REVIEW: Nuclear Melatonin Receptors
A. N. Smirnov
Laboratory of Endocrinology, School of Biology, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (7-095) 939-4309; E-mail: smirnov_an@hotmail.com
Received June 26, 2000; Revision received September 6, 2000
Current opinions on the potential role of orphan nuclear retinoid receptors of the ROR/RZR subfamily in regulatory activities of the pineal gland hormone melatonin are reviewed. The mechanisms of receptor-DNA interactions and known coactivators, tissue peculiarities of the expression of different receptor isoforms, and its regulation are described. The spectrum of probable targets for regulation by the receptors, the most promoted of which in this aspect being the immune and central nervous systems, is traced. It is clear that for final “adoption” of the orphan ROR/RZR receptors, there is need for a full collaboration of endocrinologists for solution of the still debatable questions whether and under which situations melatonin does serve as a physiological modulator of the activities of these receptors.
KEY WORDS: melatonin, orphan nuclear receptors, ROR/RZR, RORE hormone response elements, differentiation, regulation, immune system, nervous system, lipid metabolism
The history of investigations of nuclear receptors covers more than three decades. The first such proteins identified were steroid hormone receptors, which for a long time could be detected only by their ability to selectively and with high affinity bind radioactively labeled hormone ligands. In the early eighties, many endocrinologists engaged in studies of these receptors began re-qualification as molecular biologists; this led to the discovery of cDNA structure and respectively of the amino acid sequence of the first member of the nuclear receptor superfamily in 1986. Just 1-2 years later the amino acid sequences of receptors for all steroid hormones, the insect molt hormone ecdysone, dihydroxyvitamin D3, thyroid hormones, and retinoic acid were known. Intensive studies on the structural-functional organization of these proteins, the mechanisms of their interactions with DNA, and their participation in the regulation of transcription were initiated. In parallel, based on the conservativeness of the DNA-binding domain of hormone receptors, studies on the cloning of new members of the family, whose ligands were previously unknown, have appeared. Thus, such proteins were called “orphan” receptors. Accidentally or not, the first revealed and the first cloned nuclear receptor was the estrogen receptor (ER), and the first orphan receptors were estrogen receptor related proteins (ERR) [1-3]. The history of the development and current state of studies on nuclear receptors can be taken from a number of monographs and symposia [4-6]. The nuclear receptor superfamily now includes about 200 members, the vast majority of which are considered as orphans, though the ligands for some of them have already been found. Both hormone compounds, such as 9-cis-retinoic acid for retinoid receptors of the RXR group, and usual low-molecular-weight metabolites, for example hydroxy-derivatives of cholesterol for steroidogenic factor 1 (SF-1) and liver-specific receptor (LXR), can serve as such ligands. The discovery of orphan receptors brought serious corrections to the paradigm operating for decades on the signal functions of different metabolites. Moreover, it was shown that the same metabolite, including the compounds of hormonal nature, enables the use in parallel of two main paths of signal transduction: through membranes and through nuclear receptors. In this way, for example, some eicosanoids act (through G-protein-coupled membrane receptors and through peroxisome proliferator-activated receptors, PPAR). In such fashion act also the hormone of pineal gland melatonin whose nuclear receptors belong to the subfamily ROR/RZR.
The subfamily of retinoid Z receptors (RZR) or retinoid orphan receptors (ROR) includes the products of three genes: splicing variants of RORalpha (RORalpha1, RORalpha2, RORalpha3, RZRalpha) which differ in the N-terminal domain, and RZRbeta, and RORgamma, which contain 523, 556, 548, 468, 459, and 560 amino acid residues in human beings, respectively, and which have a domain organization typical for nuclear receptors [7-9] (Fig. 1). The structure includes an N-terminal variable domain A/B with a constitutive transactivation function 1 (AF1), conservative DNA-binding domain C, variable hinge domain D, relatively conservative ligand-binding domain E containing a ligand-dependent transactivation function 2 (AF2), and an optional short domain F. Detailed description of the structure and functioning of the domains are given in symposium [6]. In mice, a thymocyte-specific variant of RORgamma (RORgamma-t) of 495 amino acid residues has been found [10]. The chromosomally mapped genes of these receptors (RORalpha: human: 15q21-q22, mouse: 9 36,0 cM; RORbeta/RZRbeta: human: 9q22, mouse: 4; RORgamma: human: 1q21, mouse: 3 F2) have structure typical for nuclear receptor genes, which includes 11 exons, among them separate exons for each of two “zinc fingers” of the DNA-binding domain [11, 12]. The members of the subfamily possess constitutive transcriptional activities (positive or negative, depending on the promoter and cell type), and this activity can be intensified by a ligand.
Fig. 1. Domain organization of nuclear receptors.
ROR/RZR RECEPTOR LIGANDS
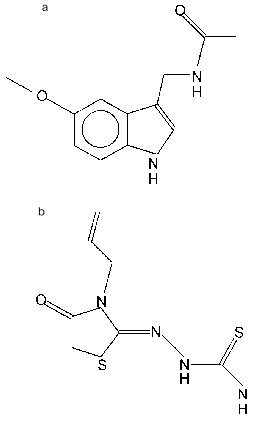
Receptors of the ROR/RZR subfamily include a few orphan receptors for which the existence of a natural regulatory ligand has been unambiguously established, this ligand having hormonal nature (i.e., it is secreted into blood and able to act remote from the place of its synthesis). This ligand is melatonin, a product of the pineal gland (epiphysis) (Fig. 2a). The constants of melatonin binding to nuclear receptors and the melatonin concentration that induces through these receptors a half-maximal effect in vitro are in the nano- or sub-nanomolar region, similar to the concentration of melatonin itself in the blood [13-16]. Thus, the expressed in transfected HeLa cells rat RZRbeta bound 2-iodomelatonin and with Kd = 5 nM, while half-maximal activation of a reporter gene was observed at hormone concentration (EC50) of 3 nM [14]. According to analysis at equilibrium, the binding of 2-[125I]iodomelatonin to cell nuclei from rat spleen and thymus was characterized by Kd values of 0.068 and 0.102 nM; according to studies on binding kinetics, these values were 0.166 and 0.537 nM, respectively [13]. Melatonin concentrations in the blood vary over a broad range depending on a time of day and season, for example, in rams from 0.14 to 2.3 nM [16]. Though retinoid hormones give the name to this receptor subfamily, they are not the ligands of these receptors. The question whether different members of the ROR/RZR subfamily differ in their hormonal binding specificity is of significant interest. Unfortunately, detail analysis of ligand determinants has not been performed yet, and the ligand-binding properties of different members of the subfamily have not been compared.
In addition to the influence on intracellular localization of a pigment in the skin (which is reflected by the name of the hormone), which is mediated by membrane hormone receptors, melatonin plays a significant role in the regulation of sexual maturation and seasoning of reproduction, shows anti-proliferative activity in a number of cells including tumor cells, serves as a trap for free radicals, and acts in many other ways [16-18]. In experiments with transfection of RORE-containing reporter constructions, melatonin and its analogs that interact predominantly with nuclear receptors of the ROR/RZR subfamily significantly increased the transactivating effects of these receptors [19, 20], though not in all studies [21]. It is possible that the stimulation of transcriptional activity of these receptors observed in the presence of fetal calf serum is due to the effect of melatonin [22, 23]. Nevertheless, experiments with melatonin administration in vivo did not yet answer the question whether melatonin does serves as a physiological regulator of the activities of receptors of the ROR/RZR group and which physiological functions are indeed regulated by melatonin through these receptors [20]. Presumably, the use of classic endocrinological approaches (epiphysisectomy, substitutive therapy, administration of anti-hormones) will answer these questions more definitely.
The discovery of the interaction of ROR/RZR group receptors with melatonin stimulated the search for other ligands of these receptors, and such ligands were found among drugs of the thiazolidinedione group (Fig. 2b) that possess anti-arthritic activity. A high correlation between the ability of a number of thiazolidinediones to activate RORalpha_and to produce preventive and therapeutic effects against adjuvant-induced arthritis in rats was found. However, the melatonin analog S20098, which is active with membrane hormonal receptors, had no ability to activate RORalpha and to prevent the development of arthritis [20].
Fig. 2. Ligands for ROR/RZR receptors: a) melatonin (N-acetyl-5-methoxytryptamine); b) CGP 52608 (1-(3-allyl-4-oxothiazolidine-2-ylidene)-4-methylthiosemicarbazone).
OTHER POSSIBLE REGULATORS FOR ROR/RZR RECEPTORS
The activity of receptors of the ROR/RZR subfamily, as well as the activity of a number of other nuclear receptors (see, for example, [24]), in addition to their own ligands, can perhaps also be regulated by kinases. Ca2+/calmodulin-dependent protein kinase IV was shown to potentiate significantly the transcriptional activities of RORalpha1, RORalpha2, and RORgamma. It is astonishing, however, that the effect of this kinase is not related to receptor phosphorylation; it is perhaps due to the effect of the kinase as a regulator of interactions with coactivators, this being indicated by the inhibition of the kinase by peptides containing the LXXLL motif (characteristic for coactivators that interact with the AF2 function of nuclear receptors) [25]. It is possible that the known antitumor activity of melatonin is partially due to the observed ability of RZRbeta and RORalpha to interact with nucleoside diphosphate kinase-2 (NDPK-2, synonyms: NM23-2, c-myc regulatory factor PuF, differentiation inhibitory factor) [26].
INTERACTIONS OF ROR/RZR RECEPTORS WITH DNA
The subfamily members bind to DNA as monomers and recognize hormone response elements (ROREs) represented by 5´-terminal- extended half-sites with the A/GGGTCA motif [7, 9, 19, 21, 22, 27, 28]. Besides an authentic DNA-binding domain of the receptor (RORalpha1 as an example), flanking sequences of the N-terminal regulatory domain and of the hinge region also take a part in RORE recognition; the hinge region, according to deletion and mutation analysis, plays a leading role in recognition of the 5´-extension of the binding site that is characteristic also for other monomeric nuclear receptors. The both sequences flanking the DNA-binding domain, according to data with hybrid receptors, are able to act as autonomous elements of discrimination between different ROREs [7, 27]. The mechanism of the participation of the sequence flanking the DNA-binding domain from the N-terminus in receptor recognition of different ROREs probably relates to the appearance of DNA bending by means of the hinge region. The N-terminal sequence is suggested to impart a proper mutual orientation of zinc fingers and the flanking C-terminal sequence. Though a functional role of DNA bending induced by nuclear receptors and other factors of transcription is still unclear, it is noteworthy that the bend angle induced by monomeric RORalpha1 and RORalpha2 appears significantly higher than is observed in the presence of dimeric receptors [29]. The equilibrium constant of dissociation of a number of studied natural ROREs for RZRalpha/RORalpha is in the range from 0.6 to 3.2 nM [19].
Using analysis of the influence of DNA methylation on the protein-nucleic acid interaction, RORalpha1 was shown to contact three guanines at the major groove of the 3´-half-site of RORE and with three adenines at the minor groove of a hexanucleotide 5´-extension of the site. The results of experiments with receptor deletion variants suggest that the receptor is oriented along one face of the DNA helix in such a way that zinc fingers of the receptor interact with the major groove of the 3´-half-site, while the flanking sequence of the hinge region interacts with the minor groove of the 5´-extension of the RORE [27, 29]. Experiments with ethylation of phosphate residues in the RORE showed that in addition to specific interactions with bases, the phosphodiester backbone of DNA also participates in receptor binding, the receptor having contacts with phosphates of both the transcribed DNA strand (at the center of the RORE) and the complementary strand (at the borders of the RORE) [27].
In certain cases a homodimer may be a functionally preferable active form of the receptor, because RZRbeta, though bound to monomeric RORE, increased transcription only in the presence of two half-sites with 5´-extensions (direct repeats with a spacer of 6-9 bases or palindromes without a spacer) in a promoter. Only a spacer with a proper distance between two ROREs provided the binding of the second receptor molecule. It should be noted, however, that no cooperation in the binding of the two RZRbeta molecules was observed, and experiments with hybrid receptors in CV1 cells did not reveal the formation of homodimers in vivo [21, 22]. The existence of a paired RORE was found in the promoter of the oxytocin gene centered at -180 and -160, both sites of RORalpha binding being significant for the stimulatory effect of the receptor on transcription [30]. The cause of the functional activity of paired ROREs in the absence of physically detected dimerization remains unclear.
The AT-base enriched 5´-extension plays a role in discrimination of signals of different subfamily members [7]. Thus, for example, the RORE of the 5-lipoxygenase gene promoter represented by the sequence CAAAATGGGTCA binds to RZRalpha and RORalpha1, but not to RORalpha2 and RORalpha3 [31]. A similar situation occurs in the case of rat apolipoprotein A-1 gene promoter, where AT-enriched 5´-extension of the RORE overlaps a TATA box [32]. Substitutions in the basic hexanucleotide sequence, as takes place in ROREs of the same gene in mice, significantly reduce the affinity for receptor and transactivating efficacy [32].
ROREs are not absolutely selective for receptors of the ROR/RZR group. For example, orphan receptors Rev-erb alpha and Rev-erb beta interact as monomers with a RORE for RORalpha1 and block the transactivating effect of the later, though being inactive by themselves [33]. Similarly, orphan receptor RVR, which does not have its own intrinsic transcriptional activity in the system used, competitively inhibited the activating effect of RORalpha2 by binding to the same RORE [34]. In a number of genes, ROREs are included in the response elements for other nuclear receptors (for example, (COUP-TF)2, PPARalpha/RXRalpha, RAR/RXR), this significantly increasing the possibilities for regulation of the respective genes, including cooperative, competitive, and mutual substitutive effects in the action of different receptors and other transcriptional factors [19, 28, 35-37].
COACTIVATORS AND COREPRESSORS OF ROR/RZR RECEPTORS
As with other nuclear receptors, the C-terminal sequence corresponding to the activation function 2 (AF2), which provides for the interaction of receptors with coactivators, is needed for manifestation of the functional activity (transactivation) of the members of the ROR/RZR subfamily [38]. AF2 regions in RZRbeta and retinoic acid receptor RARalpha are mutually substitutive, pointing to the similarity of spectra of coactivators interacting with these receptors. (However, unlike AF2 of RAR and thyroid hormone receptors (T3R), AF2 of RZRbeta cannot functioning autonomously, suggesting an influence of another part of the receptor molecule on the conformation of the AF2 region or insufficiency of separate AF2 for the composition of a fully functional surface for interactions with coactivators [21]. However, the AF2 region of RORalpha can act autonomously [39].) Within the subfamily, RORalpha1 and RORgamma also interact with similar spectra of coactivators: on combined transfection of hybrid receptor Gal4(DBD)-RORgamma and full size RORalpha1 and RORgamma, the later inhibited the activity of the hybrid protein [9]. Adjacent to the AF2 region, the C-terminal fragment of the ligand-binding domain of receptors perhaps enables the interaction with corepressors, as indicated by repression of transcription of a reporter gene by the variant of RZRbeta with deleted AF2, withdrawal of repressor activity with further deletion of the ligand-binding domain at the C-terminus, and repression of transcription by a hybrid protein containing the DNA-binding domain of Gal4 and a fragment of RZRbeta with alpha-helixes 6-11 of the ligand-binding domain [21].
The binding of coactivator p300 and other transcriptional factors, MyoD and bHLH, to receptors of the subfamily has been established, these interactions probably occurring on different surfaces of a receptor [40]. Using a system of double hybrids, the interaction of RORalpha with coactivators GRIP-1 (glucocorticoid receptor-interacting protein-1) and PBP (peroxisome proliferator-activated receptor binding protein, synonyms: TRAP220, DRIP205) was shown. Interestingly, unlike receptors for steroid, thyroid, and retinoid hormones, the interaction of RORalpha with coactivators can also occur in the absence of a ligand [39], this corresponding to constitutive activity of ROR/RZR.
TISSUE SPECIFICITY OF ROR/RZR RECEPTOR EXPRESSION
The expression of subfamily members significantly varies in different tissues. Expression of RZRbeta is the most limited. RZRbeta is detected almost exclusively in neuronal tissue related to the sensory, neuroendocrine, and limbic systems, but not to locomotion [41, 42]. RORgamma is most actively expressed in skeletal muscles and in liver, kidneys, and adipocytes [8, 9, 43] (cited by abstract). An RORgamma variant, RORgamma-t (synonym: TOR, thymus orphan receptor), is specific for thymus, where it is expressed mainly in CD4+/CD8+ double positive thymocytes [10, 44, 45]. Isoforms RZRalpha/RORalpha are expressed in a significantly larger number of tissues: pars tuberalis and pars distalis of the pituitary [46], thalamus, olfactory bulbs, cerebellum (particularly intensively in Purkinje cells) [23, 47], adipose tissue, liver, cartilage [48], skin, testes, etc.
ROR/RZR receptors are targets for regulation by factors that determine cell differentiation and functional activity of differentiated cells. Thus, RORgamma is significantly induced in differentiating adipocytes by ligands of peroxisome proliferator-activated receptor gamma (PPARgamma) and by overexpression of PPARgamma itself [43] (cited by abstract). Similar data were obtained for RORalpha [49]. RORalpha begins to be intensively expressed on differentiation of P19 cells into neuronal, but not into muscle cells [23]. In the pineal gland, the expression of RZRbeta is under control of the same mechanisms (adrenergic stimulation --> cyclic AMP) that regulate photoperiodic biosynthesis of melatonin, a ligand of this receptor [50]. Expression of different isoforms of RORalpha in developing murine brain varies in time and in different structures as indicated by non-coincidence of signals detected with the use of ribonucleic probes corresponding to the 3´- and 5´-terminal sequences of RORalpha cDNA [51]. The data on ROR/RZR expression in the course of embryonic and postnatal life are extremely fragmentary and relate mainly to nervous system structures. Thus, the level of RORalpha mRNA in Purkinje cells of murine cerebellum was shown to reach a maximum by day 15 of embryonic life [47]. In the postnatal development of a mouse, RORalpha expression in the cerebellum as well as in the thalamic and olfactory bulb neurons is maximal at day 16 of life and persists in adults. In hippocampus, maximal expression of RORalpha was observed at day 7 of life [51]. Unlike in the central nervous system, RORalpha expression in murine testes (in peri-tubular cells) could be detected only after sexual maturation [52]. RZRbeta mRNA was revealed in the dorso-medial part of the supra-chiasmic nucleus in rats at day 20 of embryonic life, and it persisted in the course of postnatal life up to day 60 [41]. In rat retinal cells, RZRbeta mRNA also appeared in the second half of embryogenesis [53]. The data suggest that receptors of the ROR/RZR group play a significant role both in processes of differentiation and in functioning of differentiated cells and tissues.
PARTICIPATION OF ROR/RZR RECEPTORS IN REGULATION OF PHYSIOLOGICAL
PROCESSES
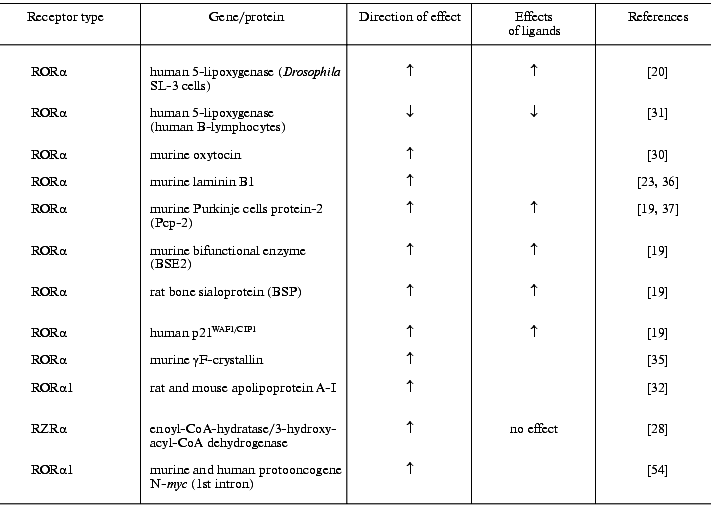
The discovery of ROREs in promoters of genes encoding proteins with very different functions (table), differential expression of different members of the subfamily and its regulation, as well as regulation of ROR/RZR by ligands suggests that the regulatory activity of these receptors encompasses a broad spectrum of physiological processes. The most studied and documented is the participation of ROR/RZR in the regulation of immune processes, central nervous system differentiation, and, possibly, the modulation of lipid metabolism.
Genes whose promoters contain ROREs providing transactivating action of
ROR/RZR receptors and effects of their ligands
Expression of RORgamma-t was found to protect hybridoma T-cells against death on their activation by means of inhibition of Fas ligand and interleukin-2 formation, this suggesting the participation of RORgamma-t in the immune response [45]. The expression of this receptor isoform rises in thymocytes before their activation through gene cluster TCR-Jalpha, the regulatory region of which was found to contain RORE. It is suggested that RORgamma-t takes part in a cascade of events leading to the maturation of alpha/beta T-cells at a stage that provides access to DNA locus TCR-Jalpha [10]. The participation of RORalpha1 and RZRalpha in the regulation of the immune response is assumed based on inhibition of 5-lipoxygenase, one of the key enzymes of the biosynthesis of pro-inflammatory leukotrienes, in B-lymphocytes by these receptors [31]. Unlike T-cells, in human peripheral blood mononuclear cells and in monocyte cultures, melatonin and thiazolidinedione agonists of RORalpha produce stimulatory effects on the formation of interleukins-2 and -6. In these cells, melatonin specifically binds both to cell membranes and to nuclei. RORalpha agonists bind only to nuclei, while a membrane melatonin receptor agonist S20098 binds only to membranes. It is important that effects of thiazolidinediones and S20098 were found to be synergistic, suggesting a double pathway in melatonin action on the immune system [55]. (Interestingly, synergism between nuclear and membrane melatonin receptors is observed also in unicellular organisms [56].)
RORalpha gene mutations providing a receptor form truncated at the C-terminus (staggerer mice) or mRNA form with deleted DNA-binding domain (in this case no protein is expressed), a number of neurological and other abnormalities (tremor, body imbalance, small size) was noted in the experimental animals. The cerebellum of such animals (where normally two RORalpha isoforms, RORalpha1 and RORalpha4/RZRalpha, are expressed [57]) is underdeveloped, Purkinje cells are fewer [58], and their electrophysiological characteristics significantly differ from the wild-type animals [52]. In heterozygous mice, a partial degeneration of Purkinje cells is observed, but it takes place in adulthood, and the degeneration rate is higher in males [59]. Degenerative processes in the cerebellum are assumed to relate to accelerated local production of pro-inflammatory cytokines [60]. In homozygous staggerer, a number of morphological-functional abnormalities were also noticed in the olfactory bulb [61]. These data unequivocally prove the participation of RORalpha in the regulation of the development of the central nervous system. Based on results of experiments with propylthiouracil-induced suppression of thyroid gland functions and substitutive therapy with thyroxin, which showed that thyroid hormones determine the time but not the final level of expression of RORalpha (as well as of a number other markers of development [62, 63]), and similarity between a number of neurological symptoms characteristic for staggerer mutation (see above) and for perinatal hypothyroidism, the hypothesis that RORalpha can serve as one of messengers in the action of thyroid hormones on the developing brain was postulated [64]. Data on a stimulatory effect of RORalpha on transcription of the Purkinje cell marker PCP-2 [37], whose expression time depends on thyroid hormones, is consistent with this conception.
RORbeta expression in progenitors of retina cells (which are of neuronal origin) is significantly reduced in the null mutation of the Chx10 gene, which induces a retardation in eye development, while transfection of RORbeta into progenitor cells significantly accelerates their proliferation, suggesting the participation of RORbeta in eye development [53]. Direct evidence for this statement was obtained using knockout of the RORbeta gene. In RORbeta-/- mice, a significant degeneration of the retina leading to a blindness in adult animals was observed. In addition, abnormality in circadian behavior and the appearance of a duck-like gait were registered in such animals [65]. The data confirm the hypothesis of a role of RZRbeta in the formation of structures engaged in sensory information processing.
The use of the staggerer mouse genetic model possessing a deletion in RORalpha [66] seems to be rather promising for studies on the role of this receptor for differentiation not only of brain, but also of many other structures and functions. Thus, apolipoprotein A-1 expression in the small intestine of animals homozygous for this mutation was found to be significantly decreased, suggesting the participation of RORalpha in apolipoprotein A-1 regulation and, consequently, in lipid metabolism in spite of relative weakness of the RORE of the gene promoter in mice [32]. As a result, in such animals an insufficiency of anti-atherogenic lipoproteins and a predisposition to atherosclerosis develop [67].
Mutant variants of receptors can serve as a useful tool for the study of ROR/RZR functions in experiments in vitro as well. Thus, transfection of mutant RORalpha1 with deleted ligand-binding domain devoid of its own intrinsic transactivation activity into proliferating myoblasts differentiating into myotubules induced a decrease in expression of endogenous RORalpha1, markers of differentiation of MyoD, myogenin (bHLH protein), and p21 (kinase cdk inhibitor), and retardation of manifestation of morphological indicators of differentiation and further development of multinuclear myotubules [40], this serving as an important indication of the participation of RORalpha1 in muscle tissue differentiation.
Accumulated experimental data unequivocally point to an important role of members of the ROR/RZR subfamily in differentiation and regulation of functions of mature cells. It would seem that the discovery of melatonin and thiazolidinediones as ligands of these receptors would stimulate the interest of investigators in questions of refining the spectrum of ROR/RZR functions and forms of melatonin activities mediated by these receptors, creation of ligands selective for separate members of the subfamily and their usage as immunomodulators, lipid metabolism regulators, etc. Surprisingly, no boom in these directions is observed. Moreover, there are only a few studies in vivo on the role of nuclear receptors in the realization of the multiple functions of melatonin [20, 68]. Perhaps this is partly due to the double nature type of ROR/RZR activity: constitutive and ligand-dependent. For most investigators, ligands serve only as tools for the study of receptors, while in many works the ligand theme is not touched upon at all. The discovery of orphan receptors, ROR/RZR in particular, with the following discovery of their ligands, is “reverse” endocrinology, including a distribution of roles between molecular biologists and endocrinologists. The absence of a rapid progress in this field can be considered as a prospective structural crisis. Perhaps the time is coming when physiological approaches, of course together with the use of the accumulated richest arsenal of methods for analysis of receptor activity, and intimate cooperation with chemists should be called on again for the final “adoption” of orphan receptors, including those of ROR/RZR subfamily.
REFERENCES
1.Gorski, J., Toft, D., and Shyamala, L. (1968)
Recent. Progr. Horm. Res., 24, 45-72.
2.Kumar, V., Green, S., Staub, A., and Chambon, P.
(1986) EMBO J., 5, 2231-2236.
3.Giguere, V., Yang, N., Segui, P., and Evans, R. M.
(1988) Nature, 331, 91-94.
4.Rosen, V. B., and Smirnov, A. N. (1981)
Receptors and Steroid Hormones [in Russian], Moscow University
Press, Moscow.
5.Carlstedt-Duke, J., Eriksson, H., and Gustafsson,
J.-A. (eds.) (1989) The Steroid/Thyroid Hormone Receptor Family and
Gene Regulation, Birkhäuser Verlag, Basel.
6.Freedman, L. P. (ed.) (1998) Molecular Biology
of Steroid and Nuclear Hormone Receptors, Birkhäuser,
Boston.
7.Giguere, V., Tini, M., Flock, G., Ong, E., Evans,
R. M., and Otulakowski, G. (1994) Genes Dev., 8,
538-553.
8.Hirose, T., Smith, R. J., and Jetten, A. M. (1994)
Biochem. Biophys. Res. Commun., 205, 1976-1983.
9.Medvedev, A., Yan, Z. H., Hirose, T., Giguere, V.,
and Jetten, A. M. (1996) Gene, 181, 199-206.
10.Villey, I., de Chasseval, R., and de Villartay,
J. P. (1999) Eur. J. Immunol., 29, 4072-4080.
11.Giguere, V., Beatty, B., Squire, J., Copeland, N.
G., and Jenkins, N. A. (1995) Genomics, 28, 596-598.
12.Medvedev, A., Chistokhina, A., Hirose, T., and
Jetten, A. M. (1997) Genomics, 46, 93-102.
13.Rafii-El-Idrissi, M., Calvo, J. R., Harmouch, A.,
Garcia-Maurino, S., and Guerrero, J. M. (1998) J. Neuroimmunol.,
86, 190-197.
14.Becker-Andre, M., Wiesenberg, I.,
Schaeren-Wiemers, N., Andre, E., Missbach, M., Saurat, J. H., and
Carlberg, C. (1994) J. Biol. Chem., 269, 28531-28534.
15.Carlberg, C., and Wiesenberg, I. (1995) J.
Pineal Res., 18, 171-178.
16.Lincoln, G. A. (1987) in Hormonal Control of
Reproduction (Austin, C. R., and Short, R. V., eds.) [Russian
translation], Mir, Moscow, pp. 71-99.
17.Reiter, R. J., and Robinson, J. (1995)
Melatonin, Bantam Books, N. Y.
18.Mor, M., Plazzi, P. V., Spadoni, G., and Tarzia,
G. (1999) Curr. Med. Chem., 6, 501-518.
19.Schrader, M., Danielsson, C., Wiesenberg, I., and
Carlberg, C. (1996) J. Biol. Chem., 271, 19732-19736.
20.Missbach, M., Jagher, B., Sigg, I., Nayeri, S.,
Carlberg, C., and Wiesenberg, I. (1996) J. Biol. Chem.,
271, 13515-13522.
21.Greiner, E. F., Kirfel, J., Greschik, H.,
Dorflinger, U., Becker, P., Mercep, A., and Schule, R. (1996) Proc.
Natl. Acad. Sci. USA, 93, 10105-10110.
22.Carlberg, C., Hooft van Huijsduijnen, R., Staple,
J. K., DeLamarter, J. F., and Becker-Andre, M. (1994) Mol.
Endocrinol., 8, 757-770.
23.Matsui, T., Sashihara, S., Oh, Y., and Waxman, S.
G. (1995) Brain Res. Mol. Brain Res., 33,
217-226.
24.Garabedian, M. J., Rogatsky, I., Hittelman, A.,
Knoblauch, R., Trowbridge, J. M., and Krstic, M. D. (1998) in
Molecular Biology of Steroid and Nuclear Hormone Receptors
(Freedman, L. P., ed.) Birkhäuser, Boston, pp. 237-260.
25.Kane, C. D., and Means, A. R. (2000) EMBO
J., 19, 691-701.
26.Paravicini, G., Steinmayr, M., Andre, E., and
Becker-Andre, M. (1996) Biochem. Biophys. Res. Commun.,
227, 82-87.
27.Giguere, V., McBroom, L. D. B., and Flock, G.
(1995) Mol. Cell. Biol., 15, 2517-2526.
28.Winrow, C. J., Capone, J. P., and Rachubinski, R.
A. (1998) J. Biol. Chem., 273, 31442-31448.
29.McBroom, L. D. B., Flock, G., and Giguere, V.
(1995) Mol. Cell. Biol., 15, 796-808.
30.Chu, K., and Zingg, H. H. (1999) J. Mol.
Endocrinol., 23, 337-346.
31.Steinhilber, D., Brungs, M., Werz, O.,
Wiesenberg, I., Danielsson, C., Kahlen, J.-P., Nayeri, S., Schrader,
M., and Carlberg, C. (1995) J. Biol. Chem., 270,
7037-7040.
32.Vu-Dac, N., Gervois, P., Grotzinger, T., de Vos,
P., Schoonjans, K., Fruchart, J.-C., Auwerx, J., Mariani, J., Tedgui,
A., and Staels, B. (1997) J. Biol. Chem., 272,
22401-22404.
33.Forman, B. M., Chen, J., Blumberg, B., Kliewer,
S. A., Henshaw, R., Ong, E. S., and Evans, R. M. (1994) Mol.
Endocrinol., 8, 1253-1261.
34.Retnakaran, R., Flock, G., and Giguere, V. (1994)
Mol. Endocrinol., 8, 1234-1244.
35.Tini, M., Fraser, R. A., and Giguere, V. (1995)
J. Biol. Chem., 270, 20156-20161.
36.Matsui, T. (1996) Biochem. Biophys. Res.
Commun., 220, 405-410.
37.Matsui, T. (1997) Genes Cells, 2,
263-272.
38.Bagchi, M. K. (1998) in Molecular Biology of
Steroid and Nuclear Hormone Receptors (Freedman, L. P., ed.)
Birkhäuser, Boston, pp. 159-189.
39.Atkins, G. B., Hu, X., Guenther, M. G., Rachez,
C., Freedman, L. P., and Lazar, M. A. (1999) Mol. Endocrinol.,
13, 1550-1557.
40.Lau, P., Bailey, P., Dowhan, D. H., and Muscat,
G. E. (1999) Nucleic Acids Res., 27, 411-420.
41.Park, H. T., Baek, S. Y., Kim, B. S., Kim, J. B.,
and Kim, J. J. (1996) Neurosci. Lett., 217, 17-20.
42.Park, H. T., Kim, Y. J., Yoon, S., Kim, J. B.,
and Kim, J. J. (1997) Brain Res., 747, 332-337.
43.Kurebayashi, S., and Hirose, T. (1998) Nippon
Rinsho, 56, 1729-1733.
44.Ortiz, M. A., Piedrafita, F. J., Pfahl, M., and
Maki, R. (1995) Mol. Endocrinol., 9, 1679-1691.
45.He, Y. W., Deftos, M. L., Ojala, E. W., and
Bevan, M. J. (1998) Immunity, 9, 797-806.
46.Hazlerigg, D. G., Barrett, P., Hastings, M. H.,
and Morgan, P. J. (1996) Mol. Cell. Endocrinol., 123,
53-59.
47.Nakagawa, S., Watanabe, M., and Inoue, Y. (1997)
Neurosci. Res., 28, 177-184.
48.Bordji, K., Grillasca, J. P., Gouze, J. N.,
Magdalou, J., Schohn, H., Keller, J. M., Bianchi, A., Dauca, M.,
Netter, P., and Terlain, B. (2000) J. Biol. Chem., 275,
12243-12250.
49.Austin, S., Medvedev, A., Yan, Z. H., Adachi, H.,
Hirose, T., and Jetten, A. M. (1998) Cell. Growth Differ.,
9, 267-276.
50.Baler, R., Coon, S., and Klein, D. C. (1996)
Biochem. Biophys. Res. Commun., 220, 975-978.
51.Sashihara, S., Felts, P. A., Waxman, S. G., and
Matsui, T. (1996) Brain Res., 42, 109-117.
52.Steinmayr, M., André, E., Conquet, F.,
Rondi-Reig, L., Delhaye-Bouchaud, N., Auclair, N., Daniel, H.,
Crépel, F., Mariani, J., Sotelo, C., and Becker-André, M.
(1998) Proc. Natl. Acad. Sci. USA, 95, 3960-3965.
53.Chow, L., Levine, E. M., and Reh, T. A. (1998)
Mech. Dev., 77, 149-164.
54.Dussault, I., and Giguere, V. (1997) Mol.
Cell. Biol., 17, 1860-1867.
55.Garcia-Maurino, S., Gonzalez-Haba, M. G., Calvo,
J. R., Goberna, R., and Guerrero, J. M. (1998) J. Neuroimmunol.,
92, 76-84.
56.Tslm, S. T., Wong, J. T., and Wong, Y. H. (1996)
J. Peneal. Res., 21, 101-107.
57.Matysiak-Scholze, U., and Nehls, M. (1997)
Genomics, 43, 78-84.
58.Dussault, I., Fawcett, D., Matthyssen, A., Bader,
J. A., and Giguere, V. (1998) Mech. Dev., 70,
147-153.
59.Doulazmi, M., Frederic, F., Lemaigre-Dubreuil,
Y., Hadj-Sahraoui, N., Delhaye-Bouchaud, N., and Mariani, J. (1999)
J. Comp. Neurol., 411, 267-273.
60.Vernet-der Garabedian, B., Lemaigre-Dubreuil, Y.,
Delhaye-Bouchaud, N., and Mariani, J. (1998) Brain Res. Mol. Brain
Res., 62, 224-227.
61.Monnier, Z., Bahjaoui-Bouhaddi, M., Bride, J.,
Bride, M., Math, F., and Propper, A. (1999) Biol. Cellulaire,
91, 29-44.
62.Oppenheimer, J. H., Schwartz, H. I., and Strait,
K. A. (1994) Eur. J. Endocrinol., 130, 15-24.
63.Koibuchi, N., and Chin, W. W. (2000) Trends
Endocr. Metab., 11, 123-128.
64.Koibuchi, N., and Chin, W. W. (1998)
Endocrinology, 139, 2335-2341.
65.Andre, E., Conquet, F., Steinmayr, M., Stratton,
S. C., Porciatti, V., and Becker-Andre, M. (1998) EMBO J.,
17, 3867-3877.
66.Hamilton, B. A., Frankel, W. N., Kerrebrock, A.
W., Hawkins, T. L., FitzHugh, W., Kusumi, K., Russell, L. B., Mueller,
K. L., van Berkel, V., Birren, B. W., Krugiyak, L., and Lander, E. S.
(1996) Nature, 379, 736-739.
67.Mamontova, A., Seguret-Mace, S., Esposito, B.,
Chaniale, C., Bouly, M., Delhaye-Bouchaud, N., Luc, G., Staels, B.,
Duverger, N., Mariani, J., and Tedgui, A. (1998) Circulation,
98, 2738-2743.
68.Weisenberg, I., Chiesi, M., Missbach, M., Spanka,
C., Pignat, W., and Carlberg, C. (1998) Mol. Pharmacol.,
53, 1131-1138.