REVIEW: Polyphosphates and Enzymes of Polyphosphate Metabolism in Escherichia coli
M. A. Nesmeyanova
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Moscow Region, 142292 Russia; fax: (095) 923-3602; E-mail: aniram@ibpm.serpukhov.su
Received December 15, 1999
This review summarizes the results of our study of polyphosphate and enzymes of polyphosphate metabolism in E. coli and their regulation by exogenous orthophosphate and other physiological and genetic factors.
KEY WORDS: E. coli, polyphosphate, enzymes, regulation, orthophosphate
Most microorganisms synthesize different reserve polymers with polyphosphate (polyP) being of special interest. This could be anticipated, since polyP contains phosphorus, one of the most important vital elements that is a constituent of all major subunits of life: amino acids, nucleotides, sugars, phospholipids, and recently has been quite obviously shown to be a “molecule for many reasons” of living cells [1]. The accumulation of polyP in bacteria is not only a natural phenomenon, but also a finely regulated process. It can be used as an energy source, a phosphate donor for sugars, and a chelator for divalent cations. Moreover, related molecules (e.g., guanosine tetraphosphate) can be involved as regulatory molecules in the functioning of global regulatory systems. The bacteria have been known to accumulate polyP for a comparatively long time [2, 3]. However the regular absence of this polymer in exponentially growing bacterial cultures resulted in a wrong notion of its absence in some bacteria. The gram-negative bacterium E. coli was also attributed to the latter [4]. Meanwhile, it is just the cells of E. coli where the most important enzyme of polyP synthesis--ATP-polyphosphate phosphotransferase, or polyphosphate kinase [5, 6]--was first found and identified, and recently purified to a homogenous state [7].
This circumstance determined our search of polyP in this bacterium, which was interesting first of all due to a very intensive investigation of its phosphorus metabolism both on the biochemical and genetic levels as a complex two-component regulatory system (Pho regulon). Analogs of such a system were later revealed in many metabolic pathways. However the place of this regulatory system in polyP metabolism was not determined. Our study of polyP and enzymes of polyP metabolism in Escherichia coli resulted in the following conclusions.
ACCUMULATION OF POLYPHOSPHATE IN E. coli CELLS IS A
FUNCTION OF CULTURE GROWTH STAGE AND REGULATED BY EXOGENOUS BUT NOT
INTRACELLULAR ORTHOPHOSPHATE
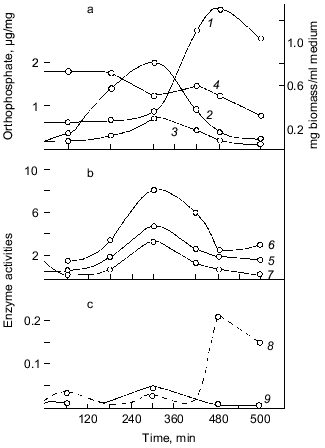
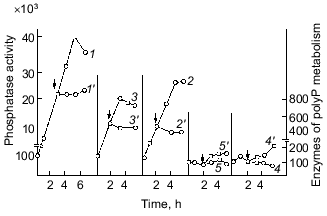
The earliest works with bacteria showed that polyP occurs in them in extremely low amounts, if at all, and is present in bacterial cells not continually and often under specific conditions of bacterial growth, usually under growth limitation by some nutrient sources [8]. Indeed, our study of the dynamics of polyP accumulation during the growth of the wild-type strain E. coli K12 on a mineral medium supported such a suggestion, simultaneously closing the question of polyP occurrence in E. coli [9]. We found in this bacterium a high-molecular-weight acid-insoluble polyP that was identified by a specific product of its partial acidic hydrolysis, trimethophosphate, using the method of Thilo and Wieker [10]. Much later, direct methods confirmed that polyP in E. coli may have a length of up to 1000 phosphate groups [11]. We showed that the highest amount of this high-polymer polyP, reaching 0.2-0.4% of dry bacterial weight (in yeast its amount may reach 20%), occurs in the cells of this bacterium only in the end of latent and beginning of logarithmic growth phase. When the culture passes to exponential growth, the level of intracellular polyP dramatically decreases 5-10-fold (Fig. 1). Thus, the accumulation of polyP precedes intensive culture growth, and then it is utilized by growing cells [9]. The above peculiarity explains unsuccessful attempts to reveal polyP in E. coli, though it was observed earlier in other bacteria as well [12]. The synchronization of bacterial cultures, displaying noticeable fluctuations in the polyP content during its growth, showed [13, 14] that its highest level is observed immediately before cell division, which seems to be very important. The content of DNA increased in the course of the whole cycle after cell division. The dynamics of accumulation and utilization of polyP undoubtedly indicates first of all the phosphogenic and probably energy source function of polyP in bacteria. It should be mentioned that in our experiments intracellular orthophosphate was accumulated less intensively than polyP under cell growth with an excess of exogenous orthophosphate. Besides, orthophosphate remained actually on the same level, which also favored the involvement of polyP in the maintenance of its constant intracellular level.
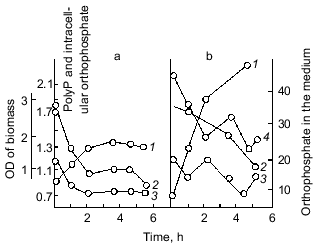
Important observations of the dynamics of polyP accumulation in bacteria described in other works [15] are concerned with certain conditions of bacterial growth limitations, e.g., nitrogen or sulfur deficiency or unfavorable pH conditions. As a rule, the accumulation of polyP under the above limitations was due to the excess of energy sources and the availability of phosphate, as well as K and Mg ions. In essence, the polyP biosynthesis responds to different external factors, whether they are components of the nutrient medium, or its physical and chemical conditions. However, polyP was utilized in most cases when bacterial cells were placed in a medium enabling cell growth. This supports polyP functioning as a phosphogen or an energy source. We showed that polyP was utilized at the same rate when cells were subjected to phosphate starvation. In this case, the level of intracellular orthophosphate remained actually the same, again supporting the regulatory role of polyP in the maintenance of the phosphate level in the cell (Fig. 2).Fig. 1. PolyP and enzymes of polyP metabolism in the process of E. coli growth. 1) Ñulture growth (mg dry biomass/ml medium); 2) polyP (µg Pi/mg dry biomass); 3) intracellular orthophosphate (µg Pi/mg dry biomass); 4) orthophosphate of the medium (µg Pi/ml medium); 5) polyphosphatase (mU/mg dry biomass, the same as 6-9); 6) tripolyphosphatase; 7) alkaline phosphatase; 8) polyphosphate kinase; 9) 1,3-diphosphoglycerate polyphosphate phosphotransferase.
The best known exclusion from the described conditions of polyP accumulation in bacteria appeared to be for several strains of Micrococcus lysodeikticus, where polyP granules accumulated during exponential growth and disappeared only in the stationary phase [16]. On the contrary, some Acinetobacter strains are able to accumulate polyP in the end of log phase or even in the stationary phase [17], and in mycobacteria the rate of polyP accumulation was the highest in the stationary phase [18].Fig. 2. The effect of exogenous orthophosphate on the level of intracellular orthophosphate and polyP in E. coli under orthophosphate starvation (a) and orthophosphate excess (b): 1) cell growth; 2) polyP content (µg Pi/mg dry biomass); 3) intracellular orthophosphate (µg Pi/mg dry biomass); 4) orthophosphate in the medium (µg Pi/ml medium).
THE ACTIVITY OF ENZYMES OF POLYPHOSPHATE METABOLISM IS ALSO A
FUNCTION OF CULTURE GROWTH STAGE
The accumulation of polyP in cells results from the two processes: synthesis and utilization of polyP. The dynamics of the known polyP-synthesizing enzymes, polyphosphate kinase [6] and 1,3-diphosphoglycerol-polyphosphate phosphotransferase [19], analyzed in our experiments showed that these enzyme activities weakly correlate with the dynamics of polyP accumulation (Fig. 1). The main peak of polyphosphate kinase activity was observed, for instance, during the stationary culture growth, when the content of polyP in cells significantly decreased. On the contrary, the activity of enzymes of polyP degradation, polyphosphatase and tripolyphosphatase in E. coli, directly correlated with polyP accumulation and with the activity of alkaline phosphatase (nonspecific phosphohydrolase) of E. coli. The polyP phosphohydrolases studied here were first found in E. coli in our experiments, and the dynamics of their activity appeared to be very interesting from the standpoint of possible participation of the enzymes of polyP metabolism in the catalysis of both forward and back reactions. For polyphosphate kinase, the forward (polyP-synthesizing) and back (polyP-degrading) reactions were established previously, and the back polyphosphate-ATP-phosphotransferase reaction [6] was shown to be more characteristic of the enzyme. As regards polyphosphatases, our data first gave us grounds to assume that polyP phosphohydrolase can catalyze also the back reaction--polyP synthesis. The same conclusion suggested itself on the basis of earlier works with other microorganisms, where the activity of polyphosphatase also directly correlated with the polyP content [20, 21]. However, no proper attention was paid to this fact, and evidently up to now the question about the bidirectional character of reactions catalyzed by polyphosphatases has not been solved. It is interesting that the polyphosphatase activity in E. coli is several orders higher than the activity of polyP-synthesizing enzymes. This suggests that the E. coli polyphosphatase may be involved rather in the synthesis than utilization of polyP under metabolic conditions with a constantly high level of exogenous orthophosphate and its transport into the cell. As we have shown [9], the utilization of polyP during the exponential growth phase correlates not with the peak of polyphosphate kinase activity but with polyphosphatase, whose activity decreases in parallel with reducing polyP content. We cannot exclude another explanation of this correlation. For instance, polyphosphatase can be activated by an increasing amount of its polyP substrate (high-molecular-weight polyP) but not be involved in its degradation until its length reaches a certain value. This period may coincide with the beginning of intensive cell growth and the resulting acute need of orthophosphate and(or) energy. In this case, the factor regulating the activity and even the direction of polyphosphatase functioning in a bacterial cell will probably be a ratio of orthophosphate to polyP in the cell and the size of polyP.
E. coli POLYPHOSPHATASES ARE CO-REGULATED ON THE METABOLIC
AND GENETIC LEVELS WITH ALKALINE PHOSPHATASE
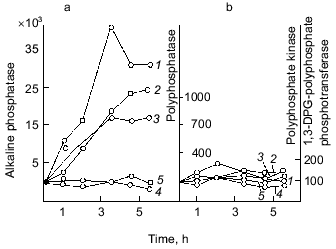
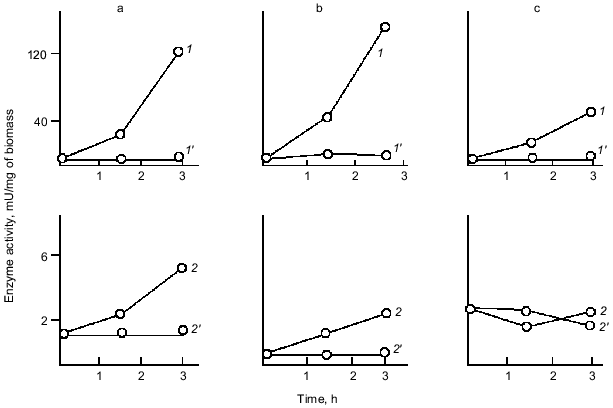
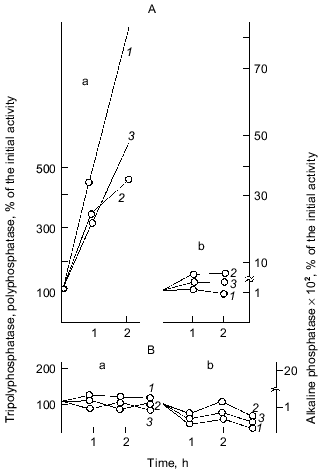
As Fig. 1 shows, polyphosphatase activities correlate during the culture growth with the activity of alkaline phosphatase--a nonspecific phosphomonoesterase whose function is to supply the cell with orthophosphate from all available exogenous sources. This enzyme is regulated by orthophosphate, which represses its synthesis when occurring in the medium in a high concentration. Under phosphate starvation, the enzyme synthesis is derepressed, and it is accumulated in high quantities. The polyphosphatase function may be similar to the function of alkaline phosphatase, which provides the cell with orthophosphate. Therefore, the regulation of these and other enzymes of polyP metabolism by exogenous orthophosphate was studied. Phosphohydrolases (alkaline phosphatase and polyphosphatases) appeared to be co-regulated by orthophosphate (Fig. 3). Their activities appreciably increased under phosphate starvation, whereas polyphosphate synthetase activities did not depend on the content of exogenous orthophosphate in the medium. The greatest derepression was observed in cells during the exponential and even the latent growth under phosphate starvation (Fig. 4). Cells from the stationary growth phase showed actually no derepression of polyphosphatase under the same conditions [22]. If orthophosphate was added to the medium during the synthesis of phosphohydrolases, the latter completely stopped (Fig. 5). It is evident that polyphosphatases are strictly regulated by exogenous orthophosphate. The character of regulation of polyphosphatase synthesis by exogenous orthophosphate suggests that the phosphohydrolase function is the most typical for this enzyme. Obviously, this very enzyme plays the leading role in polyP utilization under phosphate starvation, which proceeds much quicker than on the medium with orthophosphate.
Fig. 3. Effect of exogenous orthophosphate on the activity of enzymes of phosphorus metabolism in E. coli under orthophosphate starvation (a) and orthophosphate excess (b): 1) alkaline phosphatase; 2) tripolyphosphatase; 3) polyphosphatase; 4) polyphosphate kinase; 5) 1,3-diphosphoglycerate (DPG)-polyphosphate phosphotransferase. The enzyme activities are calculated in mU/mg dry biomass and expressed in percentage of the initial activity.
Fig. 4. Effect of culture growth stage under phosphate starvation (1, 2) and orthophosphate excess (1´, 2´) on the level of derepression of alkaline phosphatase (1) and polyphosphatase (2) in the wild-type E. coli strain: a) latent; b) log-mid; c) stationary stages.
To obtain an evidence of the co-regulation of polyphosphatases with the E. coli alkaline phosphatase not only on the metabolic but also on the genetic level, we have used mutants in the Pho regulon that controls the biosynthesis of a number of orthophosphate-dependent enzymes and other proteins. The mutation of the regulatory gene phoR resulting in a non-inducible synthesis of proteins of the Pho regulon allowed us to reveal that polyphosphatases are co-regulated with alkaline phosphatase under the control of the same regulatory system (Fig. 6). Neither alkaline phosphatase nor polyphosphatases were induced under phosphate starvation in this mutant strain [23]. About 30 other proteins participating in phosphorus metabolism are controlled by the Pho regulon. Among them there is a pore protein of outer membranes, PhoE, which forms a specific anionic pore facilitating the uptake of phosphate and phosphorylated components, and probably polyP [24]. A very low level of orthophosphate in the medium induces the Pho regulon, which comprises the Pst-pathway of using orthophosphate and the PhoB regulator of response, which induces the genes of proteins of this regulon also participating in phosphorus metabolism. However, it should be noted that the interrelations between polyP accumulation and the Pho regulon are rather complicated [25]. It was shown that polyP accumulation does not occur in mutants lacking the phoB gene but is recovered on the introduction of a plasmid with the phoB gene into this mutant strain [26].Fig. 5. Effect of orthophosphate added to a culture growing under phosphate starvation on the activity of enzymes of phosphorus metabolism in E. coli. Symbols are as in Fig. 3. Primed numbers indicate after the addition of orthophosphate at the time indicated by an arrow.
The similar function and regulation of polyphosphatase and alkaline phosphatase suggest their similar localization. Alkaline phosphatase is a typical secreted protein localized in periplasm. The localization of polyphosphatase was first established in our work [27].Fig. 6. Effect of exogenous orthophosphate on the biosynthesis of phosphohydrolases by wild-type E. coli strain (A) and uninducible mutant strain (phoR-) (B) under orthophosphate starvation (a) and orthophosphate excess (b): 1) alkaline phosphatase; 2) polyphosphatase; 3) tripolyphosphatase. Enzyme activites are calculated in mU per mg of dry biomass and expressed in percentage of the initial activity.
Fractionation of E. coli cells after cell wall lysis by lysozyme in the presence of EDTA using alkaline phosphatase as a marker showed that E. coli polyphosphatase is a surface protein of the cytoplasmic membrane, localized on its periplasmic side. In contrast to alkaline phosphatase, which is completely released from cells during cell wall lysis, polyphosphatase remains membrane-bound and is released from the membrane only after its long washing with buffer. Here it turned out that the strength of polyphosphatase binding with the membrane depends on the presence of orthophosphate in the medium. Under phosphate starvation, i.e., under derepression of the enzyme, this binding is weaker and the enzyme is more easily released to the periplasm. Probably, this very exopolyphosphatase was later isolated and purified to a homogenous state. It was a dimer of subunits with the molecular mass of 58 kD [28]. It was suggested that the high-polymer polyP hydrolyzed by the enzyme during the culture growth or at its starvation by orthophosphate is partly localized also on the outer surface of the cytoplasmic membrane of E. coli. We have found a polyphosphatase also in the ribosome-membrane complex, where it is evidently present in the course of its translocation from cytoplasm to periplasm across the cytoplasmic membrane [29]. However, if polyP is localized on the outer membrane surface, a question arises about the place of its synthesis and about the polyP-synthesizing function of polyphosphatase, if the latter is at all possible. Recently data have emerged on the ability of polyP to be secreted from the cell [30]. In case it is synthesized in cytoplasm, it has to be translocated across the cytoplasmic membrane. This suggests that polyphosphatase with a synthetic function is involved in polymerization and extension of the polyP chain in cytoplasm. The size of the latter can be a signal for a change in the direction of the catalyzed reaction, on the one hand, and in the localization of polyP and polyphosphatase, on the other hand. Both of them are secreted into the periplasm; therefore, the degradation of high-polymer polyP to orthophosphate occurs beyond the cytoplasm. The above forms the conditions for a strictly regulated utilization of released orthophosphate by the Pho regulon system, which controls not only orthophosphate transport but also the expression of numerous proteins participating in phosphorus metabolism.
At present a number of polyP-degrading enzymes are known: polyphosphate depolymerase, polyP-dependent NAD kinase, polyphosphate fructokinase, polyphosphate mannokinase, and polyphosphate glucokinase [25]. This fact once more confirms the diversity of polyP functions in cells.
I would like to touch briefly upon polyP function, probably associated with the adaptation reactions of a microbial cell. It is well known that the adaptive response in bacteria is provided by the so-called two-component regulatory systems, each of them being a family of two proteins: a membrane-bound sensor (histidine protein kinase) accepting external signals, and a cytoplasmic regulator of response interacting with the regulatory elements of DNA [31]. The Pho regulon involved in the transduction of Pi signal and in the regulation of phosphorus metabolism also belongs to such regulatory systems [24]. The association of polyP accumulation with the limitation by some compounds and even with stress conditions, accompanied by abrupt changes in their concentrations, probably makes polyP a link between these situations and the global regulatory systems. PolyP-related compounds, guanosine tetra- or pentaphosphate ((p)ppGpp), may provide this function. Its most typical involvement in the global regulatory system is associated with the deficiency of amino acids. The resulting deficiency in amino acyl tRNA stopping protein translation initiates the accumulation of ppGpp, a direct consequence of which is the inhibition of RNA polymerase, rRNA and tRNA synthesis, and activation of transcription for biosynthetic and catabolic operons. The secondary effect of ppGpp is the inhibition of the synthesis of ribosomal proteins, general protein, phospholipids and nucleotides, DNA replication, cell division, but at the same time the enhancement of chaperone synthesis, stress protein synthesis, protein degradation, and catabolic pathways. The interaction between the accumulation of this regulatory relative of polyP, (p)ppGpp, and a high-polymer polyP is rather complicated and cannot always be traced. It is assumed however that under some stress conditions ppGpp can inhibit the hydrolytic degradation of polyP due to the inhibition of polyphosphatase without affecting the function of polyphosphate kinase [25]. The disturbance of polyP metabolism in this case results in its accumulation, but so far it is hard to reveal whether it plays any regulatory role as does its low-molecular-weight relative. All the presently available data quite clearly indicate that polyP plays a significant role in cell metabolism and is really a “molecule for many purposes” in the cell.
The author thanks I. S. Kulaev for initiation of this work and all participants for assistance in the experimental work discussed in the review.
REFERENCES
1.Kornberg, A. (1994) in Phosphate in
Microorganisms: Cellular and Molecular Biology (Torriani-Gorini,
A., Yagil, E., and Silver, S., eds.) American Society for Microbiology,
Washington, D.C., pp. 204-208.
2.Dawes, E. A., and Senior, P. J. (1973) Adv.
Microbiol., 10, 235-266.
3.Kulaev, I. S. (1976) The Biochemistry of
Inorganic Polyphosphate, John Wiley & Sons, Inc., N. Y.
4.Pine, M. (1963) J. Bacteriol., 85,
301-306.
5.Kornberg, A., Kornberg, S., and Simms, E. (1956)
Biochim. Biophys. Acta, 20, 215-227.
6.Êîrnberg, S. R. (1957) Biochim.
Biophys. Acta, 26, 294-299.
7.Ahn, K., and Kornberg, A. (1990) J. Biol.
Chem., 265, 11734-11739.
8.Duguid, J. P., Smith, I. W., and Wilkinson, J. F.
(1954) J. Pathol., 67, 289-300.
9.Nesmeyanova, M. A., Dmitriyev, A. D., and Kulaev,
I. S. (1973) Mikrobiologiya, XLII, 213-219.
10.Thilo, E., and Wieker, W. Z. (1957) Anorgan.
Allgem. Chem., 291, 164-170.
11.Rao, N. N., Roberts, M. F., and Òorriani, A.
(1985) J. Bacteriol., 162, 242-247.
12.Harold, F. M., and Silvan, H. (1963) J.
Bacteriol., 89, 1262-1270.
13.Sall, T., Mudd, S., and Davis, J. C. (1956) J.
Bacteriol., 60, 130-146.
14.Sall, T., Mudd, S., and Takagi (1958) J.
Bacteriol., 76, 640-645.
15.Harold, F. M. (1966) Bacteriol. Rev.,
30, 772-788.
16.Friedberg, I., and Avigard, G. (1968) J.
Bacteriol., 96, 544-553.
17.Streichan, W., and Schon, G. (1991) Stammen
Wasser/Abwasser, 132, 301-308.
18.Winder, F. G., and Deneny, J. M. (1957) J.
Gen. Microbiol., 17, 573-585.
19.Kulaev, I. S., Bobyk, M. A., Nikolayev, N. N.,
Sergeyev, N. S., and Uryson, S. I. (1971) Biokhimiya, 36,
943-950.
20.Hughes, D. E., and Muhammed, D. (1962) in
Acides Ribonucleiques et Polyphosphates. Structure, Synthesis, et
Function (Coll. Int. CNRS, Strusburg), CNRS, Paris, p. 591.
21.Harold, F. M. (1963) J. Bacteriol.,
86, 216-221.
22.Nesmeyanova, M. A., Gonina, S. A., Severin, A.
I., and Kulaev, I. S. (1974) Mikrobiologiya, XLIII,
955-960.
23.Nesmeyanova, M. A., Gonina, S. A., and Kulaev, I.
S. (1975) Dokl. Akad. Nauk SSSR, 224, 710-714.
24.Torriani-Gorini, A. (1994)in Phosphate in
Microorganisms: Cellular and Molecular Biology (Torriani-Gorini,
A., Yagil, E., and Silver, S., eds.) American Society for Microbiology,
Washington, D.C., pp. 1-4.
25.Kornberg, A., Rao, N. N., and Autt-Riche, D.
(1999) Annu. Rev. Biochem., 68, 89-125.
26.Rao, N. N., Liu, S., and Kornberg, A. (1998)
J. Bacteriol., 180, 2186-2193.
27.Nesmeyanova, M. A., Maraeva, O. B., Severin, A.
I., and Kulaev, I. S. (1975) Dokl. Akad. Nauk SSSR, 223,
1266-1268.
28.Akiyama, M., Croock, E., and Kornberg, A. (1993)
J. Biol. Chem., 268, 633-639.
29.Severin, A. I., Lusta, K. A., Nesmeyanova, M. A.,
and Kulaev, I. S. (1976) Biokhimiya, 41, 357-362.
30.Hardoyo, J., Yamada, K., Shinjo, H., Kato, J.,
and Ohtake, H. (1994) Appl. Environ. Microbiol., 60,
3485-3490.
31.Rao, N. N., and Kornberg, A. (1996) J.
Bacteriol., 178, 1394-1400.