One Amino Acid Residue Cannot Determine the Differences in the Catalytic and Regulatory Properties of Monoamine Oxidases A and B
A. E. Medvedev*, A. S. Ivanov, and A. V. Veselovsky
Orekhovich Institute of Biomedical Chemistry, Russian Academy of Medical Sciences, Pogodinskaya ul. 10, Moscow, 119832 Russia; fax: (095) 245-0857; E-mail: medvedev@ibmh.msk.su* To whom correspondence should be addressed.
Received October 16, 2000
Two years ago we discussed the remarkable discovery by Ito's group of a key amino acid responsible for substrate specificity and inhibitor selectivity of monoamine oxidases A and B (MAO A and MAO B) [1]. We asked a reasonable question: “Is just one amino acid residue responsible for substrate specificity of monoamine oxidase A and B?” [2]. According to Tsugeno and Ito, mutation of Phe-208 to Ile in MAO A and mutation of Ile-199 to Phe in MAO B reversed the substrate specificity and inhibitor selectivity of the mutants towards the opposite form of the enzyme, MAO B and MAO A, respectively [1]. Analyzing their data, we made the suggestion that one amino acid may determine the specificity of binding of some selective substrates and inhibitors of MAO A and MAO B, but it cannot account for a number of other basic differences between these enzymes, such as their different affinities for oxygen and distinct effects of substrates on kinetic parameters of half-reactions [2]. Two years ago we discussed the remarkable discovery by Ito's group of a key amino acid responsible for substrate specificity and inhibitor selectivity of monoamine oxidases A and B (MAO A and MAO B) [1]. We asked a reasonable question: “Is just one amino acid residue responsible for substrate specificity of monoamine oxidase A and B?” [2]. According to Tsugeno and Ito, mutation of Phe-208 to Ile in MAO A and mutation of Ile-199 to Phe in MAO B reversed the substrate specificity and inhibitor selectivity of the mutants towards the opposite form of the enzyme, MAO B and MAO A, respectively [1]. Analyzing their data, we made the suggestion that one amino acid may determine the specificity of binding of some selective substrates and inhibitors of MAO A and MAO B, but it cannot account for a number of other basic differences between these enzymes, such as their different affinities for oxygen and distinct effects of substrates on kinetic parameters of half-reactions [2].
Nevertheless, Jean Shih's group continued the search for such a key amino acid, and in the September issue of the Journal of Neurochemistry (2000), Geha et al. published a paper entitled “Phe208 and Ile199 in human monoamine oxidase A and B do not determine substrate and inhibitor specificities as in rat” [3]. Using human MAO A and MAO B for mutation analysis (amino acid sequences of human and rat MAOs share 88% identity), they found that switching Phe-208-->Ile in MAO A was accompanied by 4-fold increase in Km for both serotonin (a selective substrate of MAO A) and 2-phenylethylamine (PEA, a preferred substrate of MAO B). The reciprocal point mutation Ile199-->Phe in MAO B had no effect on substrate affinity [3]. In contrast to data by Ito's group, these authors did not find reciprocal switching in the sensitivity of the mutant enzymes to diagnostic inhibitors of MAO A and MAO B, clorgyline and deprenyl, respectively.
Two months earlier at the 9th Amine Oxidase Workshop in Barcelona, R. Geha, K. Chen, and J. Shih reported the identification of one corresponding amino acid pair (one in MAO A and one in MAO B) responsible for affinity for selective substrates (serotonin and PEA) and sensitivity to diagnostic inhibitors (clorgyline and deprenyl) [4]. The Ile335-->Tyr substitution in human MAO A caused a dramatic (more than three orders of magnitude) increase in IC50 (concentration required for 50% inhibition of enzymatic activity) for clorgyline, from 1.2 nM to 7.1 µM! The mutant was also characterized by higher sensitivity for deprenyl than the wild form (IC50 of 0.12 and 1.6 µM, respectively). This point mutation also dramatically increased (by more than 30-fold) the Km for serotonin and caused some reduction in the Km for PEA. The reciprocal point mutation Tyr326-->Ile in MAO B had an opposite effect: reduction of IC50 for clorgyline (from 0.63 µM to 28 nM) and increase in IC50 for deprenyl (from 4.3 to 180 nM); it also caused 7-fold decrease in Km for serotonin and 5-fold increase in Km for PEA. However, in contrast to the preferred substrates traditionally used for analysis of MAO A and MAO B activities, the Km value for dopamine (a common substrate for MAO A and MAO B) was increased in both mutants (R. Geha and K. Chen, personal communication). The fact that “the kinetic constants of the mutants were not identical to those of the opposing wild types suggests that other amino acids are also involved in substrate and inhibitor preference” [4].
It should be noted that besides data on molecular biological manipulations with the mutant forms, old and recent results of biochemical studies obtained on wild-type MAO provide convincing evidence for the involvement of several amino acid residues in the interaction of these enzymes with substrates and inhibitors. For example, analysis of covalent adducts formed during MAO-dependent conversion of various mechanism-based inhibitors [5] suggests involvement of different components of the active site in their formation (figure). Of course this fact as such may reflect not only various reactivity of metabolically activated inhibitors with respect to flavin component and/or amino acid residues; it may be a consequence of different binding sites of these reactive species in the active site. We found [6] that some competitive reversible inhibitors (e.g., isatin) have different protective effects on MAO A and MAO B against irreversible inhibition caused by inhibitors forming different covalent adducts with different components of the active site (table). The latter is inconsistent with the hypothesis of the involvement of the same amino acid residues in the interaction of MAO with numerous substrates and inhibitors.
If the contradiction between the data of Tsugeno and Ito [1] and Geha et al. [3, 4] reflects slight species-specific differences in MAO structures (corresponding rat and human MAOs A have sequence homology of 92-93% [3]), we suggest that the “critical” amino acids found by the two groups are involved in the formation of a hydrophobic site responsible for binding of hydrophobic and aromatic groups of substrates and inhibitors in the active site. The existence of such a binding site is quite possible [7, 8]. MAOs are enzymes with board substrate specificity. They catalyze oxidative deamination of amines of various length and 3-D structure and, therefore, the existence of such site would be an effective way for “lowering substrate specificity”. So, playing with the modification of “corresponding amino acid pairs” may result in “switching of specificity and selectivity” of these enzymes with respect of some substrates and inhibitors.
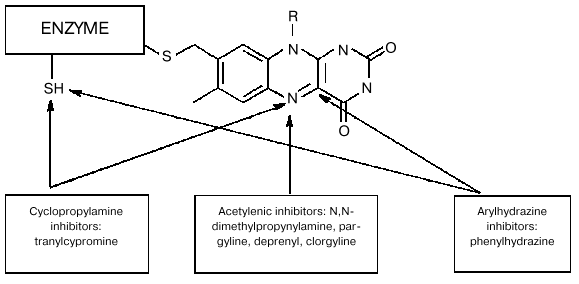
Scheme of covalent adduct formation with active site elements of MAO
Effect of isatin on irreversible inhibition of rat liver mitochondrial
MAO A and MAO B by pargyline and phenelzine (modified from [6])
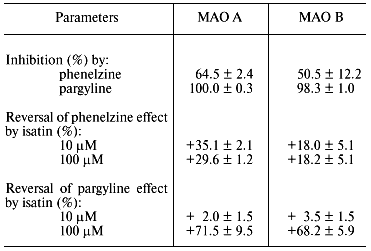
Note: After washing of mitochondria the remaining inhibition of MAO A
and B by 10 and 100 µM isatin did not exceed 5 and 15%,
respectively.
Thus, the results obtained using wild-type and mutant MAOs give an unequivocally negative answer to the question: “Is one amino acid residue responsible for substrate specificity of monoamine oxidase A and B?” For enzymes like MAO characterized by board substrate specificity, one amino acid residue (among several residues constituting the active site structure) may determine preferential binding of several (but not all!) substrates and inhibitors.
REFERENCES
1.Tsugeno, Y., and Ito, A. (1997) J. Biol.
Chem., 272, 14033-14036.
2.Veselovsky, A. V., Ivanov, A. S., and Medvedev, A.
E. (1998) Biochemistry (Moscow), 63, 1441-1446.
3.Geha, R., Chen, K., and Shih, J. (2000) J.
Neurochem., 75, 1304-1309.
4.Geha, R., Chen, K., and Shih, J. (2000) in 9th
Int. Amine Oxidase Workshop, the Millenium Meeting,
Universitat Autonoma, Barcelona, p. 59.
5.Singer, T. P. (1985) in Structure and Functions
of Amine Oxidases (Mondovi, B., ed.) CRC Press, Boca Raton,
Florida, pp. 219-230.
6.Panova, N. G., Zemskova, M. A., Axenova, L. N., and
Medvedev, A. E. (1997) Neurosci. Lett., 233, 58-60.
7.Severina, I. S. (1980) Biokhimiya,
45, 1897-1908.
8.Efange, S. M., and Boudreau, R. J. (1991) J.
Comput. Aided Mol. Des., 5, 405-417.