REVIEW: Microbial Selection of Polyphosphate-Accumulating Bacteria in Activated Sludge Wastewater Treatment Processes for Enhanced Biological Phosphate Removal
T. Mino
Department of Environmental Studies, The University of Tokyo 7-3-1, Hongo, Bunkyo-ku, Tokyo 113-8656; fax: +81-3-5841-8531; E-mail: mino@k.u-tokyo.ac.jp
Received December 7, 1999
Activated sludge processes with alternating anaerobic and aerobic conditions (the anaerobic-aerobic process) have been successfully used for enhanced biological phosphate removal (EBPR) from wastewater. It is known that polyphosphate-accumulating bacteria (PAB) play an essential role for EBPR in the anaerobic-aerobic process. The present paper reviews limited information available on the metabolism and the microbial community structure of EBPR, highlighting the microbial ecological selection of PAB in EBPR processes. Exposure of microorganisms to alternate carbon-rich anaerobic environments and carbon-poor aerobic environments in the anaerobic-aerobic process induces the key metabolic characteristics of PAB, which include organic substrate uptake followed by its conversion to stored polyhydroxyalkanoate (PHA) and hydrolysis of intracellular polyphosphate accompanied by subsequent Pi release under anaerobic conditions. Intracellular glycogen is assumed to function as a regulator of the redox balance in the cell. Storage of glycogen is a key strategy for PAB to maintain the redox balance in the anaerobic uptake of various organic substrates, and hence to win in the microbial selection. Acinetobacter spp., Microlunatus phosphovorus, Lampropedia spp., and the Rhodocyclus group have been reported as candidates of PAB. PAB may not be composed of a few limited genospecies, but involve phylogenetically and taxonomically diverse groups of bacteria. To define microbial community structure of EBPR processes, it is needed to look more closely into the occurrence and behavior of each species of PAB in various EBPR processes mainly by molecular methods because many of PAB seem to be impossible to culture.
KEY WORDS: Acinetobacter, activated sludge, anaerobic-aerobic process, ecological selection, enhanced biological phosphate removal (EBPR), glycogen, Lampropedia, microbial community, Microlunatus phosphovorus, polyhydroxyalkanoates (PHAs), polyphosphate-accumulating bacteria, Rhodocyclus, wastewater treatment
Abbreviations: DAPI) 4´,6-diamidino-2-phenylindole; DGGE) denaturing gradient gel electrophoresis; EBPR) enhanced biological phosphate removal; MK) menaquinone; 3HB) 3-hydroxybutyrate; 3HV) 3-hydroxyvalerate; 3H2MB) 3-hydroxy-2-methylbutyrate; 3H2MV) 3-hydroxy-2-methylvalerate; PAB) polyphosphate-accumulating bacteria; PCR) polymerase chain reaction; PHA) polyhydroxyalkanoate; PHB) polyhydroxybutyrate; polyP) polyphosphate; Q) ubiquinone; T-RFLP) terminal restriction fragment length polymorphisms; TCA cycle) tricarboxylic acid cycle.
Phosphate can cause eutrophication (extraordinary growth of algae) when
it is excessively discharged into closed natural water bodies like
lakes and inland seas. To control eutrophication, phosphate removal
from wastewater is often required before wastewater is discharged to
the receiving water bodies. Activated sludge processes with alternating
anaerobic and aerobic conditions have been successfully used for
enhanced biological phosphate removal (EBPR) from wastewater [1, 2]. This anaerobic-aerobic
alternation can be achieved either by spatial configuration of
anaerobic and aerobic zones in series in continuous flow systems with
sludge recycle or by temporal arrangement of anaerobic and aerobic
periods in sequence batch reactors. Such EBPR processes are referred to
as the anaerobic-aerobic or anaerobic-oxic process. It has been shown
in previous studies [2-6] that
polyphosphate-accumulating bacteria (PAB) play an essential role for
EBPR in the anaerobic-aerobic process. To achieve high and stable EBPR
performance, it is essential to maintain PAB in the system.
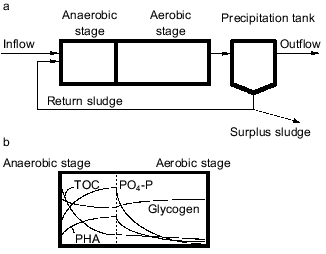
A basic configuration of the anaerobic-aerobic process is schematically shown in Fig. 1a. This process is structurally characterized by the presence of an anaerobic stage in which absolute anaerobic conditions are kept with neither oxygen nor NO2-/NO3- available as electron acceptor for activated sludge bacteria. Organic substrates are supplied from influent wastewater into the anaerobic stage and the return sludge comes into contact with the carbon source only in the anaerobic stage. Faster uptake of organic substrates in the anaerobic stage is the key for bacteria to win in the microbial selection in the EBPR process. The mechanism of proliferation of PAB can be described as follows [3, 7-9]. It is typically observed in the anaerobic stage that the activated sludge releases Pi to the bulk solution with concomitant uptake of organic substrates. In the subsequent aerobic stage, it takes up more Pi than has been released in the previous anaerobic stage. The Pi removed from the wastewater is accumulated in the cell as polyP [5]. Polyphosphate is a high-energy compound and its hydrolysis can supply energy to various biochemical reactions in the cell. In the anaerobic stage, the hydrolysis of intracellular polyP enables PAB to obtain the energy they need to take up organic substrates. Without electron acceptors (oxygen, NO2-/NO3-), aerobic bacteria and denitrifying bacteria are unable to obtain the energy required for the utilization of organic substrates, and they are thus unable to compete with PAB. Therefore, the introduction of the anaerobic stage leads to the precedence of PAB and to a rise in phosphorus content of the sludge. By withdrawing the phosphorus-rich sludge from the system as excess sludge, high phosphate removal efficiency can be achieved.
Although the anaerobic-aerobic process for EBPR is an established process from an engineering point of view, it has not been clearly defined in microbiological terms [10, 11]. For example, the phylogenetic or taxonomic groups responsible for EBPR have not been identified, and general structures of the EBPR microbial community have not been successfully described yet. Very few pure cultures have been isolated as candidates of PAB playing a key role in EBPR processes. Studies on metabolic aspects of EBPR have been mainly done based on enriched mixed cultures but not on pure cultures. This has resulted in lack of definitive and conclusive information about the microbiology and biochemistry of EBPR [11]. Thus, the mechanism of microbial ecological selection of PAB in EBPR processes has been understood very poorly. The present paper reviews limited information available on the metabolism and the microbial community structure of EBPR, highlighting the selection of PAB in EBPR processes.Fig. 1. a) Basic concept of anaerobic-aerobic process for EBPR. b) Behavior of the basic substances in EBPR. TOC, total organic carbon present in the bulk solution; PO4-P, orthophosphate present in the bulk solution; glycogen, glycogen stored in the cells; PHA, polyhydroxyalkanoates stored in the cells.
CARBON METABOLISM ADOPTED BY POLYPHOSPHATE-ACCUMULATING BACTERIA
IN EBPR PROCESSES
In terms of microbial metabolism, the anaerobic stage involves the uptake of organic substrates from wastewater by bacteria. Since the sludge comes into contact with organic substrates under anaerobic conditions, organisms that can utilize organic substrates more rapidly in an anaerobic environment gain precedence. Therefore, the reason why PAB can achieve a very high rate of organic substrate uptake under anaerobic conditions has been a major subject of concern. It has been well known that short chain fatty acids like acetate are favorable carbon sources for EBPR [12, 13], and acetate metabolism has been intensively studied as a model carbon metabolism substrate in EBPR [7, 9, 14]. A critical problem in such studies lays in the fact that none of the bacteria isolated from EBPR processes have exhibited all the key characteristics of the EBPR sludge and that any isolated pure cultures had never been verified to be primarily responsible for EBPR in an anaerobic-aerobic system until recently [11]. This is the reason that metabolic aspects of EBPR have been studied using mixed cultures enriched with PAB. Such PAB-enriched cultures have usually been obtained from lab-scale activated sludge reactors simulating the anaerobic-aerobic process fed with synthetic wastewater.
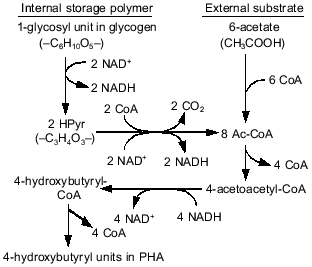
In anaerobic batch experiments with acetate as the carbon source, the activated sludge enriched with PAB typically take up acetate rapidly, accumulate PHAs in the cell, consume previously stored intracellular carbohydrate, and release Pi as a result of utilization of stored polyP [7, 9]. These typical behaviors of key substances involved in EBPR are graphically shown in Fig. 1b. The acetate taken up is converted to and accumulated as PHAs. Satoh et al. [15] found that the PHAs accumulated in the PAB-enriched sludge are composed of four monomeric units: 3HB, 3HV, 3H2MB, and 3H2MV. Inoue et al. [16] analyzed the chemical structure of these PHAs by NMR and verified that they are co-polymers composed of the above four monomeric units. As for carbohydrate, Liu et al. [17] proved enzymologically that the carbohydrate stored in the anaerobic-aerobic sludge is a polymer of glycosyl units with the alpha-1,4- and the alpha-1,6-linkages, or glycogen. When acetate is fed as the carbon source, 3HB-rich PHAs are formed in the anaerobic stage [15]. The conversion of acetate to PHA requires reducing power, because PHA is a more reduced compound than acetate. To explain the source of the reducing power under the conditions without electron acceptors, a hypothetical model was proposed by Mino et al. [9, 18] and Arun et al. [7]. In that model, anaerobic degradation of stored glycogen to acetyl-CoA as well as its partial oxidation to CO2 is assumed to account for the generation of the reducing power for PHA synthesis. This model is now called the Mino model, and its relevance has been confirmed by several researchers [19-21]. The outlines of the model are shown in Fig. 2. The theoretical stoichiometry based on the model could quantitatively explain the observed conversions of acetate and glycogen to PHA by PAB-enriched sludges, as shown in the table. Satoh et al. [15] successfully applied a similar concept to explain the anaerobic uptake of propionate in EBPR processes (see the table).
Comparison of theoretical and observed stoichiometry for anaerobic acetate or propionate uptake by PABs (after reference [23])Fig. 2. Anaerobic conversion of acetate and glycogen to PHA in polyphosphate-accumulating bacteria in the EBPR process (modified from reference [15]).
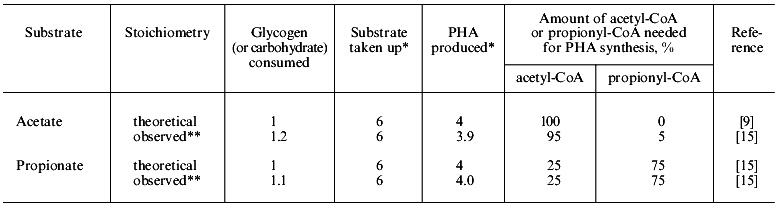
*Relative valued based on 6 moles of substrate uptake.
**Percentage of acetyl-CoA and propionyl-CoA is calculated from the data given in the paper cited.
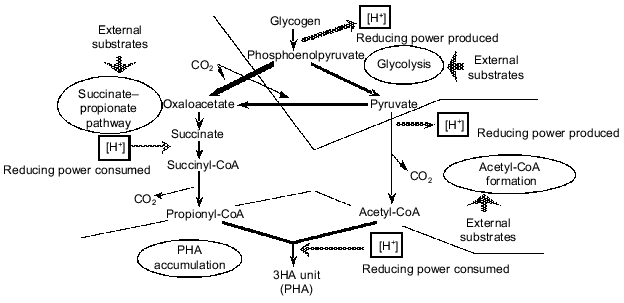
When lactate was used anaerobically by a PAB-enriched sludge, a significant amount of 3HV was detected in the PHA produced [15]. A 3HV unit of PHA is made from an acetyl-CoA molecule and a propionyl-CoA molecule. Therefore, this experimental result strongly implied that there should be a pathway to convert lactate to propionyl-CoA. To explain this result, Satoh et al. [15] and Liu et al. [17] assumed partial conversion of pyruvate to propionyl-CoA through the succinate-propionate pathway. The succinate-propionate pathway is used by certain anaerobic bacteria to metabolize pyruvate to propionate through reverse reactions of a part of the tricarboxylic acid (TCA) cycle [22]. This conversion consumes reducing power (NADH). This means that glycogen can either produce reducing power if metabolized through acetyl-CoA or consume reducing power if metabolized through propionyl-CoA. Based on such an argument, a comprehensive concept is proposed to describe a key strategy of PAB to win in the selection in EBPR processes; this is summarized in Fig. 3 ([23-25]). According to this concept, glycogen stored in the cell functions as a regulator of the redox balance in the cell. Conversion of glycogen to acetyl-CoA and CO2 generates reducing power, whereas conversion to propionyl-CoA via the succinate-propionate pathway consumes reducing power. It is essential for PAB to be able to take up carbon sources faster than other bacteria in the anaerobic stage, no matter what kinds of substrates are fed from the influent wastewater. The above mechanism can provide PAB with a capability to take up various kinds of reduced or oxidized organic substrates in the anaerobic phase without disturbing the redox balance in the cell. The glycogen storage is a key strategy for PAB to maintain the redox balance in the anaerobic uptake of various organic substrates, and hence to win in the selection in the EBPR process [11, 24].
Another hypothesis was postulated by Matsuo et al. [26], Comeau et al. [27, 28], and Wentzel et al. [13, 29] to account for the source of the reducing power in anaerobic acetate metabolism. According to this hypothsis, the TCA cycle is assumed to operate under anaerobic conditions in order to oxidize a part of acetate to CO2 and to generate reducing power in the form of NADH. This model is referred to as the Comeau-Wentzel model. Usually the TCA cycle operates only under aerobic or anoxic conditions. The oxidation of succinate to fumarate in the TCA cycle requires a terminal electron acceptor with a redox potential (E0´) more positive than that of fumarate/succinate couple (+32 mV). Only O2 (O2/H2O, E0´ = +818 mV), NO3- (NO3-/NO2-, E0´ = + 433 mV), and NO2- (NO2-/N2-, E0´ = +970 mV) appear to meet these conditions [30]. Contradictory to this thermodynamic theory, a complete TCA cycle has been found to operate in some anaerobic eubacteria [31] or archae [32]. These microorganisms can oxidize acetate completely to CO2 via the TCA cycle by utilizing elemental sulfur, thiosulfate, or sulfate as electron acceptor. It is believed, however, that major part of the required reducing power should be generated through the glycogen metabolism as described in the Mino model rather than through the TCA cycle as predicted by the Comeau-Wentzel model. The reasons are as follows: 1) the theoretical stoichiometry for the glycogen metabolism can explain very well the experimentally observed anaerobic acetate uptake, PHA formation, glycogen utilization, and CO2 production by PAO-enriched sludges [15, 33, 34]; 2) a 13C tracer experiment using NMR [15] indicated that the acetate taken up by the sludge anaerobically was not oxidized to CO2 and thus not metabolized through the TCA cycle, and 3) experiments using 13C-NMR [19, 20] demonstrated that glycogen is involved in the anaerobic metabolism of EBPR sludges.Fig. 3. A conceptual model for anaerobic carbon metabolism in an EBPR process (after references [23, 25]).
On the other hand, there is evidence that indicates the possibility of partial involvement of the TCA cycle in the generation of reducing power by PAB in the anaerobic stage of the EBPR process [20]. Namely, 13C-labeled carbon in the acetate fed to a PAB-enriched sludge was found to be released as CO2 under absolute anaerobic conditions. So far, this is the only experimental result indicating the possible functioning of the TCA cycle in the anaerobic phase of the EBPR process. The function of the TCA cycle in the anaerobic uptake of carbon sources by PAB as well as its contribution to the microbial selection in the EBPR process remains to be further investigated.
In the EBPR process, microorganisms are exposed to alternate carbon-rich anaerobic environments and carbon-poor aerobic environments. By this alternation, synthesis and degradation of three kinds of biopolymers (polyP, PHA, and glycogen) are induced and metabolic cycling through these biopolymers is established in microorganisms. Such metabolic cycling is energy consuming and not favorable for microorganisms in terms of energy utilization efficiency. Ecologically, however, this metabolic cycling enables PAB to win in the microbial selection in the anaerobic-aerobic process [11]. To explain how this metabolic cycling is regulated in the cell, a metabolic flux model was developed by Pramanik et al. [35]. This model contains a complete set of metabolic pathways involved in biosynthesis and energy production and accounts for energy requirements for macromolecule synthesis and metabolite transport across the cell membrane. The model not only supports the hypothesis that the biopolymer metabolism provides a means for PAB to balance intracellular energy supplies, but also suggest pathways at which metabolic regulation should occur [35].
CANDIDATES FOR POLYPHOSPHATE-ACCUMULATING BACTERIA
Acinetobacter spp. Acinetobacter spp. were first proposed as the bacteria responsible for EBPR [2]. Subsequently many researchers reported its predominance in EBPR processes based on culture-dependent identification methods [3, 4, 36]. Later, different culture-independent methods like fluorescent antibody staining [37], quinone profile measurement [38, 39], or fluorescent in situ hybridization with an oligonucleotide probe specific for Acinetobacter [40] were applied to investigate the relevance of Acinetobacter in EBPR. It has now been demonstrated that the classical culture-dependent methods for bacterial counting and isolation are strongly selective for Acinetobacter spp. [40-43] and that Acinetobacter spp. is not primarily responsible for EBPR [11]. Since Acinetobacter spp. were once believed to be “the” candidate of PAB, intensive studies on their physiology and genetics were carried out [44-48]. Results of such studies were reviewed elsewhere [10, 49]. Some isolated strains of Acinetobacter spp. accumulate polyP and PHA under aerobic conditions. But none of them has been demonstrated to possess the typical metabolic characteristics of the PAB-enriched sludge, namely: 1) acetate uptake and its conversion to PHA, and 2) hydrolysis of polyP and subsequent Pi release under the anaerobic conditions (for details, see [10, 11, 50]). A recent study [51] reported that two strains of Acinetobacter isolated from EBPR processes were able to accumulate polyP aerobically, grew in an anaerobic-aerobic chemostat, but were unable to assimilate acetate for production of PHA in the anaerobic stage. So there is still no evidence that Acinetobacter spp. behave as typical PAB.
Microlunatus phosphovorus. Nakamura et al. [52, 53] isolated a new polyphosphate-accumulating bacterium from a laboratory-scale EBPR process and proposed the name Microlunatus phosphovorus for it. Microlunatus phosphovorus is physiologically close to PAB found in enriched sludges. It accumulates large amounts of polyP under aerobic conditions, which is then consumed along with the anaerobic uptake of carbon sources like glucose and casamino acids. However, it lacks key metabolic characteristics; it neither takes up acetate nor accumulates PHA under anaerobic conditions. Recently, a 16S rRNA-targeted oligonucleotide probe for Microlunatus phosphovorus was developed and applied to quantify the bacterium in an EBPR process [54]. By the use of the probe, Microlunatus phosphovorus was found to constitute about 2.7% of the total bacteria, when the percentage of PAB detected by DAPI stain for polyP was about 9% of the total bacteria. The relevance of Microlunatus phosphovorus to EBPR still remains to be clarified. Ubukata and Takii [55] isolated a bacterium that morphologically and physiologically resembles Microlunatus phosphovorus and demonstrated that the bacterium exhibited the anaerobic utilization and aerobic accumulation of polyP only after alternating anaerobic and aerobic conditions were applied a few times. This result implies that the enzyme system for the polyP metabolism is not constitutive but inducible.
Lampropedia spp. A polyP and PHB storing bacterial strain was isolated from an SBR system designed for EBPR and identified as Lampropedia spp. [56, 57]. This isolate has the ability to take up acetate and store it as PHA with concomitant polyP degradation and Pi release under anaerobic conditions. Functionally it possesses the key metabolic characteristics of PAB, but morphologically it has a very unique sheet-like cell arrangement that is not common in EBPR processes. It is not certain at this moment that Lampropedia spp. surely plays a significant role in EBPR in general.
Rhodocyclus. The presence of the Rhodocyclus group in EBPR processes was first reported by Bond et al. [58]. When the PCR cloning approach was applied to two EBPR reactors, 15 out of 189 isolated clones phylogenetically belonged to the group Rhodocyclus [58]. Recently, fluorescent in situ hybridization showed that an EBPR reactor fed with an acetate-containing substrate was dominated by the Rhodocyclus group up to 81% of the DAPI-stained cells [59]. A type clone was isolated from this community. Type-specific in situ hybridization and direct rRNA sequencing revealed that this type was the dominant bacterium in the community. Staining of intracellular polyP and PHB confirmed that the dominant bacterium in this community performs the key metabolism of PAB, namely acetate uptake accompanied by PHA accumulation and Pi release along with polyP utilization under anaerobic conditions [59]. This result strongly suggests that certain species within the Rhodocyclus group should be mainly responsible for EBPR at least under certain circumstances.
MICROBIAL COMMUNITY STRUCTURE OF ENHANCED BIOLOGICAL PHOSPHATE
REMOVAL PROCESS
When Acinetobacter was first proposed as PAB [2], there were very few researchers who raised the question of whether Acinetobacter is the only bacterium responsible for EBPR. It may have been somehow assumed that EBPR sludges with high phosphate removal capability were dominated by a single group of microorganisms, and few attempts were made to find candidates of PAB other than Acinetobacter. Now, new and powerful tools for the analysis of microbial community structures have been developed and used to analyze EBPR sludges. They include chemotaxonomic methods like quinone profiling [38] and molecular methods like the fluorescent in situ hybridization (FISH) [60], the clone library approach, denaturing gradient gel electrophoresis (DGGE) [61], terminal restriction fragment length polymorphisms (T-RFLP) [62], etc.
High microbial diversity of the EBPR sludge has been demonstrated by using these new techniques. Quinone profiling was applied to characterize activated sludge community structures [38, 63, 64]. The type of quinone in biological samples can be quantitatively determined, and the quinone patterns should explicitly reflect the chemotaxonomic composition of the examined samples. It was suggested that EBPR sludges consist of several different chemotaxonomic groups. The most abundant quinone, Q-8, accounts for only about 31% of total quinone in a PAB-enriched sludge (phosphorus content, 1.94 mmoles or 60 mg/g of suspended solids); the second most abundant one, Q-10, accounts for 8.5%; the third, MK-8(H4), 6.5% (calculated from Hiraishi et al. [64]). The FISH technique with group-specific oligonucleotide probes targeting rRNA showed that an EBPR sludge contained the alpha-, beta-, and gamma-subclasses of proteobacteria, the cytophage group, and Gram-positive bacteria with high G+C DNA contents (13, 25-33, 10-12, 1, and 17-27% of the total cell count, respectively) [41]. In other words, the EBPR sludge phylogenetically consisted of several different microbial groups, which has been confirmed using FISH by other researchers as well [42, 65]. The T-RFLP of PCR-amplified 16S rDNA more directly showed [66] the high population diversity; about 19-24 different numerically dominant ribotypes were observed in a highly PAB-enriched sludge (phosphorus content, 12% of suspended solids). The DGGE technique also showed that PCR-amplified 16S rDNA fragments of EBPR sludges contained a number of dominant DNA fragments with different sequences, implying that the examined EBPR communities had high genotypical diversity [67]. All these results strongly suggest that PAB are not a single genotype or a few limited genospecies, but involve phylogenetically and taxonomically diverse groups of bacteria.
Bond et al. [58] applied PCR cloning to two activated sludges, one with high phosphate removal performance as well as the typical metabolism of PAB and the other without. They found that the Rhodocyclus group belonging to the beta-subclass of proteobacteria was present in significantly higher numbers in the high-phosphate sludge than in the low-phosphate sludge. This result suggests that the Rhodocyclus group may have a specific role in EBPR. However, the Rhodocyclus group occupied only 14% of the total isolated clones in the high-phosphate sludge. It is not certain that this can account for the observed high phosphate removal performance. As discussed before, an EBPR community was reported in which a type of Rhodocyclus was overwhelmingly dominating (81% of total bacteria) [59]. For the time being, this is the only case where a single dominant bacterium was shown to be essentially responsible for EBPR. Kawaharasaki et al. [68] used dual staining with DAPI for polyP and rRNA-targeted oligonucleotide probes specific to different bacterial groups to identify PAB in situ. Many of the Gram-positive bacteria with high G+C DNA content and the alpha-subclass of proteobacteria gave the fluorescent DAPI signal of polyP. Therefore, these two groups were considered to accumulate polyP in the EBPR sludge examined. Christensson et al. [69] reported that the Gram-positive bacteria with high G+C DNA content was suspected to play an important role in EBPR because relatively high occurrence of this bacterial group was observed in a clone library from an EBPR process. A significant portion of the clones of Gram-positive bacteria with high G+C DNA content was phylogenetically close to Terrabacter tumescens. Based on comparison of the electrophoregrams of T-RFLP of 16S rDNA fragments from a highly PAB-enriched sludge (phosphorus content, 12% of suspended solids) and a sludge with very low phosphate content (2% of suspended solids), Liu et al. [62] concluded that the Gram-positive bacteria with high G+C DNA contents are not the only major component of PAB. Hiraishi et al. [64] showed using the quinone profiling approach that the quinone patterns of activated sludges treating municipal sewage were similar to each other irrespective of the type of activated sludge process; sludges from EBPR processes and conventional processes had very similar quinone patterns. The comparison of quinone patterns from different activated sludges suggested that introduction of the anaerobic stage into the fully aerobic conventional activated sludge process does not result in significant population shift. These findings described above may again lead to the conclusion that PAB, which physiologically possess unique metabolic characteristics, should include different phylogenetic and taxonomic bacterial groups: most probably the alpha-, beta-, and gamma-subclasses of proteobacteria and the Gram-positive bacteria with high G+C DNA contents are the candidates.
FUTURE PERSPECTIVES
The present review shows that PAB are not composed of a few limited genospecies, but involve phylogenetically and taxonomically diverse groups of bacteria. The type of bacteria responsible for EBPR may vary among different situations. To clearly define the microbial community structure of EBPR processes and to describe mechanism of ecological selection for PAB in EBPR processes, a closer look into occurrence and behavior of individual species of PAB in various EBPR processes will be needed. Since many of PAB seem to be impossible to culture, molecular methods are surely powerful tools for this purpose. A common EBPR metabolism seems to exist in phylogenetically diverse microbial populations of PAB. This suggests the possibility that the key genes of the EBPR metabolism are common among different bacteria. It is important and interesting to determine such key genes and to find how they are regulated genetically or enzymologically.
REFERENCES
1. Barnard, J. L. (1975) Water Res., 9,
485-490.
2. Fuhs, G. W., and Chen, M. (1975) Microb.
Ecol., 2, 119-138.
3. Buchan, L. (1983) Wat. Sci. Tech.,
15, 87-103.
4. Lotter, L. H. (1985) Wat. Sci. Tech.,
17, 127-138.
5. Mino, T., Kawakami, T., and Matsuo, T. (1984)
Wat. Sci. Tech., 17, 93-106.
6. Mino, T., Kawakami, T., and Matsuo, T. (1985)
Wat. Sci. Tech., 17, 11-21.
7. Arun, V., Mino, T., and Matsuo, T. (1988) Water
Res., 22, 565-570.
8. Marais, G. v. R., Lowenthal, R. E., and Siebritz,
I. (1982) Proc. Post Conf. Seminar on Phosphate Removal in
Biological Treatment Processes, Vol. 2,pp. 5-6.
9. Mino, T., Tsuzuki, Y., and Matsuo, T. (1987)
Proc. IAWPRC Int. Conf. on “Biological Phosphate Removal from
Wastewaters”, Adv. Wat. Pollut. Cont. (Ramadori, R., ed.)
Pergamon Press, Rome,pp. 27-38.
10. Jenkins, D., and Tandoi, V. (1991) Water
Res., 25, 1471-1478.
11. Mino, T., van Loosdrecht, M. C. M., and Heijnen,
J. J. (1998) Water Res., 32, 3193-3207.
12. Rabinowitz, B., Koch, F. A., Vassos, T. D., and
Oldham, W. K. (1987) Proc. IAWPRC Int. Conf. on “Biological
Phosphate Removal from Wastewaters”, Adv. Wat. Pollut.
Cont. (Ramadori, R., ed.) Pergamon Press, Rome, pp. 349-352.
13. Wentzel, M. C., Ekama, G. A., Loewenthal, R. E.,
Dold, P. L., and Marais, G. v. R. (1989) Water SA, 15,
89-102.
14. Smolders, G. J. F., van Loosdrecht, M. C. M.,
and Heijnen, J. J. (1996) Water Res., 30, 2748-2760.
15. Satoh, H., Mino, T., and Matsuo, T. (1992)
Wat. Sci. Tech., 26, 933-942.
16. Inoue, Y., Sano, F., Nakamura, K., Yosie, N.,
Saito, Y., Satoh, H., Mino, T., Matsuo, T., and Doi, Y. (1996)
Polymer Int.,39, 183-189.
17. Liu, W. T., Mino, T., Nakamura, K., and Matsuo,
T. (1994) J. Ferment. Bioeng., 77, 535-540.
18. Mino, T., and Matsuo, T. (1984) Japan. J.
Water Pollut. Res., 7, 605-609 (in Japanese).
19. Maurer, M., Gujer, W., Hany, R., and Bachmann,
S. (1997) Water Res., 31, 907-917.
20. Pereira, H., Lemos, P. C., Reis, M. A., Crespo,
J. P. S. G., Carrondo, M. J. T., and Santos, H. (1996) Water
Res., 30, 2128-2138.
21. Smolders, G. J. F., van der Meij, J., van
Loosdrecht, M. C. M., and Heijnen, J. J. (1995) Biotech.
Bioeng., 47, 277-287.
22. Gottschalk, G. (1986) Bacterial
Metabolism,2nd ed., Springer-Verlag, N. Y.
23. Mino, T., Satoh, H., and Matsuo, T. (1994)
Wat. Sci. Tech., 29, 67-70.
24. Mino, T., Liu, W. T., Satoh, H., and Matsuo, T.
(1996) Proc. 10th Forum for Applied Biotechnology, Brugge, Vol.
1, pp. 1769-1776.
25. Mino, T., Liu, W. T., Satoh, H., and Matsuo, T.
(1997) Proc. 7th Int. Symp. on Microbial Ecology, pp.
509-616.
26. Matsuo, Y. (1985) Proc. 40th Annual Conf. of
the Jap. Soc. Civil Eng., Vol. 40, pp. 989-990.
27. Comeau, Y., Hall, K. J., Hancock, R. E. W., and
Oldham, W. K. (1986) Water Res., 20, 1511-1521.
28. Comeau, Y., Oldham, W. K., and Hall, K. J.
(1987) Proc. IAWPRC Int. Conf. on “Biological Phosphate Removal
from Wastewaters”, Adv. Wat. Pollut. Cont. (Ramadori,
R., ed.) Pergamon Press, Rome, pp. 39-55.
29. Wentzel, M. C., Lotter, L. H., Ekama, G. A.,
Loewenthal, R. E., and Marais, G. R. (1991) Wat. Sci. Tech.,
23, 567-576.
30. Thauer, R. K. (1988) Eur. J. Biochem.,
176, 497-508.
31. Shmitz, R. A., Bonch-Osmolovskaya, E. A., and
Thauer, R. K. (1990) Arch. Microbiol., 154, 274-279.
32. Selig, M., and Schonheit, P. (1994) Arch.
Microbiol., 162, 286-294.
33. Satoh, H., Ramey, W. D., Koch, F. A., Oldham, W.
K., Mino, T., and Matsuo, T. (1996) Wat. Sci. Tech., 34,
8-15.
34. Smolders, G. J. F., van Loosdrecht, M. C. M.,
and Heijnen, J. J. (1994) Biotech. Bioeng., 43,
461-470.
35. Pramanik, J., Trelstad, P. L., Schuler, A. J.,
Jenkins, D., and Keasling, J. D. (1999) Water Res., 33,
462-476.
36. Wentzel, M. C., Loewenthal, R. E., Ekama, G. A.,
and Marais, G. v. R. (1988) Water SA, 14, 81-92.
37. Cloete, T. E., and Steyn, P. L. (1987) Proc.
IAWPRC Int. Conf. on “Biological Phosphate Removal from
Wastewaters”, Adv. Wat. Pollut. Cont. (Ramadori, R.,
ed.) Pergamon Press, Rome, pp. 335-338.
38. Hiraishi, A., Masamune, K., and Kitamura, H.
(1989) Appl. Environ. Microbiol., 30, 197-210.
39. Hiraishi, A., and Morishita, Y. (1990) J.
Ferment. Bioeng., 69, 368-371.
40. Wagner, M., Erhart, R., Manz, W., Amann, R.,
Lemmer, H., Wedi, D., and Schleifer, K.-H. (1994) Appl. Environ.
Microbiol., 60, 792-800.
41. Kampfer, P., Erhart, R., Beimfohr, C.,
Bohringer, J., Wagner, M., and Amann, R. (1996) Microb. Ecol.,
32, 101-121.
42. Snaidr, J., Amann, R., Huber, I., Ludwig, W.,
and Schleifer, K. H. (1997) Appl. Environ. Microbiol.,
63,2884-2896.
43. Wagner, M., Amann, R., Kampfer, P., Assumus, B.,
Hartmann, A., Hutzler, P., and Schleifer, K.-H. (1993) Appl.
Environ. Microbiol., 59, 1520-1525.
44. Bonting, C. F. C., van Veen, H. W., Taverne, A.,
Kortstee, G. J. J., and Zehnder, A. J. B. (1992) Arch.
Microbiol., 158, 139-144.
45. Bonting, C. F. C., Willemsen, B. M. F., Vliet,
W. A., Bouvet, P. J. M., Kortstee, G. J. J., and Zehnder, A. J. B.
(1992) FEMS Microbiol. Ecol., 102, 57-64.
46. Knight, G. C., Seviour, R. J., Soddell, J. A.,
McDonnell, S., and Bayly, R. C. (1995) Water Res.,
29,2081-2084.
47. Van Groenestijn, J. W., Deinema, M. H., and
Zehnder, A. J. B. (1987) Arch. Microbiol., 148,
14-19.
48. Van Groenestijn, J. W., Bentvelsen, M. M. A.,
Deinema, M. H., and Zehnder, A. J. B. (1989) App. Environ.
Microbiol., 55, 219-223.
49. Kortstee, J. J., Appeldoorn, K. J., Bonting, C.
F. C., van Niel, E. W. J., and van Veen, H. W. (1994) FEMS
Microbiol. Rev.,15, 137-153.
50. Van Loosdrecht, M. C. M., Smolders, G. J., Kuba,
T., and Heijnen, J. J. (1997) Antonie van Leewenhoek, 71,
109-116.
51. Tandoi, V., Majone, M., May, J., and Ramadori,
R. (1998) Water Res., 32, 903-2912.
52. Nakamura, K., Masuda, K., and Mikami, E. (1991)
J. Ferment. Bioeng., 71, 258-263.
53. Nakamura, K., Hiraishi, A., Yoshimi, Y.,
Kawarasaki, M., Masuda, K., and Kamagata, Y. (1995) Int. J. Syst.
Bacteriol., 45, 17-22.
54. Kawaharasaki, M., Kanagawa, T., Tanaka, H., and
Nakamura, K. (1998) Wat. Sci. Tech., 37, 481-484.
55. Ubukata, Y., and Takii, S. (1994) Water
Res., 28, 247-249.
56. Stante, L., Cellamare, C. M., Mamaspina, F.,
Bortone, G., and Tilche, A. (1997) Water Res., 31,
1317-1324.
57. Stante, L., Cellamare, C. M., Mamaspina, F., and
Tilche, A. (1996) Med. Fac. Landboww. Univ. Gent, 61/4b.
58. Bond, P. L., Hugenholtz, P., Keller, J., and
Blackall, L. L. (1995) Appl. Environ. Microbiol., 61,
1910-1916.
59. Hesselmann, R. P. X., Werlen, C., Hahn, D., van
der Meer, J. R., and Zehnder, A. J. B. (1999) System. Appl.
Microbiol., accepted.
60. Amann, R. I., Krumholz, L., and Stahl, D. A.
(1990) J. Bacteriol., 172, 762-770.
61. Muyzer, G., de Waal, E. C., and Uitterlinden, A.
G. (1993) Appl. Environ. Microbiol., 59, 695-700.
62. Liu, W. T., Marsh, T. L., Cheng, H., and Forney,
L. J. (1997) Appl. Environ. Microbiol.,63,4516-4522.
63. Hirashi, A., and Ueda, Y. (1997) Jpn. J.
Water Treat. Biol.,33,137-149.
64. Hiraishi, A., Ueda, Y., and Ishihara, J. (1998)
Appl. Environ. Microbiol., 64, 992-998.
65. Sudiana, I. M., Satoh, H., Mino, T., and Matsuo,
T. (1998) Wat. Sci. Tech., 38, 69-76.
66. Liu, W. T., Marsh, T., and Forney, J. L. (1998)
Wat. Sci. Tech., 37, 417-422.
67. Brdjanovic, D., van Loosdrecht, M. C. M.,
Hooijmans, C. M., Alaerts, G. J., and Heijnen, J. J. (1997) J.
Environ. Eng., 123, 144-153.
68. Kawaharasaki, M., Tanaka, H., Kanagawa, T., and
Nakamura, K. (1999) Water Res., 33, 257-265.
69. Christensson, M., Blackall, L. L., and Welander,
T. (1998) Appl. Microbiol. Biotechnol.,49,226-234.