Some Properties of Caldesmon and Calponin and the Participation of These Proteins in Regulation of Smooth Muscle Contraction and Cytoskeleton Formation
N. B. Gusev
Department of Biochemistry, School of Biology and School of Fundamental Medicine, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-2747; E-mail: NBGusev@mail.ru
Received March 19, 2001; Revision received May 3, 2001
The interaction of caldesmon with different Ca2+-binding proteins has been analyzed, and it is supposed that one of the conformers of calmodulin might be an endogenous regulator of caldesmon. The arrangement of caldesmon and Ca2+-binding proteins within their complexes has been analyzed by different methods. The central helix of calmodulin is supposed to be located near the single Cys residue in the C-terminal domain of caldesmon. The N-terminal globular domain of calmodulin interacts with sites A and B´ of caldesmon, whereas the C-terminal globular domain of calmodulin binds to site B of caldesmon. The complex of calmodulin and caldesmon is very flexible; therefore, both parallel and antiparallel orientation of polypeptide chains of the two proteins is possible in experiments with short fragments of caldesmon and calmodulin. The length, flexibility, and charge of the central helix of calmodulin play an important role in its interaction with caldesmon. Phosphorylation of caldesmon by different protein kinases in vitro has been analyzed. It was shown that phosphorylation catalyzed by casein kinase II of sites located in the N-terminal domain decreases the interaction of caldesmon with myosin and tropomyosin. Caldesmon and calponin may interact with phospholipids. The sites involved in the interaction of these actin-binding proteins with phospholipids have been mapped. It is supposed that the interaction of calponin and caldesmon with phospholipids may play a role in the formation of cytoskeleton. Calponin interacts with 90-kD heat shock protein (hsp90) that may be involved in transportation of calponin and its proper interaction with different elements of cytoskeleton. Calponin, filamin, and alpha-actinin can simultaneously interact with actin filaments. Simultaneous binding of two actin-binding proteins affects the structure of actin bundles and their mechanical properties and may be of great importance in formation of different elements of cytoskeleton.
KEY WORDS: actin, calmodulin, caldesmon, calponin, phospholipids, protein kinases, phosphorylation
In 1939 Sergei Evgenievich Severin founded the Department of Biochemistry at the School of Biology of Moscow State University, and for many years was the head of this department. Muscle biochemistry was one of the traditional lines of investigation in this department. Drs. N. P. Meshkova, I. M. Bocharnikova, and E. V. Petushkova performed successful investigations of some properties of myosin and actomyosin. Drs. A. A. Boldyrev, V. A. Tkachuk, O. D. Lopina, and A. M. Rubtsov analyzed the structure and properties of transporting ATPases involved in the regulation of striated muscle contraction. S. E. Severin was especially interested in the problem of regulation of different biochemical processes. Therefore, regulatory proteins of the contractile apparatus became a subject of inquiry at the Department of Biochemistry. This paper is devoted to a critical review of the data in the literature concerning structure, properties, and mechanism of functioning of some regulatory actin-binding proteins and summarizing experimental data obtained at the Department of Biochemistry in this field.
REGULATION OF FUNCTIONAL ACTIVITY OF CALDESMON BY DIFFERENT
Ca2+-BINDING PROTEINS AND PHOSPHORYLATION
The structure of caldesmon and its interaction with different Ca2+-binding proteins. Caldesmon is a ubiquitous actin-binding protein involved in the regulation of smooth muscle contraction and cytoskeleton formation. The intron-exon structure of the caldesmon gene and the domain structure of the protein are now well investigated. These data as well as data concerning the interaction of caldesmon with different proteins are thoroughly discussed in a number of reviews [1-4]. Therefore, we will only briefly delineate the domain structure of caldesmon and will pay the main attention to the interaction of caldesmon with different Ca2+-binding proteins and phosphorylation of caldesmon by different protein kinases.
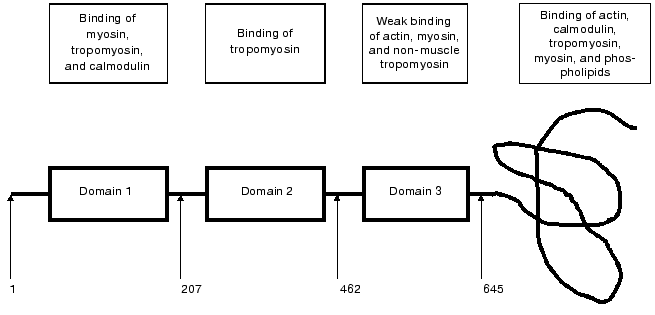
Caldesmon consists of four domains. The first, N-terminal domain (about 210 residues) provides for the interaction of caldesmon with tropomyosin and myosin. It has been shown that this domain also contains weak binding sites for calmodulin. The second domain (about 250 residues) characteristic for smooth muscle caldesmon is mainly helical and participates in the interaction with tropomyosin. The third, also helical and comparably short domain (about 180 residues) is distantly similar to certain sites in the troponin T structure and is involved in the interaction of caldesmon with myosin, tropomyosin, and probably with actin. The fourth, C-terminal domain has a compact structure with low helical content. This domain provides for the interaction of caldesmon with actin, Ca2+-binding proteins, myosin, and tropomyosin (Fig. 1). The fourth domain plays the most important role in the functioning of caldesmon. In the absence of Ca2+ and Ca2+-binding proteins, this domain tightly interacts with actin and inhibits actomyosin ATPase activity either by itself or through its action on tropomyosin [6, 7]. Binding of Ca2+-saturated Ca2+-binding proteins to the C-terminal domain affects the interaction of caldesmon with actin and tropomyosin and in this way reverses the inhibitory action of caldesmon on the actomyosin ATPase activity [1-5]. This rather simple scheme of regulation of actomyosin activity by caldesmon is complicated by certain circumstances. First, there are two [8] or even three [9] calmodulin-binding sites in the C-terminal domain of caldesmon. Second, under in vitro conditions reversion of the inhibitory action of caldesmon on the actomyosin ATPase is achieved only at a very high excess of calmodulin over caldesmon and only under very specific conditions [3]. Therefore, the question arises whether calmodulin is the only protein involved in the regulation of caldesmon action and if so how calmodulin and caldesmon interact with each other.
Trying to answer these questions, we analyzed the interaction of caldesmon with several Ca2+-binding proteins. We found that parvalbumin, lactalbumin, and calcimedin do not interact with caldesmon [10, 11] and therefore cannot be considered as Ca2+-dependent regulators of caldesmon activity. However, the low molecular weight dimeric Ca2+-binding protein S100 formed tight complexes with caldesmon in vitro and effectively reversed the inhibitory action of caldesmon on the ATPase activity of actomyosin [10]. Later these finding were confirmed by Japanese and English colleagues [12, 13]. It is worthwhile to mention that two other proteins, although belonging to the family of S100 proteins, namely caltropin and calcyclin, are also able to interact with caldesmon. However, caltropin regulates caldesmon functioning [14], whereas calcyclin was completely ineffective [15].Fig. 1. Scheme of smooth muscle caldesmon structure. The three first domains are shown as rectangular, the forth domain is depicted as a curving line. Numbers correspond to the amino acid residues located at the border of the neighboring domains. The frames in the upper part of the figure show proteins interacting with the corresponding domains of caldesmon (after Huber, 1997 [5] with modifications).
S100 protein effectively reversed the inhibitory action of caldesmon in the actomyosin system reconstructed from skeletal actin and myosin [10, 13]. At the same time, S100 was much less effective in the system reconstructed from smooth muscle actin [13]. Since neither of the Ca2+-binding proteins tested was effective in reversing the inhibitory action of caldesmon in the system reconstructed from smooth muscle actin, S. B. Marston suggested the presence of a special Ca2+-binding protein participating in the regulation of caldesmon. In a series of investigations performed in Marston's laboratory, a special method was developed for isolation of this protein [16, 17], and some properties of this protein were analyzed in our joint investigation [17]. The data of mass-spectroscopy and partial sequencing reveal that this Ca2+-binding protein is indistinguishable from calmodulin. However, the N-terminal domain of this Ca2+-binding protein was more flexible than the corresponding domain of calmodulin. This protein differs from calmodulin in the kinetics of its proteolysis and the mechanism of interaction with caldesmon [17]. Since the molecular weight of this smooth muscle protein was completely identical to that of calmodulin and this protein does not contain any tightly bound ligands, we suppose that the protein under investigation is calmodulin fixed in an unusual conformation. It is interesting that this conformation is rather stable and seems not to be in equilibrium with other probable conformations of calmodulin. Many questions concerning the structure of this unusual calmodulin conformer remain unanswered and the existence of a stably fixed conformation of calmodulin with special properties requires further experimental support.
In order to understand the mechanism of calmodulin-caldesmon interaction, we performed experiments with proteolytic fragments of calmodulin and its mutants. Under certain conditions, limited proteolysis results in the cleavage of calmodulin into two peptides having very similar size. Both N-terminal (residues 1-77) and C-terminal (residues 78-148) peptides of calmodulin interact with caldesmon, with the C-terminal fragments having higher affinity to caldesmon [18]. Similar results were obtained with several other proteolytic fragments of calmodulin [18]. Based on these results, we concluded that both the N- and C-terminal domains of calmodulin are involved in the interaction with caldesmon, i.e., there are multiple contacts between these two proteins.
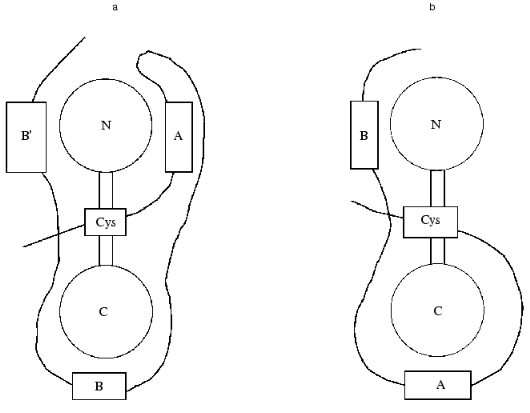
Calmodulin has a dumb-bell structure. The two spherical parts of this dumb-bell each contain two Ca2+-binding sites, and the “handle” formed by a long alpha-helix can melt and bent in its central part [19]. Calmodulin can interact with its protein targets either in the bent or in extended conformation [19]. In the first case the melting of the central helix leads to the movement of the two globular domains of calmodulin to each other and calmodulin encompasses the amphiphilic alpha-helix of the protein target [20, 21]. In the second case, the central helix does not melt and calmodulin interacts with its target protein in extended conformation [19]. The experimental data indicate that calmodulin interacts with caldesmon in the extended conformation [22, 23]. We tried to analyze the topography of the caldesmon-calmodulin complex and to determine the sites involved in the interaction of these proteins. For this purpose, we used proteolytic fragments of calmodulin with different length and deletion mutants of caldesmon containing calmodulin-binding sites. It is supposed that the C-terminal domain of caldesmon contains three calmodulin-binding sites: center A (close to W716/658), center B (close to W749/692), and center B´ (close to W779/722) (the first figure corresponds to the number of the Trp residue in human caldesmon and the second one to the number of Trp residue in chicken caldesmon). In our investigation, we used deletion mutants H2 (containing sites A and B) and H9 (containing sites B and B´). Analyzing the interaction of the deletion mutants of caldesmon with proteolytic fragments of calmodulin and taking into account that site B´ has a rather low affinity to calmodulin, we came to certain conclusions about the spatial organization of the calmodulin-caldesmon complex. The N-terminal proteolytic fragment of calmodulin (residues 1-77) can be chemically cross-linked to both H2 and H9 deletion mutants of caldesmon. Using native electrophoresis, we found that the N-terminal fragment of calmodulin (residues 1-77) interacts with higher affinity with the H2 deletion mutant of caldesmon. The C-terminal fragment of calmodulin (residues 78-148) has higher affinity to the H9 deletion mutant of caldesmon [24]. Based on these experimental data, we supposed that the N-terminal domain of calmodulin interacts with sites A and B´ of caldesmon, whereas the C-terminal domain of caldesmon forms contacts with site B of caldesmon (Fig. 2a).
A little bit later Wang et al. [25] performed similar experiments using shorter and longer deletion fragments of caldesmon and intact calmodulin. For measuring the distance between Ca2+-binding sites of calmodulin and calmodulin-binding sites of caldesmon, these authors used the method of intrinsic Trp fluorescence as well as the method of migration of energy from Trp to Tb3+ that was used as an analog of Ca2+. Wang et al. [25] concluded that the N-terminal domain of calmodulin is located near site B of caldesmon, whereas the C-terminal domain of calmodulin forms contacts with site A of caldesmon (Fig. 2b). However, these authors mentioned that under certain conditions reversed orientation of calmodulin and caldesmon is also possible. In this latter case, the orientation of the two proteins will be identical to that postulated in our investigation. We think there are several controversial points in the paper of Wang et al. [25]. For example, these authors do not consider the probable effect of site B´ on the interaction of calmodulin and caldesmon. They also postulate that the order of Tb3+ binding is identical for isolated calmodulin and calmodulin bound to caldesmon, although this is not proved experimentally. However, we suppose that the main reason for the contradiction between our conclusions [24] and those of Wang et al. [25] is the utilization of fragments different in size. It is possible that the structure of short fragments of caldesmon in the isolated form will be different from the corresponding structure inside the intact protein, and therefore the above-mentioned discrepancy can be solved only after crystallization of the calmodulin-caldesmon complex.Fig. 2. Two alternative models of spatial organization of the complex of calmodulin and the C-terminal fragment of caldesmon. Calmodulin is shown as a dumb-bell with N and C corresponding to the N- and C-terminal globular domains of calmodulin. The C-terminal domain of caldesmon is shown as a bold curved line. Calmodulin-binding sites A, B, and B´ are shown as rectangles. The position of a single cysteine residue located in the third domain of caldesmon is marked by a rectangular Cys. a) Model postulated on the base of experimental data of Medvedeva et al. [24]; b) model suggested by Wang et al. [25].
Valuable information on the topography of interaction of two proteins can be obtained using point mutations. The point mutations “switching off” the first (E31A) or the second (E67A) Ca2+-binding sites lead to a dramatic decrease of the affinity of calmodulin to caldesmon [26]. A rather significant decrease of calmodulin-caldesmon interaction was observed after immobilization of the N- or C-terminal domains by means of introducing disulfide bridges preventing Ca2+ binding [25, 27]. An important role in the interaction with caldesmon is played by the central alpha-helix of calmodulin. Replacing the acidic cluster (82EEE84) in the central helix by three Lys residues or deletion of residues 82-84 leads to a significant decrease of the affinity of calmodulin to caldesmon [26]. To obtain more detailed information on the involvement of the central helix of calmodulin in its interaction, we obtained point mutations of calmodulin where Lys75 was replaced by proline (K75P), glutamic acid (K75E), alanine (K75A) or valine (K75V). Mutations K75E and K75P lead to partial destabilization of the central helix, whereas mutation K75A and K75V, on the contrary, result in formation of additional hydrophobic contacts and in this way stabilize the extended central helix of calmodulin [28]. It turns out that the mutants K75E and K75P not only have higher affinity to caldesmon, but they also are the most effective in reversing caldesmon-induced inhibition of actomyosin ATPase activity [29]. The mutants K75A and K75V had lower affinity to caldesmon and were unable to completely abolish inhibition of actomyosin ATPase activity induced by caldesmon [29]. Troponin C is a structural analog of calmodulin; however, the central helix of troponin C contains an additional tripepetide (KGK) and therefore is one turn longer than the central helix of calmodulin [30]. Troponin C interacts with caldesmon with lower affinity than calmodulin [31].
Summarizing, we conclude that both the globular N- and C-terminal domains and the central helix of calmodulin are involved in the interaction with caldesmon. Shortening or lengthening of the central helix as well as increasing its rigidity decrease the interaction of calmodulin with caldesmon [26, 29, 31]. Analyzing the orientation of Ca2+-binding proteins in their complex with caldesmon, it is worthwhile to mention that Cys98, located in the central helix of troponin C, can be crosslinked to the single Cys residue located in the C-terminal domain of caldesmon (C636/C580) [31]. Thus, the central helix of troponin C (and therefore probably of calmodulin) is located near C636/580 of caldesmon that according to the literature [27, 32] is close to sites A, B, and B´ of caldesmon involved in the interaction with the globular domains of calmodulin (Fig. 2). It is worth mentioning that the dimeric Ca2+-binding protein S100, whose three-dimensional structure [33] is different from that of calmodulin and troponin C, also interacts with caldesmon and rather effectively reverses the inhibitory action of caldesmon on actomyosin ATPase activity. S100 can also be crosslinked to the C636/580 of caldesmon via a disulfide bridge [34].
The data presented mean that all the above-mentioned Ca2+-binding proteins are located near a single Cys (C638/580) in the C-terminal domain of caldesmon. This orientation provides for approaching of Ca2+-binding sites of calmodulin, troponin C and probably of S100 with three Trp residues located in the C-terminal domain of caldesmon (Fig. 2) and changes of the structure of caldesmon. The conformation changes induced by different Ca2+-binding proteins are different. Conformational changes induced in the caldesmon structure by troponin C and probably by S100 lead to decrease of its interaction with actin, dissociation of caldesmon from actin, and by this means reversal of inhibition of actomyosin ATPase activity. Calmodulin (or one of its conformers) affects the structure of caldesmon in such a way that its interaction with actin is modified and therefore reversal of inhibition of actomyosin ATPase activity is achieved without dissociation of caldesmon from actin. This fine mechanism of regulation is possible only in the case if the structure of calmodulin (or one of its conformers) ideally corresponds to the binding sites located in the C-terminal domain of caldesmon. Probably this is the reason why an even very subtle mutation in far-removed parts of calmodulin strongly affects its interaction with caldesmon.
Phosphorylation and regulation of the functional activity of caldesmon. The contractile activity of smooth muscle as well as motility of non-muscle cells can be regulated by different external factors (such as extension), hormones, and physiologically active compounds. Many of these factors affect cell motility via phosphorylation of certain contractile and regulatory proteins. Under in vitro conditions, caldesmon is phosphorylated by a number of protein kinases [35, 36].
Casein kinase of the second type (CK2) phosphorylates Ser26, Ser73, and Thr83, i.e., the sites located only in the first N-terminal domain of caldesmon (see Fig. 1) [38-40]. The question of which of these sites is predominantly phosphorylated by CK2 remains unanswered [40]. Nevertheless, all investigators agree that phosphorylation catalyzed by CK2 leads to decrease of interaction of caldesmon with smooth muscle myosin [39-41] and tropomyosin [41]. This is probably because the sites of phosphorylation are close to the sites of interaction of caldesmon with myosin and tropomyosin [40, 42]. The central and the C-terminal parts of caldesmon are tightly bound to actin, whereas the N-terminal part is more or less free and may contact myosin [1-5, 36]. Under certain conditions, caldesmon may tether actin and myosin filaments and in this way regulate tonic contraction [36, 42]. In this case, phosphorylation by CK2 will prevent tethering of myosin and actin filaments and developing of tonic contraction. The question of participation of casein kinase 2 in caldesmon phosphorylation in vivo remains unanswered [36, 40]. However, in smooth muscle extracts casein kinase 2 possesses the highest caldesmon kinase activity both for isolated caldesmon and for caldesmon inside actin filaments [43].
Several protein kinases phosphorylate different sites located in the C-terminal domain of caldesmon. Cdc2 protein kinase phosphorylates Ser and Thr residues in the C-terminal domain of caldesmon [44, 45]. This phosphorylation leads to a significant reduction of binding of caldesmon to actin and dissociation of caldesmon from actin filaments. This process may be of great importance during reorganization of cytoskeleton in the case of mitosis. MAPK p42/p44 also phosphorylates several sites in the C-terminal domain of caldesmon. The sites phosphorylated by MAPK in vitro coincide with the sites phosphorylated in caldesmon isolated from smooth muscle [35, 36, 46, 47]. Phosphorylation of Ser759/702 of caldesmon by MAPK or replacement of these residues by Asp results in a decrease of inhibitory action of caldesmon on the actomyosin ATPase activity [48]. At the same time, actin and calmodulin partially inhibit phosphorylation of caldesmon by MAPK and stimulation of smooth muscle is not accompanied by significant increase of phosphorylation of the sites potentially phosphorylated by this enzyme [49]. Therefore a hypothesis is put forward that smooth muscles contain another yet unidentified protein kinase able to phosphorylate caldesmon and by this means affecting its inhibitory action on actomyosin ATPase.
Intact protein kinase C and endogenous proteolytic products of protein kinase C are also able to phosphorylate isolated caldesmon. The sites phosphorylated by protein kinase C and its proteolytic products are located mainly in the third and fourth domains of caldesmon [36, 37, 50, 51]. Phosphorylation by protein kinase C decreases the interaction of caldesmon with actin and by this means decreases the inhibitory action of caldesmon on the actomyosin ATPase activity [50-52]. At present it is difficult to give an unambiguous answer whether protein kinase C participates in caldesmon phosphorylation in vivo. However, taking into account a rather high content of this enzyme in the cell, the existence of a number of isoforms of this enzyme, and participation of protein kinase C in a number of protein kinase cascades one cannot exclude direct or indirect involvement of protein kinase C in caldesmon phosphorylation.
PARTICIPATION OF CALDESMON AND CALPONIN IN CYTOSKELETON
FORMATION
Interaction of caldesmon and calponin with phospholipids. As already mentioned, caldesmon is a typical actin-binding protein located on actin filaments involved in the interaction with myosin and generation of movement and on microfilaments building cytoskeleton. In a number of cells caldesmon is located on actin filaments just under the external membrane [53]. Caldesmon seems to be involved in receptor capping [54] and exocytosis [55]. We supposed that being located near the membrane caldesmon might interact with phospholipids. Using light scattering, gel filtration, and electrophoresis, we found that indeed caldesmon interacts with acidic phospholipids [56]. We found that Ca2+-saturated calmodulin interferes with the binding of caldesmon to phospholipids. A similar effect was observed after phosphorylation of caldesmon by protein kinase C [57]. These results were later confirmed by Czurylo et al. [58]. Using proteolytic fragments and deletion mutants of caldesmon, we have shown that the phospholipid-binding sites are located in the C-terminal domain of caldesmon close to the sites of interaction with calmodulin and the sites phosphorylated by protein kinase C [57, 59]. There is a short amphiphilic alpha-helix (residues 702/645-717/660) in the C-terminal domain of caldesmon. This helix seems to be able to form complexes with acidic phospholipids [60]. These theoretical predictions completely agree with experimental data obtained in different laboratories [57-59, 61, 62]. Experiments with the model phospholipids were extended by experiments performed on natural biological membranes [61]. Moreover, it has been shown that caldesmon can fix actin filaments to the surface of phospholipid vesicles [62]. Thus, the C-terminal site of caldesmon may interact with phospholipids and caldesmon may function as an anchor fixing actin filaments to biological membranes. It is important that this interaction is modulated by calmodulin and phosphorylation by protein kinase C. Probably this is the reason why caldesmon is involved in exocytosis, receptor capping, and fixing of cytoskeleton to membranes.
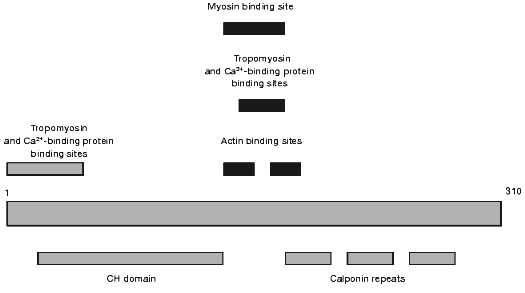
Another actin-binding protein that might participate in the regulation of actin-myosin interaction and in the organization of cytoskeleton was discovered at the end of the 80s by Takahashi et al. [63] and was named calponin. Calponin is now thoroughly investigated and many of its properties are described in a number of review articles [4, 64-66]. A schematic of the calponin structure is presented in Fig. 3. The N-terminal part contains the sites of weak interaction with Ca2+-binding proteins and tropomyosin. This part also contains the so-called CH-domain (calponin homology domain) that is found in the structure of many other actin-binding proteins. The central part of calponin contains two actin-binding sites. These sites partially overlap with the sites involved in the interaction with Ca2+-binding proteins, myosin, and tropomyosin (see Fig. 3). The C-terminal part of calponin contains three homologous calponin repeats (Fig. 3). Thus, calponin like caldesmon may interact with actin, tropomyosin, myosin, and Ca2+-binding proteins. It is supposed that binding to actin calponin inhibits actomyosin ATPase and that this inhibition can be reversed after its interaction with Ca2+-binding proteins or after phosphorylation of calponin by different protein kinases [64, 67]. The calponin content in smooth muscle and non-muscle cells is very high. Calponin is located predominantly on actin filaments involved in formation of cytoskeleton. Smaller quantities of calponin were found on actin filaments located in the so-called contractile domain where actin co-localizes with myosin and is involved in active generation of movement [64, 65, 68]. Therefore, the hypothesis is put forward that calponin is mainly involved in the formation of cytoskeleton, but not in the regulation of actin-myosin interaction. It is worth mentioning that calponin, like caldesmon, is often found on actin bundles located just under cell membranes [53]. Moreover, after smooth muscle cell stimulation calponin migrates into the perimembrane area and can serve as an original adapter carrying MAP-kinase and protein kinase C to the membranes [69-71]. Therefore, it seems reasonable to analyze the interaction of calponin with phospholipids.
Theoretical analysis of the primary structure of calponin revealed short amphiphilic helices in the N-terminal part of the molecule. These helices may interact with phospholipids [60]. Subsequent experimental checking of this hypothesis showed that calponin indeed can interact with acidic phospholipids and that both electrostatic and hydrophobic interactions play important roles in the binding of calponin to phospholipids. Phospholipid-binding sites are located in the N-terminal domain of calponin, and Ca2+-saturated calmodulin decreases the interaction of calponin with phospholipids [72, 73]. In this respect, the properties of calponin are similar to those of caldesmon, and this suggests that calponin, like caldesmon, may be involved in fixation of cytoskeleton to membrane.Fig. 3. Scheme of the structure of smooth muscle calponin.
Interaction of calponin with other cytoskeleton proteins and with 90-kD heat shock protein. If calponin indeed plays an important role in cytoskeleton formation, one can suppose that it will also be able to interact not only with the proteins of microfilaments, but with different proteins of microtubules and intermediate filaments. It has been shown that under certain conditions calponin can interact with smooth muscle desmin, affect desmin polymerization, and cosediment with intermediate filaments [74, 75]. These experimental data might mean that calponin can simultaneously interact with both intermediate filaments and microfilaments and in this way stabilize cytoskeleton. Moreover, the data of Japanese authors indicate that calponin interacts with tubulin, and that this interaction is regulated by phosphorylation of calponin and calmodulin [76].
As already mentioned, the intracellular concentration of calponin is very high [67]; this protein tends to aggregate and can interact with a number of different intracellular proteins [4, 64-66, 74-76]. It is most amazing that the stimulation of cells is accompanied by migration of calponin from the contractile apparatus towards the membrane [69, 70] and that calponin can find proper places of its localization. We supposed that the heat shock proteins could participate in the interaction with calponin and its transportation to proper alignment in the cytoskeleton. We analyzed the interaction of calponin with 90-kD heat shock protein (hsp90) and found that the heat shock protein forms tight complexes with calponin [77]. The N-terminal domain of calponin and two sites in the structure of hsp90 are involved in formation of this complex [78, 79]. Probably the most important finding is that hsp90 prevents calponin-induced polymerization of actin and decreases calponin induced bundling of actin filaments [79]. Thus, we can suggest that hsp90 somehow participates in transporting of calponin to the places of its binding and provides for proper alignment of the cytoskeleton. Future experimental work in this direction will help to test this assumption.
Effect of different actin-binding proteins on the interaction of calponin with actin. As already mentioned, calponin and caldesmon belong to the group of actin-binding proteins participating in cytoskeleton formation. In this respect, it seems reasonable to analyze simultaneous binding of caldesmon and calponin with actin filaments. Makuch et al. [80] found that titration by caldesmon partially displaced calponin from thin filaments, whereas calponin practically completely prevents the interaction of caldesmon with actin. This means that calponin and caldesmon cannot be simultaneously bound to actin filaments. This conclusion agrees with cytochemical results that indicate that caldesmon is predominantly bound to actin filaments located in the so-called contractile domain, whereas calponin is predominantly bound to actin filaments in the so-called cytoskeleton domain [65, 68].
Many intracellular structures formed by actin filaments contain a number of different actin-binding proteins. For instance, dense bodies and the structures providing for anchoring of actin filaments to membrane contain calponin, filamin, and alpha-actinin [65, 68]. The structural data indicate that filamin, calponin, and alpha-actinin interact with the same or closely located sites of actin [81, 82]. This fact apparently means that these three proteins should compete with each other and should not be located in the same cell compartments. However, as already mentioned these proteins may be located on the same actin filaments. To solve this apparent contradiction we analyzed simultaneous binding of calponin, filamin, and alpha-actinin to actin filaments. It turns out that calponin and filamin may independently bind to the bundles of actin filaments and the binding of one actin-binding protein has practically no effect on the binding of the second protein [83, 84]. Insignificant hindrances were observed during simultaneous binding of calponin and alpha-actinin to the bundles of actin filaments [84, 85]. The largest competition was observed in the case of filamin and alpha-actinin. However, even in this case the presence of one of the proteins causes no more than 40-50% decrease in the binding of another actin-binding protein [83, 84]. According to the data of electron microscopy, the structure of actin bundles formed in the presence of single actin-binding proteins was different from that formed in the presence of two different proteins. For example, in the presence of alpha-actinin we observed ladder-like structure where alpha-actinin forms perpendicular rungs connecting two actin filaments distantly separated from each other [84]. If actin was polymerized in the presence of both calponin and alpha-actinin, we observed formation of tightly and parallel arranged bundles of actin filaments [84]. The change in the structure correlates with the change in mechanical properties of actin bundles [83]. This seems to be of great importance in formation of different elements of cytoskeleton such as dense bodies or the sites of fixing of actin filaments to the membrane. The data obtained mean that the sites of actin involved in the binding of calponin, filamin, and alpha-actinin only partially overlap with each other. Probably this is the reason why in the case of calponin, filamin, and alpha-actinin the total stoichiometry of actin-binding proteins/actin may be higher than 0.5 [84]. Taking into account that each of these proteins may simultaneously interact with two actin monomers [81, 82], we conclude that the investigated proteins weakly compete with each other and that at saturation practically each actin monomer may contact with the same or with a different actin-binding protein. Probably this is the reason for the very wide diversity of different cell structures formed by actin filaments inside the cell.
The author is deeply grateful to his colleagues Drs A. V. Vorotnikov, N. V. Bogatcheva, M. V. Medvedeva, Ma YuShu, O. O. Panasenko, and A. A. Polyakov and to the students of the Department of Biochemistry directly and actively participating in investigations described in this paper. The author is thankful to Dr. D. I. Levitsky (Bach Institute of Biochemistry of the Russian Academy of Sciences), Dr. V. P. Shirinsky (Cardiological Research Center, Moscow), as well as to Prof. S. B. Marston (Imperial College, London, Great Britain), Prof. D. M. Watterson (Northwestern University, Chicago, USA), and Prof. J. H. Collins (University of Maryland, Baltimore, USA) for close and fruitful cooperation.
This investigation was supported by grants from the Russian Foundation for Basic Research (RFBR), a joint grant of RFBR and INTAS, and by a grant from the Wellcome Trust.
REFERENCES
1.Sobue, K., and Sellers, J. R. (1991) J. Biol.
Chem., 266, 12115-12118.
2.Marston, S. B., and Huber, P. A. J. (1995) in
Biochemistry of Smooth Muscle Contraction (Barany, M., ed.)
Academic Press, San Diego, CA, pp. 77-90.
3.Marston, S., Burton, D., Copeland, O., Fraser, I.,
Gao, Y., Hodgkinson, J., Huber, P., Levine, B., El-Mezgueldi, M., and
Notarianni, G. (1998) Acta Physiol. Scand., 164,
401-414.
4.Bogatcheva, N. V., and Gusev, N. B. (1997)
Uspekhi Biol. Khim., 37, 3-48.
5.Huber, P. A. (1997) Int. J. Biochem. Cell
Biol., 29, 1047-1051.
6.Marston, S. B., and Redwood, C. S. (1993) J.
Biol. Chem., 268, 12317-12320.
7.Chalovich, J. M., Sen, A., Resetar, A., Leinweber,
B., Fredricksen, R. S., Lu, F., and Chen, Y.-D. (1998) Acta Physiol.
Scand., 164, 427-435.
8.Wang, Z., Yang, Z. Q., and Chako, S. (1997) J.
Biol. Chem., 272, 16896-16903.
9.Marston, S. B., Fraser, I. D. C., Huber, P. A. J.,
Pritchard, K., Gusev, N. B., and Torok, K. (1994) J. Biol.
Chem., 269, 8134-8139.
10.Skripnikova, E. V., and Gusev, N. B. (1989)
FEBS Lett., 257, 380-382.
11.Bogatcheva, N. V., Panaiotov, M. P., Vorotnikov,
A. V., and Gusev, N. B. (1993) FEBS Lett., 335,
193-197.
12.Fujii, T., Machino, K., Andoh, H., Sato, T., and
Kondo, H. (1990) J. Biochem., 107, 133-137.
13.Pritchard, K., and Marston, S. B. (1991)
Biochem. J., 277, 819-824.
14.Mani, R. S., McCubbin, W. D., and Kay, C. M.
(1992) Biochemistry, 31, 11896-11901.
15.Filipek, A., Zasada, A., Wojda, U., Makuch, R.,
and Dabrowska, R. (1996) Comp. Biochem. Physiol., 113B,
745-752.
16.Pritchard, K., and Marston, S. B. (1993)
Biochem. Biophys. Res. Commun., 190, 668-673.
17.Notarianni, G., Gusev, N., Lafitte, D., Hill, T.
J., Cooper, H. S., Derrick, P. J., and Marston, S. B. (2000) J.
Muscle Res. Cell Motil., 21, 537-549.
18.Medvedeva, M. V., Kolobova, E. A., Wang, P., and
Gusev, N. B. (1996) Biochem. J., 315, 1021-1026.
19.Crivici, A., and Ikura, M. (1995) Annu. Rev.
Biophys. Biomol. Struct., 24, 85-116.
20.Meador, W. E., Means, A. R., and Quiocho, F. A.
(1992) Science, 257, 1251-1255.
21.Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu,
G., Klee, C. B., and Bax, A. (1992) Science, 256,
632-638.
22.Mabuchi, K., Wang, C.-L. A., and Grabarek, Z.
(1995) Biophys. J., 68, A359.
23.Krueger, J. K., Wang, C.-L. A., and Trewhella, J.
(1999) Biophys. J., 76, A282.
24.Medvedeva, M. V., Kolobova, E. A., Huber, P. A.
J., Fraser, I. D. C., Marston, S. B., and Gusev, N. B. (1997)
Biochem. J., 324, 255-262.
25.Wang, E., Zhuang, S., Kordowska, J., Grabarek,
Z., and Wang, C.-L. A. (1997) Biochemistry, 36,
15026-15034.
26.Medvedeva, M. V., Bushueva, T. L., Shirinsky, V.
P., Lukas, T. J., Watterson, D. M., and Gusev, N. B. (1995) FEBS
Lett., 360, 89-92.
27.Huber, P. A. J., El-Mezgueldi, M., Grabarek, Z.,
Slatter, D. A., Levine, B. A., and Marston, S. B. (1996) Biochem.
J., 316, 413-420.
28.Medvedeva, M. V., Polyakova, O. V., Watterson, D.
M., and Gusev, N. B. (1999) FEBS Lett., 450, 139-143.
29.Medvedeva, M. V., Djemuchadze, D. R., Watterson,
D. M., Marston, S. B., and Gusev, N. B. (2001) Biochim. Biophys.
Acta, 1544, 143-150.
30.Houdusse, A., Love, M. L., Dominguez, R.,
Grabarek, Z., and Cohen, C. (1997) Structure, 5,
1695-1711.
31.Polyakov, A. A., and Gusev, N. B. (1997)
Biochem. J., 321, 873-878.
32.Mornet, D., Bonet-Kerrache, A., Strasburg, G. M.,
Patchell, V. B., Perry, S. V., Huber, P. A. J., Marston, S. B.,
Slatter, D. A., Evans, J. S., and Levine, B. A. (1995)
Biochemistry, 34, 1893-1901.
33.Kilby, P. M., van Eldik, L., and Roberts, G. C.
K. (1996) Structure, 4, 1041-1052.
34.Polyakov, A. A., Huber, P. A. J., Marston, S. B.,
and Gusev, N. B. (1998) FEBS Lett., 422, 235-239.
35.Gerthoffer, W. T., and Pohl, J. (1994) Can. J.
Physiol. Pharmacol., 72, 1410-1414.
36.Shirinsky, V. P., Vorotnikov, A. V., and Gusev,
N. B. (1999) in Molecular Mechanisms of Smooth Muscle
Contraction (Kohama, K., and Sasaki, Y., ed.) R. G. Landes Company,
TX, USA, pp. 59-79.
37.Vorotnikov, A. V., Shirinsky, V. P., and Gusev,
N. B. (1988) FEBS Lett., 236, 321-324.
38.Wawrzynow, A., Collins, J. H., Bogatcheva, N. V.,
Vorotnikov, A. V., and Gusev, N. B. (1991) FEBS Lett.,
289, 213-216.
39.Sutherland, C., Renaux, B. S., McKay, D. J., and
Walsh, M. P. (1994) J. Muscle Res. Cell Motil., 15,
440-456.
40.Wang, Z., and Yang, Z.-Q. (2000)
Biochemistry, 39, 11114-11120.
41.Bogatcheva, N. V., Vorotnikov, A. V., Birukov, K.
G., Shirinsky, V. P., and Gusev, N. B. (1993) Biochem. J.,
290, 437-442.
42.Vorotnikov, A. V., Marston, S. B., and Huber, P.
A. (1997) Biochem. J., 328, 211-218.
43.Vorotnikov, A. V., Gusev, N. B., Hua, S.,
Collins, J. H., Redwood, C. S., and Marston, S. B. (1993) FEBS
Lett., 334, 18-22.
44.Yamashiro, S., Yamakita, Y., Hosoya, H., and
Matsumura, F. (1991) Nature, 349, 169-172.
45.Yamashiro, S., Yamakita, Y., Yoshida, K.,
Takiguchi, K., and Matsamura, F. (1995) J. Biol. Chem.,
270, 4023-4030.
46.Adam, L. P., and Hathaway, D. R. (1993) FEBS
Lett., 322, 56-60.
47.Childs, T. J., Watson, M. H., Sanghera, J. S.,
Campbell, D. L., Pellech, S. L., and Mak, A. S. (1992) J. Biol.
Chem., 267, 22853-22859.
48.Redwood, C. S., Marston, S. B., and Gusev, N. B.
(1993) FEBS Lett., 327, 85-89.
49.Krymsky, M. A., Chibalina, M. V., Shirinsky, V.
P., Marston, S. B., and Vorotnikov, A. V. (1999) FEBS Lett.,
452, 254-258.
50.Ikebe, M., and Hornick, T. (1991) Arch.
Biochem. Biophys., 288, 538-542.
51.Vorotnikov, A. V., Gusev, N. B., Hua, S.,
Collins, J. H., Redwood, C. S., and Marston, S. B. (1994) J. Muscle
Res. Cell Motil., 15, 37-48.
52.Tanaka, T., Ohta, H., Kanda, K., Tanaka, T.,
Hidaka, H., and Sobue, K. (1990) Eur. J. Biochem., 188,
495-500.
53.Takeuchi, K., Takahashi, K., Abe, M., Nishida,
W., Hiwada, K., Nabeya, T., and Maruyama, K. (1991) J. Biochem.,
109, 311-316.
54.Walker, G., Kerrick, W. G. L., and Bourguignon,
L. Y. W. (1989) J. Biol. Chem., 264, 496-500.
55.Bourgoyne, R. D., Cheek, T. R., and Norman, K. M.
(1986) Nature, 319, 68-70.
56.Vorotnikov, A. V., and Gusev, N. B. (1990)
FEBS Lett., 277, 134-136.
57.Vorotnikov, A. V., Bogatcheva, N. V., and Gusev,
N. B. (1992) Biochem. J., 284, 911-916.
58.Czurylo, E. A., Zborowski, J., and Dabrowska, R.
(1993) Biochem. J., 291, 403-408.
59.Bogatcheva, N. V., Huber, P. A. J., Fraser, I. D.
C., Marston, S. B., and Gusev, N. B. (1994) FEBS Lett.,
342, 176-180.
60.Bogatcheva, N. V., and Gusev, N. B. (1995)
FEBS Lett., 363, 269-272.
61.Makowski, P., Makuch, R., Sikorski, A. F.,
Jezierski, A., Pikula, S., and Dabrowska, R. (1997) Biochem. J.,
328, 505-509.
62.Makuch, R., Zasada, A., Mabuchi, K., Krauze, K.,
Wang, C.-L. A., and Dabrowska, R. (1993) Biophys. J., 73,
1607-1616.
63.Takahashi, K., Hiwada, K., and Kokubu, T. (1986)
Biochem. Biophys. Res. Commun., 141, 20-26.
64.Gimona, M., and Small, V. J. (1996) in
Biochemistry of Smooth Muscle Contraction (Barany, M., ed.)
Academic Press, San Diego, pp. 91-103.
65.Small, J. V., and Gimona, M. (1998) Acta
Physiol. Scand., 164, 341-348.
66.Churylo, E. A. (2000) Tsitologiya,
42, 7-18.
67.Walsh, M. P. (1994) Can. J. Physiol.
Pharmacol., 72, 919-936.
68.Mabuchi, K., Li, Y. T., Tao, T., and Wang, C.-L.
A. (1996) J. Muscle Res. Cell Motil., 17, 243-260.
69.Parker, C., Takahashi, K., Tao, T., and Morgan,
K. G. (1994) Am. J. Physiol., 267, C1261-C1270.
70.Menice, C. B., Hulvershorne, J., Adam, L. P.,
Wang, C.-L. A., and Morgan, K. G. (1997) J. Biol. Chem.,
272, 25157-25161.
71.Leinweber, B., Parissenti, A. M., Gallant, C.,
Gangopadhyay, S. S., Kirwan-Rhude, A., Leavis, P., and Morgan, K.
(2000) J. Biol. Chem., 275, 40329-40336.
72.Bogatcheva, N. V., and Gusev, N. B. (1995)
FEBS Lett., 371, 123-126.
73.Fujii, T., Yamana, K., Ogomo, Y., and Kondo, Y.
(1995) J. Biochem., 117, 999-1003.
74.Wang, P., and Gusev, N. B. (1996) FEBS
Lett., 392, 255-258.
75.Mabuchi, K., Li, B., Ip, W., and Tao, T. (1997)
J. Biol. Chem., 272, 22662-22666.
76.Fujii, T., Hiromori, T., Hamamoto, M., and
Suzuki, T. (1997) J. Biochem., 122, 344-351.
77.Ma, Y., Bogatcheva, N. V., and Gusev, N. B.
(1998) Biochemistry (Moscow), 63, 1282-1289.
78.Bogatcheva, N. V., Ma, Y. S., Urosev, D., and
Gusev, N. B. (1999) FEBS Lett., 457, 369-374.
79.Ma, Y. S., Bogatcheva, N. V., and Gusev, N. B.
(2000) Biochim. Biophys. Acta, 1476, 300-310.
80.Makuch, R., Birukov, K. G., Shirinsky, V. P., and
Dabrowska, R. (1991) Biochem. J., 280, 33-38.
81.McGough, A. (1998) Curr. Opin. Struct.
Biol., 8, 166-176.
82.Hodgkinson, J. L., El-Mezgueldi, M., Craig, R.,
Vibert, P., Marston, S. B., and Lehman, W. (1997) J. Mol. Biol.,
273, 150-159.
83.Leinweber, B., Tang, J. X., Stafford, W., and
Chalovich, J. M. (1999) Biophys. J., 77, 3208-3217.
84.Panasenko, O. O., and Gusev, N. B. (2001)
Biochim. Biophys. Acta, 1544, 393-405.
85.Panansenko, O. O., and Gusev, N. B. (2000)
IUBMB Life, 49, 277-283.