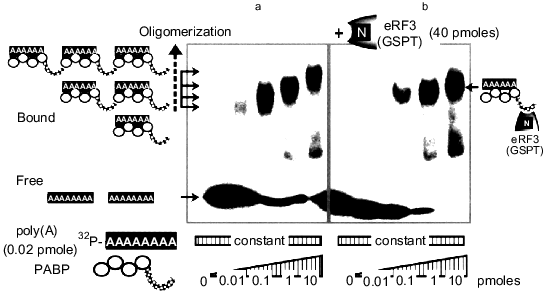
Further analysis with the varying lengths of GSPT and PABP in the two-hybrid assay system indicated that the interaction was mediated through the amino-terminal region of GSPT and the carboxy-terminal domain of PABP (see Fig. 4 later). The functional significance of this interaction was also estimated by means of in vitro binding and gel mobility-shift assays. As shown in Fig. 3, 5´-32P-labeled, synthetic poly(A) RNA binds to PABP, resulting in the formation of a retarded band. There was a progressive reduction of the mobility of the complex indicative of more than one copy of PABP on the labeled poly(A) as the amount of the RNA-binding protein was increased (Fig. 3a). However, in the presence of the amino-terminal domain of GSPT (Fig. 3b), a discrete transition rather than a gradual reduction of the mobility of the complex was observed. It thus appeared that only one complex forms at saturating concentrations of PABP in the presence of GSPT; its mobility was lower than that of single complex of poly(A)--PABP due to its association with GSPT amino-terminal domain. These results indicate that PABP could no longer interact with another PABP to form a multiple, regularly spaced complex on the poly(A) tail when its carboxy-terminus was associated with GSPT. This seems consistent with the previous finding that PABP lacking the carboxy-terminal domain displays the same properties [25].
Fig. 3. Effect of amino-terminal region of GSPT on interaction between PABP and poly(A) mRNA.The indicated amounts of PABP I (up to 10 pmoles) were incubated with (b) or without (a) the amino-terminal domain (amino acids 1-204) of GSPT2 protein in the presence of 0.02 pmole (23 mers) of 32P-labeled poly(A) RNA in 20 µl of reaction mixture. The reaction mixture was electrophoresed on 6% polyacrylamide gels under non-denaturing conditions.
POSSIBLE MECHANISM FOR THE SIGNAL TRANSDUCTION MEDIATED THROUGH GSPT/eRF3
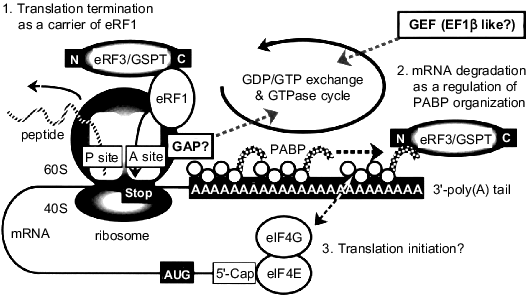
Based on these results, we would like to propose a possible mechanism for the signal transduction mediated through GSPT/eRF3 as follows (Fig. 4). A GTP-bound form of GSPT/eRF3 would bind to a ribosome with eRF1 via its carboxy-terminal EF1alpha-like domain when the A site is occupied by a termination codon (step 1). Although the precise mechanism of translation termination has not been fully elucidated, the release of synthesized polypeptide from the P site might be coupled to GTP hydrolysis on GSPT/eRF3. In this regard, the presence of an intrinsic activity of GTPase-activating protein (GAP) has recently been reported in the ribosome [18, 21]. The GDP-bound GSPT/eRF3 thus formed may dissociate from the ribosome and associate with the carboxy-terminal domain of PABP via its amino terminus. This new interaction should cause destabilization of PABP associated with the 3´-poly(A) tail, leading to the degradation by ribonuclease(s) of the mRNA recruited for the translation (step 2). In other words, a signal from each translation-termination cycle in the ribosomal A site would be transferred to the 3´-poly(A) tail of mRNA as a degradation signal by the action of GSPT/eRF3. This idea may be supported by the findings that repetitive translation shortens the 3´-poly(A) tail of mRNA and that the high expression of PABP gene can extend the lifetime of mRNA [26-29]. In this relation, Czaplinski et al. have quite recently reported another but similar function of eRF1/eRF3 [30]. They demonstrated that the product of the UPF1 gene, which is an RNA-dependent ATPase containing RNA helicase activity, interacts with eRF1/eRF3 in a nonsense mutation-mediated mRNA decay pathway. The interaction of the UPF1 gene product with the releasing factors appears to enhance the termination and degrade the aberrant mRNAs.
Moreover, it has recently been reported that the poly(A)-binding domains of PABP can associate with the translation initiation factor eIF4G, which binds to the 5´-cap structure of mRNA through eIF4E to promote the translation initiation (for review see [22, 23]). Thus, the signal from translation-termination cycle might also regulate the new initiation step via the eRF3/PABP/eIF4G-signaling cascade (step 3). Finally, the GDP-bound form of GSPT/eRF3 would be returned to its GTP-bound form probably through the action of a GDP--GTP exchange factor (GEF). This GEF may be structurally related to EF1beta because of the sequence similarity between GSPT/eRF3 and EF1alpha. Clearly, further experimentation will be necessary to show whether our current in vitro findings on GSPT/eRF3 indeed reflect its multi-functional properties in living eukaryotic cells.Fig. 4. A proposed model for the functions of GSPT/eRF3 in eukaryotic protein synthesis. When the A site of ribosome is occupied by a stop codon, GSPT/eRF3 carries eRF1 for the translation termination via its carboxy-terminal EF1alpha-like domain (step 1). eRF3/GSPT also associates with PABP via its amino-terminal region for the destabilization of mRNA (step 2) and/or the initiation of new translation cycle (step 3). See text for further explanation. GAP, GTPase-activating protein; GEF, GDP--GTP exchange factor.
This work was supported in part by research grants from the “Research for the Future” Program of the Japan Society for the Promotion of Science (JSPS-RFTF 96L00505) and the Scientific Research Fund of the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
1.Kikuchi, Y., Shimatake, H., and Kikuchi, A. (1988)
EMBO J., 7, 1175-1182.
2.Kushnirov, V. V., Ter-Avanesyan, M. D., Telkov, M.
V., Surguchov, A. P., Smirnov, V. N., and Inge-Vechtomov, S. G. (1988)
Gene, 66, 45-54.
3.Surguchov, A. P. (1988) Trends Biochem.
Sci., 13, 120-123.
4.Surguchov, A. P., Beretetskaya, Y. V., Fominykch,
E. S., Pospelova, E. M., Smirnov, V. N., Ter-Avanesyan, M. D., and
Inge-Vechtomov, S. G. (1980) FEBS Lett., 111,
175-178.
5.Eustice, D. C., Wakem, L. P., Wilhelm, J. M., and
Sherman, F. (1986) J. Mol. Biol., 188, 207-214.
6.Hoshino, S., Miyazawa, H., Enomoto, T., Hanaoka,
F., Kikuchi, Y., Kikuchi, A., and Ui, M. (1989) EMBO J.,
8, 3807-3814.
7.Hoshino, S., Imai, M., Mizutani, M., Kikuchi, Y.,
Hanaoka, F., Ui, M., and Katada, T. (1998) J. Biol. Chem.,
273, 22254-22259.
8.Hoshino, S., Imai, M., Kobayashi, T., Uchida, N.,
and Katada, T. (1999) J. Biol. Chem., 274,
16677-16680.
9.Zhouravleva, G., Frolova, L., Le Goff, X., Le
Guellec, R., Inge-Vechtomov, S., Kisselev, L., and Philippe, M. (1995)
EMBO J., 14, 4065-4072.
10.Basu, J., Williams, B. C., Li, Z., Williams, E.
V., and Goldberg, M. L. (1998) Cell Motil. Cytoskeleton,
39, 286-302.
11.Riis, B., Rattan, S. I. S., Clark, B. F. C., and
Merrick, W. C. (1990) Trends Biochem. Sci., 15,
420-424.
12.Bourne, H. R., Sanders, D. A., and McCormick, F.
(1991) Nature, 349, 117-127.
13.Ozawa, K., Murakami, Y., Eki, T., Yokoyama, K.,
Soeda, E., Hoshino, S., Ui, M., and Hanaoka, F. (1992) Somatic Cell
Mol. Genet., 18, 189-194.
14.Frolova, L., Le Goff, X., Rasmussen, H. H.,
Cheperegin, S., Drugeon, G., Kress, M., Armen, I., Haenni, A. L.,
Celis, J. E., Philippe, M., Jastesen, J., and Kisselev, L. (1994)
Nature, 372, 701-703.
15.Stansfield, I., Jones, K. M., Kushnirov, V. V.,
Dagkesamanskaya, A. R., Poznyakovski, A. I., Paushkin, S. V., Nierras,
C. R., Cox, B. S., Ter-Avanesyan, M. D., and Tuite, M. F. (1995)
EMBO J., 14, 4365-4373.
16.Merkulova, T. I., Frolova, L. Y., Lazer, M.,
Camonis, J., and Kisselev, L. L. (1999) FEBS Lett., 443,
41-47.
17.Ebihara, N., and Nakamura, Y. (1999) RNA,
5, 739-750.
18.Frolova, L. Y., Simonsen, J. L., Merkulova, T.
I., Litvinov, D. Y., Martensen, P. M., Rechinsky, V. O., Camonis, J.
H., Kisselev, L. L., and Justesen, J. (1998) Eur. J. Biochem.,
256, 36-44.
19.Nissen, P., Kjeldgaad, M., Soren, T., Galina, P.,
Reshetnikova, L., Clark, B. F. C., and Nyborg, J. (1995)
Science, 270, 1464-1472.
20.Ito, K., Ebihara, K., Uno, M., and Nakamura, Y.
(1996) Proc. Natl. Acad. Sci. USA, 93, 5443-5448.
21.Frolova, L., Le Goff, X., Zhouravleva, G.,
Davydova, E., Philippe, M., and Kisselev, L. (1996) RNA,
2, 334-341.
22.Gallie, D. R. (1998) Gene, 216,
1-11.
23.Sachs, A. B., Sarnow, P., and Hentze, M. W.
(1997) Cell, 89, 831-838.
24.Sachs, A. B., Davis, R. W., and Kornberg, R. D.
(1987) Mol. Cell. Biol., 7, 3268-3276.
25.Kuhn, U., and Pieler, T. (1996) J. Mol.
Biol., 256, 20-30.
26.Bernstein, P., and Ross, J. (1989) Trends
Biochem. Sci., 14, 373-377.
27.Jackson, R. J., and Standart, N. (1990)
Cell, 62, 15-24.
28.Bernstein, P., Peltz, S. W., and Ross, J. (1989)
Mol. Cell. Biol., 9, 659-670.
29.Wormington, M., Searfoss, A. M., and Hurney, C.
A. (1996) EMBO J., 15, 900-909.
30.Czaplinski, K., Ruiz-Echevarria, M. J., Paushkin,
S. V., Han, X., Weng, Y., Perlick, H. A., Dietz, H., Ter-Avanesyan, M.
D., and Peltz, S. W. (1998) Genes Dev., 12,
1665-1677.