REVIEW: Molecular Analysis of Polyphosphate Accumulation in Bacteria
A. Kuroda* and H. Ohtake
Department of Molecular Biotechnology, Graduate School of Advanced Sciences of Matter, Hiroshima University, Higashi-Hiroshima, Hiroshima 739-8527, Japan; fax: 81-824-22-3758; E-mail: akuroda@ipc.hiroshima-u.ac.jp* To whom correspondence should be addressed.
Received October 4, 1999
The dynamic behavior of inorganic polyphosphate (polyP), its accumulation and disappearance, is the most striking aspect of polyP metabolism in bacteria. Imbalance between polyP synthesis and degradation results in fluctuations of polyP by 100- to 1000-fold. We here review recent results with respect to this polyP metabolism in bacteria. PolyP accumulation in response to amino acid starvation, accompanied by increased levels of stringent factors, has been observed in Escherichia coli. Inhibition by stringent factors of polyphosphatase interrupts the dynamic balance between the synthesis and degradation of polyP, accounting for polyP accumulation. Polyphosphate kinase is required for activation of intracellular protein degradation, which is required for adaptation at the onset of amino acid starvation. The adaptation to amino acid starvation is mediated by the network of stringent response and polyP metabolism. PolyP accumulation independent of stringent response has also been observed. Novobiocin, an inhibitor for DNA gyrase, stimulated accumulation of polyP but not that of stringent factors. However, a temperature-sensitive DNA gyrase mutant did not exhibit polyP accumulation at the non-permissive temperature. Antagonistic relationship of polyP to nucleic acid synthesis, explored by Harold, appears to be more complicated. We discuss relationship of Pi regulation to polyP accumulation in E. coli and Klebsiella aerogenes. A function of polyP as an in vivo phosphagen affecting polyP accumulation is also discussed.
KEY WORDS: polyphosphate, polyphosphate kinase, Escherichia coli, stringent response, guanosine tetraphosphate, adaptation, amino acid starvation, protein degradation, novobiocin, Klebsiella aerogenes, phosphate starvation, overplus, activated sludge
Abbreviations: Pi) inorganic phosphate; polyP) inorganic polyphosphate; PPK) polyphosphate kinase; ppGpp) guanosine 5´-diphosphate 3´-diphosphate; pppGpp) guanosine 5´-triphosphate 3´-diphosphate; Mops) morpholinopropane sulfonate; Nov) novobiocin.
Biologically synthesized polyphosphate (polyP) is a linear polymer of
inorganic phosphate (Pi) residues linked by high-energy
phosphoanhydride bonds [1]. Many bacteria are
capable of accumulating Pi in the form of polyP. The
principal enzyme responsible for the synthesis of polyP in
Escherichia coli is polyP kinase (PPK), which uses the
gamma-Pi of ATP to extend the polymer [2]. E. coli possesses an exopolyphosphatase
(PPX) which is highly processive in catalyzing the hydrolysis of
terminal residues of long chain polyP to Pi [3]. In bacteria, it has been demonstrated that polyP
accumulation occurs under conditions of nutritional imbalance [4]. Accumulation and disappearance of polyP in E.
coli are dynamic fluctuations of 100- to 1000-fold [5]. The most striking aspect of polyP metabolism is
this variability of the polyP content, consequently affecting many
cellular functions. PPK is highly conserved in many bacterial species
[6]. Strong DNA/DNA hybridization signals can be
detected from many sludge bacteria by using the E. coli ppk gene
as a DNA probe. To understand polyP metabolism is also practically very
important. Currently available methods for removing Pi from
wastewater rely primarily on polyP accumulation of sludge bacteria [7]. However, because of the complexity of activated
sludge and lack of knowledge about polyP metabolism, these processes
operate essentially by the “black-box” principle and often
lack predictability and stability.
PolyP ACCUMULATES IN RESPONSE TO AMINO ACID STARVATION
Serine hydroxamate, an analog of serine, is used for induction of amino acid starvation [8]. Levels of polyP changed from 1 to 15 nmol/mg protein within 15 min in response to amino acid starvation [9]. This accumulation was observed in many microorganisms including Klebsiella aerogenes and Pseudomonas aeruginosa. Amino acid starvation may be a common signal for polyP accumulation. However, no significant changes in the activities of PPK or PPX were observed in crude cell extracts during amino acid starvation [9]. The stringent response is a pleiotropic physiological response elicited by a failure of the capacity for tRNA aminoacylation to keep up with the demands of protein synthesis [8]. Stringent factors, guanosine 5´-diphosphate 3´-diphosphate (ppGpp), and guanosine 5´-triphosphate 3´-diphosphate (pppGpp), accumulate during stringent response [8]. Mutants that fail to produce stringent factors (e.g., relA and spoT) are deficient in polyP accumulation in response to amino acid starvation. The relationship of stringent factors to polyP accumulation deserves special attention. The stringent factors had no influence on the activities of PPK. However, the effects of these compounds on PPX activity were strikingly inhibitory. At 100 µM, pppGpp inhibited PPX by 90% [9]. Inhibition of PPX by stringent factors was consistent with a behavior as a competitive inhibitor (Ki values of 10 µM for pppGpp and 200 µM for ppGpp) [9]. In amino acid-starved cells, the levels of pppGpp increase from 0.5 to 200 µM and those of ppGpp from 20 µM to 1 mM; these elevated levels far exceed those required to inhibit PPX in vitro [9]. Turnover of polyP (resulting from synthesis by PPK and hydrolysis by PPX) was found to be 12 min or less. This result was simulated by the accumulation of polyP in a reaction mixture containing purified PPK and PPX in the presence of pppGpp and ppGpp [9]. The profound inhibition by stringent factors of the breakdown of polyP by PPX, but without effect of polyP synthesis by PPK, could explain blockage of turnover of polyP and its accumulation (Fig. 1).
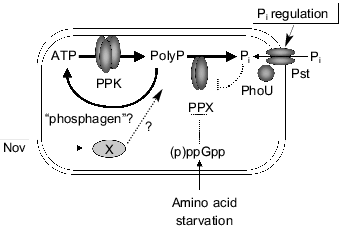
Fig. 1. A model for regulation of polyP metabolism in E. coli. PolyP accumulated in response to amino acid starvation. Stringent factors, (p)ppGpp, strongly inhibit PPX activity. Novobiocin stimulates polyP accumulation independent of stringent response, but its target remains obscure. PhoU protein negatively regulates Pi transport as well as levels of polyP. Increased only PPK activity in response to Pi starvation is responsible for polyP overplus in K. aerogenes. Potent degradative pathways of polyP as a phosphagen may affect polyP accumulation. (Pst, Pi-specific transport system).
POLYPHOSPHATE KINASE IS REQUIRED FOR ADAPTATION TO AMINO ACID STARVATION
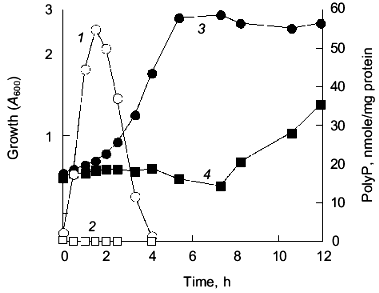
E. coli accumulates polyP in response to a nutritional downshift from rich to a minimal medium [10]. An E. coli mutant, lacking both the ppk and ppx genes, failed to accumulate polyP and exhibited an extended lag upon growth recovery to logarithmic growth (Fig. 2). Introduction of the ppk gene on a low copy plasmid into the mutant abolished the lag. This phenotype of the mutant can be attributed to an impaired adaptation to amino acid starvation [11]. Support for this hypothesis was provided by the following observations: 1) supplementation of amino acids to the Mops minimal medium abolished the extended lag phase; 2) the mutant was significantly reduced in growth rate when only amino acids were removed from the medium; 3) levels of ppGpp remained high in the mutant during the nutrient downshift. Once the mutant adapted to the Mops medium, it grew as well as did the wild type.
Upon starvation for amino acids, E. coli induces expression of amino acid biosynthetic enzymes [8]. Transcriptional lacZ fusions with hisG, ilvG, trpE, and argE were used for monitoring amino acid biosynthetic genes. The mutant failed to express beta-galactosidase from all of these fusion genes during nutritional downshift. We assumed that the mutant failed to transcribe amino acid biosynthetic genes. However, Northern analysis revealed that the mutant synthesized hisG-lacZ mRNA to a significant level, about 20% that of the wild type [11]. This result indicated that polyP accumulation might have affected the initiation of the hisG transcription, but was apparently insufficient to explain why the mutant showed no expression of beta-galactosidase from the hisG-lacZ fusion gene. At the onset of nutritional downshift, E. coli is starved for amino acids required to synthesize new polypeptides, so that an amino acid pool generated from intracellular protein degradation is very important to adapt to nutrient downshift [12]. The expression of the fusion depended on the external supplementation of amino acids in the mutant. We found that the wild type demonstrated increased rates of protein degradation in response to the nutritional downshift, while the mutant did not [11]. Metabolic network of ppGpp and polyP relates to the adaptation to the nutritional downshift. We isolated a temperature-sensitive ppk mutant. Abolishment of the growth lag was accomplished only at a permissive temperature in this mutant, but with small accumulation of polyP at a level of 1 nmol/mg protein (about 50 times less than the wild type). This result implied that PPK, rather than the amount of polyP, is important for the growth recovery.Fig. 2. Cell growth and polyP accumulation during the nutritional downshift. E. coli MG1655 (wild type) and CF5802 (ppk ppx mutant) cells were subjected to the nutritional downshift from rich (.×_Y.) to minimal (Mops) medium. PolyP accumulated in response to the downshift in the wild type (1), but not in the mutant (2). Growth was measured as the optical density at 600 nm (A600) in the wild type (3) and the ppk ppx mutant (4).
Colincinogenic factor E1 (Col E1) plasmid continues to replicate when protein synthesis is inhibited by amino acid starvation [13]. The wild type continued Col E1 plasmid replication during nutrient downshift, while the mutant did not (Kuroda and Ohtake, unpublished results). The E. coli ppk mutant fails to make adaptive changes in stationary phase needed for resistance to various stresses and for survival [5, 14].
ANTIBIOTICS TO INDUCE PolyP ACCUMULATION
The polyP accumulation is reversed if growth is allowed to resume. Harold reported that polyP was rapidly degraded and the Pi was quantitatively transferred to the nucleic acid fraction, suggesting that cessation of nucleic acid synthesis and assimilation of Pi from the medium continued resulting in polyP accumulation [4]. According to his model, inhibitors for nucleic-acid synthesis should stimulate polyP accumulation. Indeed novobiocin (a specific DNA gyrase inhibitor) stimulated polyP accumulation, while neither inhibitors for protein nor cell wall synthesis did (Kuroda and Ohtake, unpublished results). Then we used several inhibitors for DNA synthesis and measured levels of polyP. Although some of them inhibited DNA synthesis stronger than novobiocin (Nov), they did not stimulate polyP accumulation (see table). Furthermore, temperature-sensitive mutants of DNA replication and RNA synthesis did not accumulate polyP at the non-permissive temperature, and the mutants still accumulated in response to the addition of Nov. Nov inhibits DNA replication by directly binding to the gyrase B-subunit. A temperature-sensitive mutant (gerBts) did not accumulate polyP even at the non-permissive temperature. Therefore, the target of Nov seems different from the gyrase B-subunit. Several Nov-binding proteins in addition to the gyrase have been reported in E. coli. To know the antagonistic relationship of polyP to nucleic acid synthesis, we will have to determine possible targets for Nov involved in polyP accumulation (Fig. 1). PolyP accumulation was observed in both P. aerginosa and K. aerogenes by the addition of Nov into the culture. It should be noted that Nov induced polyP accumulation independent of the accumulation of the stringent factors (Kuroda and Ohtake, unpublished results).
Effect of various inhibitors on polyP accumulation and DNA synthesis
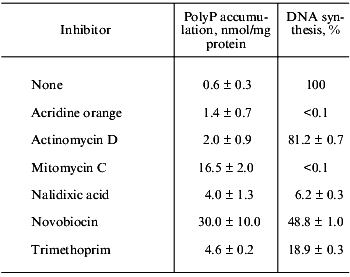
Note: Inhibitors (200 µg/ml) were added into exponentially growing
E. coli. PolyP and [3H]thymidine incorporation into
acid-insoluble fraction were measured after 70 min.
A NEGATIVE REGULATOR FOR PolyP ACCUMULATION
One of the Nov resistant E. coli mutants (MT4) exhibited 1000-fold higher levels of polyP than did the wild type under nutritionally rich conditions (Kuroda and Ohtake, unpublished results). The 30th codon of the phoU gene was changed from glycine to aspartic acid in the mutant. Introduction of the wild type phoU gene into the MT4 mutant reduced polyP to the wild type level. Therefore, the phoU gene is a negative regulator for polyP accumulation. This mutant exhibited the Pi uptake at the low concentration of Pi even when grown under Pi-rich conditions. It is known that phoU mutants express the Pi-specific transport system (Pst) even under Pi-rich conditions [15]. Introduction of the pst genes on a multicopy plasmid led to large accumulation of polyP [16]. It was reported that phoB mutants, a positive regulator for the pho regulon, did not accumulate polyP although they developed wild type levels of stringent factors upon amino acid starvation [5]. Regulation of Pi transport seems very important for polyP accumulation (Fig. 1). High concentrations of Pi inhibited PPX activity: at 20 mM, Pi inhibited PPX by 90% (Kuroda and Ohtake, unpublished results). Rao et al. reported that a phoU35 mutation did not affect polyP accumulation [5]. The mutant MT4 apparently has multiple mutation sites because the only phoU mutation does not confer Nov resistance to the wild type. It may be a possible explanation that another mutation in combination with the phoU mutation contributes to polyP accumulation in the mutant MT4 (Kuroda and Ohtake, unpublished results).
PolyP OVERPLUS IN Klebsiella aerogenes
In contrast to E. coli, K. aerogenes exhibits rapid and extensive polyP accumulation, called polyP overplus, when Pi is added to cells previously subjected to Pi starvation [4]. A ppk mutant of K. aerogenes did not show polyP overplus [4]. PPK is responsible for polyP overplus (Kuroda and Ohtake, unpublished results). The PPK shares 93% identical amino acids with the E. coli PPK [17]. Like the E. coli ppk-ppx operon, the K. aerogenes ppx gene exists immediately downstream of the ppk gene [17]. Importantly, a putative pho box exists in the promoter region of the K. aerogenes ppk-ppx operon [17]. Unlike the E. coli ppk-ppx operon, the K. aerogenes ppk-ppx operon is likely to be under control of the PhoB and PhoR proteins [17]. As expected, the PPK activity increased in response to Pi starvation and decreased upon addition of Pi. However, unlike PPK, the PPX activity did not increase, rather slightly decreased, under conditions of Pi starvation although the ppx mRNA was induced (Morohoshi, Kuroda, and Ohtake, unpublished results). It is clear that both increased polyP synthesis and decreased polyP degradation are responsible for polyP overplus in K. aerogenes.
IS PolyP USED AS A PHOSPHAGEN?
The degradative pathway presents an intriguing problem to consider polyP accumulation as well as the functions of polyP. Polyphosphatase represents the main pathway of degradation. It may be true that polyP serves as a phosphorus reservoir. The reverse reaction of PPK, in which ADP is converted to ATP by polyP, is kinetically slower than the polyP synthesis, but can be driven to completion by an excess of ADP [2]. More generally, PPK functions as a nucleoside diphosphate kinase, converting GDP, CDP, and UDP to their respective nucleoside triphosphates [18]. This may be one possible route of polyP degradation [2]. Although the level of polyP exceeds that of ATP by about 10-fold during polyP accumulation, the rapid turnover rate of ATP in the cell (0.2 sec) argues against polyP simply as an ATP store [10]. However, there have been supports of the hypothesis that polyP serves as a phosphagen. The term “phosphagen” is defined as phosphorylated compounds that function as stores of phosphate-bond energy. The phosphoryl group may be transferred to ADP to form ATP. A K. aerogenes ppx mutant rapidly degraded polyP when growth is resumed after nutritional downshift, suggesting that PPK consumed polyP by its reverse reaction and provided ATP in vivo (Fig. 1, Kuroda and Ohtake, unpublished results). A multiple complex of the RNA degradosome (RNA helicase, ribonuclease E, polynucleotide phosphorylase, and enolase) contains PPK [19]. It was considered that polyP may be used to regenerate ATP consumed by RNA helicase when it unwound RNA [19]. A function of polyP and PPK to maintain the microenvironment in such a complex should be further investigated.
PolyP is used for phosphorylation of substrate. Activated sludge processes with alternating anaerobic and aerobic conditions have been adopted widely to eliminate phosphorus from wastewater [7]. Microlunatus phosphovorus is a microorganism isolated from polyP-accumulating sludge. It accumulated polyP under aerobic conditions and degraded polyP under anaerobic conditions along with an uptake of glucose [20]. We have recently revealed that M. phosphovorus has a polyP-dependent glucose kinase. It has been reported that Mycobacteria has a polyP- and ATP-dependent glucose kinase [21]. It is noteworthy that the M. phosphovorus enzyme utilizes polyP but not ATP as a phosphorus source (Tanaka, Kuroda, and Ohtake, unpublished results). This evidence strongly supports that polyP is utilized for the direct phosphorylation of glucose. This enzyme activity has never been detected in E. coli or K. aerogenes. PolyP-accumulating sludge degraded polyP in the presence of acetate and release Pi. We have detected that a significant amount of pyrophosphate is first generated from polyP along with an uptake of acetate (Gotanda, Kuroda, and Ohtake, unpublished results). Whether polyP is directly used for activation of acetate, conversion to a thioester (acetyl-CoA), is under investigation.
These works were supported in part by Proposal-Based New Industry Creative Type Technology R&D Promotion Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, by a grant from Research Institute of Innovative Technology for the Earth (RITE), and by a grant for scientific research from the Ministry of Education, Science and Culture of Japan. A part of the work (relationship of ppGpp to polyP accumulation) was originally done by Kuroda in A. Kornberg's laboratory (Stanford University). We thank Drs. J. Kato, T. Ikeda, A. Kornberg, and Kornberg's colleagues for helpful discussions.
REFERENCES
1.Kulaev, I. S. (1979) The Biochemistry of
Inorganic Polyphosphates, John Wiley & Sons, N. Y.
2.Kornberg, A. (1995) J. Bacteriol.,
177, 491-496.
3.Akiyama, M., Crooke, E., and Kornberg, A.
(1992) J. Biol. Chem., 267, 22556-22561.
4.Harold, F. M. (1966) Bacteriol. Rev.,
30, 772-794.
5.Rao, N. N., Liu, S., and Kornberg, A.(1998) J.
Bacteriol., 180, 2186-2193.
6.Tzeng, C.-M., and Kornberg, A. (1999) Mol.
Microbiol., 29, 381-382.
7.Ohtake, H., Takahashi, K., Tsuzuki, Y., and Toda,
K. (1985) Water Res., 19, 1587-1594.
8.Cashel, M., Gentry, D. R., Hernandez, V. J., and
Vinella, D. (1996) in Escherichia coli and Salmonella:
Cellular and Molecular Biology (Neidhardt, F. C., ed.) American
Society for Microbiology, Washington, pp. 1458-1496.
9.Kuroda, A., Murphy, H., Cashel, M., and Kornberg,
A. (1997) J. Biol. Chem., 272, 21240-21243.
10.Ault-Riche, D., Fraley, C. D., Tzeng, C-.M., and
Kornberg, A. (1998) J. Bacteriol., 180, 1841-1847.
11.Kuroda, A., Tanaka, S., Ikeda, T., Kato, J.,
Takiguchi, N., and Ohtake, H. (1999) Proc. Natl. Acad. Sci. USA,
96, 14264-14269.
12.Miller, C. G. (1996) in Escherichia coli
and Salmonella: Cellular and Molecular Biology (Neidhardt, F.
C., ed.) American Society for Microbiology, Washington, pp.
938-954.
13.Clewell, D. B. (1972) J. Bacteriol.,
110, 667-676.
14.Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y.,
Kameda, A., Tanaka, K., Takahashi, H., Munekata, M., Rao, N. N., and
Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA, 94,
11210-11215.
15.Steed, P. M., and Wanner, B. L. (1993) J.
Bacteriol., 175, 6797-6809.
16.Kato, J., Yamada, K., Muramatsu, A., Hardoyo, and
Ohtake, H. (1993) Appl. Environ. Microbiol., 59,
3744-3749.
17.Kato, J., Yamamoto, T., Yamada, K., and Ohtake,
H. (1993) Gene, 137, 237-242.
18.Kuroda, A., and Kornberg, A. (1997) Proc.
Natl. Acad. Sci. USA, 94, 439-442.
19.Blum, E., Py, B., Carpousis, A. J., and Higgins,
C. F.(1997) Mol. Microbiol., 26, 387-398.
20.Nakamura, K., Masuda, K., and Mikami, E. (1991)
J. Ferment. Bioeng., 71, 258-263.
21.Hsieh, P. C., Shenoy, B. C., Samols, D., and
Phillips, N. F. B. (1996) J. Biol. Chem., 271,
4909-4915.