REVIEW: Proton Transport Mechanism of Bacteriorhodopsin as Revealed by Site-Specific Mutagenesis and Protein Sequence Variability
L. S. Brown
Department of Physiology and Biophysics, University of California, Irvine, California 92697, USA; fax: 1-949-824-8540; E-mail: lsbrown@uci.edu
Received January 31, 2001; Revision received May 3, 2001
A large share of the current ideas about the mechanism of proton transport by bacteriorhodopsin has emerged from studies of site-specific mutants. This review is an attempt to check some of these ideas against the natural variability in the primary structure of the protein.
KEY WORDS: bacteriorhodopsin, halobacteria, mutations, photocycle, proton transport, conserved residues
Bacteriorhodopsin (BR) is a halobacterial proton pump, which translocates protons from the cytoplasmic to the extracellular side of the purple membrane when under illumination. Absorption of a photon starts a photochemical cycle, causing isomerization of the BR chromophore, retinal, and the subsequent deprotonation of its Schiff base (this deprotonated state is the M intermediate). The proton is transferred to a primary acceptor, Asp-85, that causes another proton to be released to the extracellular surface. Later, the Schiff base is reprotonated from Asp-96, which in turn is reprotonated from the cytoplasmic surface and results in proton uptake. For further details of how BR functions, see recent reviews [1-4] and other articles in this issue.
In thirty years of research on BR, several hundred site-specific mutants of this protein were produced and characterized. At first, the system of choice was heterologous expression of the mutated BR in E. coli [5]. Later, this system was replaced by homologous expression in H. salinarum [6, 7]. As a result, a great deal of information was accumulated in regard to the specific roles of the amino acids important for function. In some cases, the conclusions concerning the mechanism of proton transport based on such information have been contradictory. This happened either because the different expression systems yielded different properties for the same mutant, the interpretation was based on different replacements of the same residue, or the effects of mutations were indirect and misinterpreted (for a review of problems with the interpretation of mutant phenotypes, see [8]).
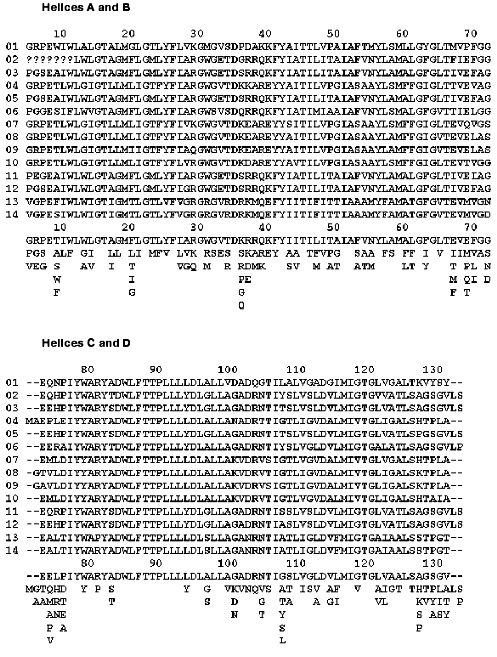
Fortunately, Nature offers a powerful tool for verifying our ideas about roles of specific amino acids by keeping the functionally important residues conserved between related species. Bacteriorhodopsin sequences from various halobacteria were analyzed before [9-12], both in terms of their overall similarity and their evolutionary significance. Fourteen different BR sequences, some of them called archaerhodopsins (AR), cruxrhodopsins (CR), or deltarhodopsins, can be found in a literature or GenBank (figure). It is instructive to view some of the non-trivial results of site-specific mutagenesis through the prism of the natural diversity of BR. To do this, we should make two basic assumptions. First, any residue important for proton pumping ought to be conserved among different BR-like proteins. The reverse of this assumption is not necessarily true, because some residues can be conserved if they are important for folding, insertion or overall structural stability of all halobacterial retinal proteins, not only BRs. Second, we must assume that all the BRs have the same function, i.e., they are proton pumps. When some of them acquired a different function in the course of evolution, the conservation pattern is changed as can be seen clearly in the sequences of halorhodopsins or sensory rhodopsins I and II [11, 12].
Alignment of known BR sequences from various halobacteria: 1) [13, 14]; 2, 3, 4) [9, 11]; 5) [15]; 6) [16]; 7) [17, 18]; 8) [19]; 9) [20]; 10) [11]; 11) [21]; 12) [22]; 13) [23]; 14) [11]. The alignment was performed using MSA [24] server v.2.1 (http://www.ibc.wustl.edu/ibc/msa.html). Numbering of residues corresponds to H. halobium (salinarum) BR. The sequences were truncated from both ends to exclude tails. Question marks signify missing data, dashes mean absence of a corresponding residue in a sequence. To visualize a consensus sequence, the residues encountered at each position were placed below the alignments, the one occurring most frequently being on top of each column and the one occurring least frequently being on the bottom.
MUTATIONS AFFECTING THE CHROMOPHORE
Almost every residue that belongs to the retinal pocket or its nearest vicinity, as well as to the complex counterion of the Schiff base, is conserved. Mutating these residues can lead to changes in such parameters of the chromophore as color, proton affinity of the Schiff base, isomeric state, and rates of photoisomerization and thermal reisomerization [25-27]. If one were to take the retinal and the side chain of Lys-216 connected to it, and search for all the amino acid side chains at a distance closer than 4.5 A, a set of conserved (with one notable exception) residues would emerge. Performing this operation on BR structure with the highest available resolution (1C3W [28]) using the WPDB program (V.2.2, ViSoft Inc.) nets the following conserved set: Met-20, Val-49 (or the similar Ile), Ala-53, Tyr-83, Asp-85, Trp-86, Thr-89, Thr-90, Leu-93, Met-118, Ile-119, Gly-122, Trp-138, Ser-141, Thr-142, Trp-182, Tyr-185, Pro-186, Trp-189, Asp-212, Ala-215, Val-217 (or the similar Ile). Besides the well-known dramatic effects of replacements of the immediate members of the Schiff base counterion (Asp-85, Asp-212 [29]), Tyr-185 [30, 31] and Pro-186 [32], mutations of some of the other residues result in photocycle changes that are more subtle. For example, the mutations Val-49-Ala and Ala-53-Val drastically change the protonation equilibrium that develops in the photocycle between the retinal Schiff base and Asp-85 [33]. Replacing Leu-93 with Ala [34] or Trp-182 with Phe [35] strongly slows down the retinal thermal reisomerization, while replacement of Trp-86 with Phe disrupts normal light adaptation due to changes in water binding [36].
There is only one non-conserved residue close to the retinal C-18 atom, Met-145. In various BR sequences it is either Leu or Met or Phe (figure). Both the BR mutant Met-145-Phe and AR-2, which has Phe at this position (sequence 8 in the figure), exhibit changed isomeric composition in the dark-adapted state and a 10 nm blue shift in the absorption maximum [37]. At the same time, the isomeric composition of the light-adapted state, as well as the proton pumping, were normal in both cases [38] implying that this rather conservative replacement of Met-145 is not deleterious for the BR functioning.
MUTATIONS AFFECTING PROTON RELEASE
Most of the residues involved in the proton release are grouped around the hydrogen-bonded cluster of amino acid side chains and water molecules near the extracellular surface of BR [39]. Three members of this cluster are indispensable for normal (early) proton release (Arg-82 [40, 41], Glu-204 [42], and Glu-194 [43]), and they are all conserved (figure). There is one exception worth mentioning, though. In the BR sequence from halobacterial strain HT [23] (sequence 13 in the figure) Arg-82 is replaced by Pro. This is unexpected, because this arginine is a superconserved residue in all halobacterial retinal proteins [11]. One possible explanation is suggested by the fact that this strain does not demonstrate any proton pump activity because the bop gene is never expressed [23]. Lack of expression would allow accumulation of mutations otherwise eliminated by natural selection. Interestingly, and for reasons that are unclear, replacement of a single residue on the cytoplasmic side of helix G (Gly-231-Cys) reverses the effect of Arg-82-Ala mutation on proton release [44]. It should be noted that Gly-231 is not a conserved residue and can be encountered as either Glu or Asp in other halobacterial species (figure).
There are two other conserved residues in this hydrogen-bonded cluster, Tyr-83, which becomes connected to it in the M intermediate of the photocycle [39], and Tyr-57. Their indispensability for proton release is not as clear-cut as that of the preceding group. Replacement of Tyr-83 with Asn changes the proton affinity of the release group but does not abolish proton release completely [45]. At the same time, the analogous replacement of Tyr-57 abolishes production of the M intermediate and proton transport altogether [46]. When Tyr-57 is exchanged for Phe, the overall changes are much smaller, and even though the rate of the proton release slows down for a substantial part of the molecules, the normal proton release is not totally blocked [47]. This can be interpreted as the effect of removal of hydrogen bond of the hydroxyl group of Tyr-57 to a water molecule [39] on proton release, while the former mutation reveals the dramatic effect of removing the bulk of the tyrosine phenyl ring on the not so distant retinal Schiff base and its counterion.
Among the groups surrounding the proton releasing cluster, five more conserved amino acid residues are implicated in regulation of its proton affinity, but without being obligatory for normal proton release. These include Thr-205 [48, 49], Arg-134 [50, 51], Ser-193 (or Thr in some species) [49, 52], Phe-208 [53], and Tyr-79 (Brown, Needleman, and Lanyi, unpublished data). Interestingly, another group from this region reported to substantially affect the proton release kinetics, Lys-129 [54], is not conserved at all. Besides Lys, it can be His, Ala, Ser, or Pro (figure). In accordance with the pattern of proton release in the Lys-129-His mutant [54], AR-1 (sequence 7 in the figure), which also has a His in this position, exhibits abnormal (late) proton release [55]. This leads to the interesting question, is early proton release important for BR functioning at all? So far, there is no evidence that delaying the early proton release until the very end of the photocycle dramatically alters the proton pumping capability of BR. The pumping efficiency may go down somewhat because of the slower photocycle turnover in most of the BR mutants discussed above. But this is probably not so important taking into account that in natural habitats sunlight intensity is the limiting factor [56]. What remains to be seen, though, is if these mutants are as efficient in pumping against the proton gradient as the wild-type BR.
Finally, two glutamic acid residues on the extracellular surface of BR (Glu-9 and Glu-74), were suggested as participants in proton releasing pathways either on structural [57, 58] or mutational [59] basis. It should be noted that several spectroscopic studies of the appropriate mutants questioned this idea [60, 61]. A look at the conservation map (figure) shows that Glu-74 is not conserved, but can also be Thr or Ala. As for Glu-9, it is preserved in all BR sequences, despite the absence of any drastic effect of its replacement by Ala [60] or Gln [59] on the photocycle. A hint to why it is conserved may lie in the fact that the stability of BR is decreased when this Glu is exchanged for Ala (Brown, Richter, Needleman, and Lanyi, unpublished data).
MUTATIONS AFFECTING REPROTONATION OF THE SCHIFF BASE AND PROTON
UPTAKE FROM THE CYTOPLASMIC SURFACE
It is more difficult to decide about the importance of a residue on the cytoplasmic side of BR than on the extracellular, in part because the mechanisms of Schiff base reprotonation and proton uptake are understood in much less structural detail. Also, several important events are coupled to one another in this region, including reprotonation of the Schiff base, retinal reisomerization, conformational change of the protein backbone, and proton uptake from the cytoplasmic surface [62]. Because of this coupling, it is difficult to decide which molecular event is the one directly affected by a mutation. All of this makes it even more interesting to look at the conservation pattern in the cytoplasmic half of BR. As one would expect from the primary proton donor, which is necessary for the fast reprotonation of the Schiff base [46, 63, 64], Asp-96 is conserved. Several residues were suggested to maintain its high proton affinity in the initial state and modulate it through the photocycle. Replacing Thr-46 with Val resulted in accelerated reprotonation of the Schiff base, but greatly slowed reprotonation of Asp-96 [48]. This residue, later found to be hydrogen-bonded to Asp-96 [28], is also conserved. A few members of the hydrophobic environment of Asp-96 (so-called leucine barrel, with phenylalanine lids) were mutagenized, and the phenotypes were similar to that of Thr-46-Val implying roles in insulating Asp-96 from the cytoplasmic aqueous medium. Among them are Phe-42 [65] (present as Phe or Tyr), Leu-100 [66], and Leu-223 (Brown, Dioumaev, Needleman, and Lanyi, unpublished data). All of these residues are conserved. Interestingly, replacements of several amino acids in this region, which are not close partners of Asp-96, produce the same distinct phenotype of fast reprotonation of the Schiff base with very slow recovery of the initial state. Those include also mutants of the conserved Phe-171 [67] and Ser-226 [48]. Conservation of these residues is not surprising, given the fact of dramatic decrease of the photocycle turnover for the respective mutants. At the same time, a few residues, which produced similar, but less dramatic changes when exchanged for Cys, are not conserved as strongly. For example, Ile-222 [66] can be also Val or Ala, and Val-167 (Brown, Needleman, and Lanyi, unpublished data) can be Leu or Thr (figure).
Another interesting residue, Arg-227, is conserved as a positively charged Arg or His, and when replaced by Gln, produces the changes in the direction opposite from the previous group of mutants, slowing down the decay of M intermediate [68]. Similar changes were observed also for Phe-156-Cys replacement [66] (this residue can be present also as Leu) and for Asp-38-Cys mutation [69] (this residue is also found to be present as Arg, Lys, or Glu). The molecular rationale for these effects is not understood, as all of these residues are located quite far from Asp-96. Clearly, their strong conservation is not as important as that of the hydrophobic environment of this aspartic acid, which is necessary to keep it protonated in the unphotolyzed protein.
The question of a role of Asp-38 is the most perplexing one. When Asp-38 is exchanged for Arg the reprotonation of the Schiff base slows down dramatically [69] and important conformational changes in the protein backbone are suppressed [70]. One could argue that such suppression in this mutant originates from the lack of accumulation of the N intermediate, which is characterized by conformational changes larger than those in the M state [71, 72] (but see evidence to the contrary in [73]). On the other hand, the Asp-38-Asn mutation does not produce such great changes, implying that the carboxylic group of Asp-38 is not indispensable for proton transfer reactions [56]. The properties of the Asp-38-Arg mutant are a real surprise, considering that at least nine BR-like pigments have a positively charged Arg or Lys at this position (figure). Does that mean that these species of halobacteria can tolerate twenty-fold slower turnover of the photocycle because the buildup of a proton gradient is limited by light intensity? Or do they have some other replacements compensating for the slowdown of the photocycle? Careful look at the sequences in the figure reveals that replacement Asp-38-Arg is always accompanied by the replacement Lys-40-Gln. Unfortunately, the phenotypes of Lys-40 mutants are unknown, so we cannot check this hypothesis for consistency. Finally, a slowly decaying M intermediate is needed if some halobacteria use a BR-like pigment as a sensory rhodopsin, not a pumping one. In this situation the long-living M state is an advantage, like in the case of halobacterial sensory rhodopsins [74].
Another contentious question is the role of carboxylic acids on the cytoplasmic surface in proton uptake. It was suggested that these carboxyls can act as a proton collecting funnel [57, 75]. Replacing some of the surface Asp by Asn (Glu by Gln), either as single mutations or in various combinations, did not produce substantial changes in proton uptake kinetics [56]. The same was true when they were replaced with Cys or Arg [69], with the above-mentioned exception of the Asp-38-Arg mutant. Only a few of these carboxylic acids are conserved, among them is Asp-36 and Glu-166 (can be also present as Asp). Because the exchange of either of these two for a non-protonatable residue produced only minimal changes in the photocycle [56], it should be concluded that the purpose for their conservation is not related to proton uptake. Other carboxylic acids include the non-conserved Asp-38 (can be also Arg, Lys, and Glu as discussed above), Asp-102 (exists also as Gly, Lys, or Asn), Asp-104 (also encountered as Asn) and Glu-161 (found also as Ala, Lys, Arg, and Ser). There are four more non-conserved acidic residues on the C-terminal tail on the cytoplasmic side (Glu-232, Glu-234, and Glu-237, as well as Asp-242), but mutational data are not available for any of those. Proteolytic removal of the C-tail slows down proton uptake less than twofold [76]. To sum up, the conservation pattern of the cytoplasmic carboxylic acids can hardly lend support to the idea about their crucial involvement in proton conduction.
Comparing the results of site-specific mutagenesis of BR with the information on its natural diversity in various halobacterial species can be instructive. Such a comparison often yields the expected result: residues that produce strongly perturbed phenotypes when replaced through mutagenesis are strictly conserved. At the same time, there are several interesting exceptions from this rule, which allow us to pose questions about the actual roles of certain amino acids in the proton transport. Absence of conservation can help in eliminating wrong choices of residues suggested to be important for BR function.
REFERENCES
1.Haupts, U., Tittor, J., and Oesterhelt, D. (1999)
Annu. Rev. Biophys. Biomol. Struct., 28, 367-399.
2.Heberle, J., Fitter, J., Sass, H. J., and
Büldt, G. (2000) Biophys. Chem., 85, 229-248.
3.Lanyi, J. K. (2000) J. Phys. Chem. B,
104, 11441-11448.
4.Kaulen, A. D. (2000) Biochim. Biophys. Acta,
1460, 204-219.
5.Dunn, R. J., Hackett, N. R., McCoy, J. M., Chao, B.
H., Kimura, K., and Khorana, H. G. (1987) J. Biol. Chem.,
262, 9246-9254.
6.Soppa, J., and Oesterhelt, D. (1989) J. Biol.
Chem., 264, 13043-13048.
7.Ni, B., Chang, M., Duschl, A., Lanyi, J., and
Needleman, R. (1990) Gene, 90, 169-172.
8.Brown, L. S. (2000) Biochim. Biophys. Acta,
1460, 49-59.
9.Otomo, J., Urabe, Y., Tomioka, H., and Sasabe, H.
(1992) J. Gen. Microbiol., 138, 2389-2396.
10.Mukohata, Y. (1994) Biophys. Chem.,
50, 191-201.
11.Ihara, K., Umemura, T., Katagiri, I.,
Kitajima-Ihara, T., Sugiyama, Y., Kimura, Y., and Mukohata,Y. (1999)
J. Mol. Biol., 285, 163-174.
12.Mukohata, Y., Ihara, K., Tamura, T., and
Sugiyama, Y. (1999) J. Biochem., 125, 649-657.
13.Ovchinnikov, Y. A., Abdulaev, N. G., Feigina, M.
Y., Kiselev, A. V., Lobanov, N. A., and Nazimov, I. V. (1978)
Bioorg. Khim., 4, 1573-1574.
14.Khorana, H. G., Gerber, G. E., Herlihy, W. C.,
Gray, C. P., Anderegg, R. J., Nihei, K., and Biemann, K. (1979)
Proc. Natl. Acad. Sci. USA, 76, 5046-5050.
15.Tateno, M., Ihara, K., and Mukohata, Y. (1994)
Arch. Biochem. Biophys., 315, 127-132.
16.Sugiyama, Y., Yamada, N., and Mukohata, Y. (1994)
Biochim. Biophys. Acta, 1188, 287-292.
17.Sugiyama, Y., Maeda, M., Futai, M., and Mukohata,
Y. (1989) J. Biol. Chem., 264, 20859-20862.
18.Soppa, J., Duschl, J., and Oesterhelt, D. (1993)
J. Bacteriol., 175, 2720-2726.
19.Uegaki, K., Sugiyama, Y., and Mukohata, Y. (1991)
Arch. Biochem. Biophys., 286, 107-110.
20.Wang, H., Zhan, S. X., Sun, Q. G., Xu, D. Q.,
Zhao, W., Huang, W. D., and Li, Q. G. (2000) Chin. Sci. Bull.,
45, 1108-1113.
21.Kitajima, T., Hirayama, J.-I., Ihara, K.,
Sugiyama, Y., Kamo, N., and Mukohata, Y. (1996) Biochem. Biophys.
Res. Commun., 220, 341-345.
22.Yatsunami, R., Kawakami, T., Ohtani, H., and
Nakamura, S. (2000) Extremophiles, 4, 109-114.
23.Kamekura, M., Seno, Y., and Tomioka, H. (1998)
Extremophiles, 2, 33-39.
24.Lipman, D., Altschul, S. F., and Kececioglu, J.
(1989) Proc. Natl. Acad. Sci. USA, 86, 4412-4415.
25.Rothschild, K. J., Braiman, M. S., Mogi, T.,
Stern, L. J., and Khorana, H. G. (1989) FEBS Lett., 250,
448-452.
26.Greenhalgh, D. A., Farrens, D. L., Subramaniam,
S., and Khorana, H. G. (1993) J. Biol. Chem., 268,
20305-20311.
27.Song, L., El-Sayed, M. A., and Lanyi, J. K.
(1993) Science, 261, 891-894.
28.Luecke, H., Schobert, B., Richter, H.-T.,
Cartailler, J.-P., and Lanyi, J. K. (1999) J. Mol. Biol.,
291, 899-911.
29.Marti, T., Rösselet, S. J., Otto, H., Heyn,
M. P., and Khorana, H. G. (1991) J. Biol. Chem., 266,
18674-18683.
30.Duñach, M., Berkowitz, S., Marti, T., He,
Y.-W., Subramaniam, S., Khorana, H. G., and Rothschild, K. J. (1990)
J. Biol. Chem., 265, 16978-16984.
31.Sonar, S., Krebs, M. P., Khorana, H. G., and
Rothschild, K. J. (1993) Biochemistry, 32, 2263-2271.
32.Ahl, P. L., Stern, L. J., Mogi, T., Khorana, H.
G., and Rothschild, K. J. (1989) Biochemistry, 28,
10028-10034.
33.Brown, L. S., Gat, Y., Sheves, M., Yamazaki, Y.,
Maeda, A., Needleman, R., and Lanyi, J. K. (1994) Biochemistry,
33, 12001-12011.
34.Subramaniam, S., Greenhalgh, D. A., Rath, P.,
Rothschild, K. J., and Khorana, H. G. (1991) Proc. Natl. Acad. Sci.
USA, 88, 6873-6877.
35.Weidlich, O., Schalt, B., Friedman, N., Sheves,
M., Lanyi, J. K., Brown, L. S., and Siebert, F. (1996)
Biochemistry, 35, 10807-10814.
36.Hatanaka, M., Kashima, R., Kandori, H., Friedman,
N., Sheves, M., Needleman, R., Lanyi, J. K., and Maeda, A. (1997)
Biochemistry, 36, 5493-5498.
37.Ihara, K., Amemiya, T., Miyashita, Y., and
Mukohata, Y. (1994) Biophys. J., 67, 1187-1191.
38.Ihara, K., Yamada, N., Itoh, S., and Mukohata, Y.
(1994) Biophys. J., 66, A112.
39.Luecke, H., Schobert, B., Richter, H.-T.,
Cartailler, J.-P., and Lanyi, J. K. (1999) Science, 286,
255-260.
40.Otto, H., Marti, T., Holz, M., Mogi, T., Stern,
L. J., Engel, F., Khorana, H. G., and Heyn, M. P. (1990) Proc. Natl.
Acad. Sci. USA, 87, 1018-1022.
41.Balashov, S. P., Govindjee, R., Kono, M.,
Imasheva, E., Lukashev, E., Ebrey, T. G., Crouch, R. K., Menick, D. R.,
and Feng, Y. (1993) Biochemistry, 32, 10331-10343.
42.Brown, L. S., Sasaki, J., Kandori, H., Maeda, A.,
Needleman, R., and Lanyi, J. K. (1995) J. Biol. Chem.,
270, 27122-27126.
43.Balashov, S. P., Imasheva, E. S., Ebrey, T. G.,
Chen, N., Menick, D. R., and Crouch, R. K. (1997) Biochemistry,
36, 8671-8676.
44.Alexiev, U., Mollaaghababa, R., Khorana, H. G.,
and Heyn, M. P. (2000) J. Biol. Chem., 275,
13431-13440.
45.Imasheva, E. S., Balashov, S. P., Ebrey, T. G.,
Chen, Y., Menick, D. R., and Crouch, R. K. (2000) Biophys. J.,
78, 475A.
46.Soppa, J., Otomo, J., Straub, J., Tittor, J.,
Meessen, S., and Oesterhelt, D. (1989) J. Biol. Chem.,
264, 13049-13056.
47.Govindjee, R., Kono, M., Balashov, S. P.,
Imasheva, E., Sheves, M., and Ebrey, T. G. (1995) Biochemistry,
34, 4828-4838.
48.Marti, T., Otto, H., Mogi, T., Rösselet, S.
J., Heyn, M. P., and Khorana, H. G. (1991) J. Biol. Chem.,
266, 6919-6927.
49.Lu, M., Govindjee, R., Balashov, S. P., Ebrey, T.
G., Chen, Y., Menick, D. R., and Crouch, R. K. (2000) Biophys.
J., 78, 476A.
50.Misra, S., Martin, C., Kwon, O.-H., Ebrey, T. G.,
Chen, N., Crouch, R. K., and Menick, D. R. (1997) Photochem.
Photobiol., 66, 774-783.
51.Lu, M., Balashov, S. P., Ebrey, T. G., Chen, N.,
Chen, Y., Menick, D. R., and Crouch, R. K. (2000) Biochemistry,
39, 2325-2331.
52.Garczarek, F., Uliczka, T., and Gerwert, K.
(2000) Abst. 9th Int. Conf. on Retinal Proteins, Szeged,
Hungary, p. 79.
53.Cao, Y., Brown, L. S., Sasaki, J., Maeda, A.,
Needleman, R., and Lanyi, J. K. (1995) Biophys. J., 68,
1518-1530.
54.Govindjee, R., Imasheva, E. S., Misra, S.,
Balashov, S. P., Ebrey, T. G., Chen, N., Menick, D. R., and Crouch, R.
K. (1997) Biophys. J., 72, 886-898.
55.Lukashev, E. P., Govindjee, R., Kono, M., Ebrey,
T. G., Sugiyama, Y., and Mukohata, Y. (1994) Photochem.
Photobiol., 60, 69-75.
56.Brown, L. S., Needleman, R., and Lanyi, J. K.
(1999) Biochemistry, 38, 6855-6861.
57.Kimura, Y., Vassylyev, D. G., Miyazawa, A.,
Kidera, A., Matsushima, M., Mitsuoka, K., Murata, K., Hirai, T., and
Fujiyoshi, Y. (1997) Nature, 389, 206-210.
58.Pebay-Peyroula, E., Rummel, G., Rosenbusch, J.
P., and Landau, E. M. (1997) Science, 277, 1676-1681.
59.Sanz, C., Lazarova, T., Sepulcre, F.,
González-Moreno, R., Bourdelande, J.-L., Querol, E., and
Padrós, E. (1999) FEBS Lett., 456, 191-195.
60.Dioumaev, A. K., Richter, H.-T., Brown, L. S.,
Tanio, M., Tuzi, S., Saitô, H., Kimura, Y., Needleman, R., and
Lanyi, J. K. (1998) Biochemistry, 37, 2496-2506.
61.Zscherp, C., Schlesinger, R., Tittor, J.,
Oesterhelt, D., and Heberle, J. (1999) Proc. Natl. Acad. Sci.
USA, 96, 5498-5503.
62.Luecke, H., Schobert, B., Cartailler, J.-P.,
Richter, H.-T., Rosengarth, A., Needleman, R., and Lanyi, J. K. (2000)
J. Mol. Biol., 300, 1237-1255.
63.Otto, H., Marti, T., Holz, M., Mogi, T., Lindau,
M., Khorana, H. G., and Heyn, M. P. (1989) Proc. Natl. Acad. Sci.
USA, 86, 9228-9232.
64.Holz, M., Drachev, L. A., Mogi, T., Otto, H.,
Kaulen, A. D., Heyn, M. P., Skulachev, V. P., and Khorana, H. G. (1989)
Proc. Natl. Acad. Sci. USA, 86, 2167-2171.
65.Dioumaev, A. K., Brown, L. S., Needleman, R., and
Lanyi, J. K. (1998) Biochemistry, 37, 9889-9893.
66.Brown, L. S., Váró, G.,
Needleman, R., and Lanyi, J. K. (1995) Biophys. J., 69,
2103-2111.
67.Kamikubo, H., Kataoka, M., Váró,
G., Oka, T., Tokunaga, F., Needleman, R., and Lanyi, J. K. (1996)
Proc. Natl. Acad. Sci. USA, 93, 1386-1390.
68.Drachev, L. A., Kaulen, A. D., Khorana, H. G.,
Mogi, T., Postanogova, N. V., Skulachev, V. P., and Stern, L. J. (1992)
Photochem. Photobiol., 55, 741-744.
69.Riesle, J., Oesterhelt, D., Dencher, N. A., and
Heberle, J. (1996) Biochemistry, 35, 6635-6643.
70.Sass, H.-J., Gessenich, R., Koch, M. H. J.,
Oesterhelt, D., Dencher, N. A., Büldt, G., and Rapp, G. (1998)
Biophys. J., 75, 399-405.
71.Vonck, J. (2000) EMBO J., 19,
2152-2160.
72.Oka, T., Yagi, N., Fujisawa, T., Kamikubo, H.,
Tokunaga, F., and Kataoka, M. (2000) Proc. Natl. Acad. Sci. USA,
97, 14278-14282.
73.Subramaniam, S., Lindahl, M., Bullough, P.,
Faruqi, A. R., Tittor, J., Oesterhelt, D., Brown, L., Lanyi, J., and
Henderson, R. (1999) J. Mol. Biol., 287, 145-161.
74.Sasaki, J., and Spudich, J. L. (2000) Biochim.
Biophys. Acta, 1460, 230-239.
75.Checover, S., Nachliel, E., Dencher, N. A., and
Gutman, M. (1997) Biochemistry, 36, 13919-13928.
76.Govindjee, R., Ohno, K., and Ebrey, T. G. (1982)
Biophys. J., 38, 85-87.