Kinetic Analysis of Maturation and Denaturation of DsRed, a Coral-Derived Red Fluorescent Protein
V. V. Verkhusha1*, N. A. Akovbian2,3, E. N. Efremenko2, S. D. Varfolomeyev2, and P. V. Vrzheshch1,3
1Center for Molecular Medicine, 2Faculty of Chemistry, and 3International Biotechnological Center, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 938-2422; E-mail: vrzh@genebee.msu.ru* To whom correspondence should be addressed.
Received May 21, 2001
The red fluorescent protein DsRed recently cloned from Discosoma coral, with its significantly red-shifted excitation and emission maxima (558 and 583 nm, respectively), has attracted great interest because of its spectral complementation to other fluorescent proteins, including the green fluorescent protein and its enhanced mutant EGFP. We demonstrated that the much slower DsRed fluorescence development could be described by a three-step kinetic model, in contrast to the fast EGFP maturation, which was fitted by a one-step model. At pH below 5.0 DsRed fluorescence gradually decreased, and the rate and degree of this fluorescence inactivation depended on the pH value. The kinetics of fluorescence inactivation under acidic conditions was fitted by a two-exponential function where the initial inactivation rate was proportional to the fourth power of proton concentration. Subsequent DsRed alkalization resulted in partial fluorescence recovery, and the rate and degree of such recovery depended on the incubation time in the acid. Recovery kinetics had a lag-time and was fitted minimally by three exponential functions. The DsRed absorbance and circular dichroism spectra revealed that the fluorescence loss was accompanied by protein denaturation. We developed a kinetic mechanism for DsRed denaturation that includes consecutive conversion of the initial state of the protein, protonated by four hydrogen ions, to the denatured one through three intermediates. The first intermediate still emits fluorescence, and the last one is subjected to irreversible inactivation. Because of tight DsRed tetramerization we have suggested that obligatory protonation of each monomer results in the fluorescence inactivation of the whole tetramer.
KEY WORDS: red fluorescent protein, chromophore, maturation, denaturation, renaturation, tetramer, kinetics
Abbreviations: DsRed) red fluorescent protein; GFP) green fluorescent protein.
Fluorescent proteins are widely used in biochemistry, biotechnology, and
cell and developmental biology [1]. A unique
property of these homologous chromoproteins is an intramolecular
autocatalytic formation of the chromophoric group. This process called
maturation proceeds through several steps. For the best studied
representative of this family, a green fluorescent protein (GFP) from
the jellyfish Aequorea victoria, the process of
post-translational maturation includes the formation of
p-hydroxybenzylideneimidazolidinone due to the reactions of
condensation between the carbon of the carboxyl group of Ser65 and the
nitrogen of the amino group of Gly67, and dehydrogenation of the
methylene bridge of Tyr66 by dissolved oxygen [2].
The necessary condition for the fluorescence of
p-hydroxybenzylideneimidazolidinone is the presence of a protein
shell covering the chromophore (beta-barrel) [3]. The beta-barrel forms an almost perfect
compact cylinder of about 2.4 nm diameter and 4.2 nm length [4]. The chromophore of GFP is located close to the
geometrical center of the cylinder, approximately in the middle of an
irregular alpha-helix. Due to the unique structure of the
protein, its chromophore is completely isolated from the outer
solution, thus making it possible to avoid access of oxygen and other
substances to the chromophore, and prevent the energy dissipation due
to isomerization of the bonds or oscillation processes [5].
The red fluorescent protein, DsRed, is a distant homolog of GFP; its gene was cloned from the coral Discosoma sp. [6]. The chromophore of DsRed is formed similarly to that of GFP by cyclization of the carbon chain between Gln66 and Gly68 and subsequent dehydrogenation of the Calpha-Cbeta bond of Tyr67 [7]. It has been suggested that the reactions of dehydration of the alphaC-N bond of Gln66 yielding an acylamide and trans-cis-isomerization of the peptide bond between Phe65 and Gln66 residues [8] are also involved in maturation of DsRed.
The process of chromophore maturation in a fluorescent protein, which can be detected by its fluorescence acquisition, must reflect the kinetic mechanism of the reactions involved. So far, the only kinetic study that has been performed is that of chromophore formation in the Aequorea GFP/Ser65Thr mutant during its renaturation from inclusion bodies of E. coli [9]. In the present study, we demonstrated that DsRed denatures in weak acidic medium, and alkalization of the medium resulted in renaturation of the protein [10]. We also investigated the kinetic mechanisms for maturation, denaturation, and renaturation of the red fluorescent protein.
MATERIALS AND METHODS
The 0.71-kb NdeI-BamHI fragment of wild type DsRed and the 0.75-kb NcoI-BamHI fragment of EGFP (the GFP/Phe64Leu/Ser65Thr mutant) were amplified from the pDsRed1-N1 and pEGFP-N1 plasmids (Clontech, USA) using PCR and cloned into the pET-11c and pET-11d vectors (Invitrogen, USA), respectively. The polyhistidine tag was added to the C-terminals of both proteins in the course of PCR-amplification. The resulting plasmids (pET-11c-DsRed-Hisx6 and pET-11d-EGFP-Hisx6) were transformed into an E. coli BL21(DE3) strain (Invitrogen). Protein expression was induced by incubation of the cells in the presence of 0.5 mM IPTG (isopropyl-beta-D-thiogalactopyranozide, Sigma, USA) at 37°C for 16 h. The recombinant DsRed was purified on Agarose Ni-NTA (Qiagen, USA). The protein solutions were concentrated to 12 mg/ml in 100 mM citrate-phosphate buffer, pH 7.6, using Ultrafree-4 centrifugal filters (Millipore, USA). The purity of the recombinant DsRed was no less than 95% as indicated by SDS-PAGE. Protein concentration was measured using a standard kit for protein assay (BioRad, USA).
For spectroscopic studies, an aliquot of DsRed stock solution was diluted 20-1200-fold with 10-100 mM phosphate or citrate-phosphate buffer of different pH values. For kinetic studies, a solution of DsRed in 100 mM citrate-phosphate buffer, pH 5.0, was acidified to a necessary pH value with 5 M phosphoric acid. If it was necessary, the pH value of the solution was further adjusted back to 5.0 with 10 M NaOH. The changes in pH were made directly in a spectrophotometric cuvette under intensive stirring. A sample for kinetic studies contained 0.01 mg/ml DsRed and was illuminated in a fluorimetric cuvette at 556 nm; the emission was recorded at 583 or 590 nm. To study the absorption and CD spectra of the renatured at pH 5.0 DsRed, the protein was purified by gel filtration on Sephadex G-75 (Pharmacia, Sweden).
To study maturation of the recombinant proteins, E. coli cells expressing EGFP or DsRed were grown for 3 days under anaerobic conditions (to prevent oxygen-dependent post-translational modifications of the chromophoric groups). Access of air oxygen was provided at the step of the cell lysis. To prepare cell lysate, the cells were precipitated by centrifugation at 0°C, resuspended at 0°C in phosphate buffer (130 mM NaCl, 7 mM Na2HPO4*12H2O, and 3 mM NaH2PO4*2H2O, pH 7.4), and then sonicated in an ice bath. The lysate was centrifuged (12,000g, 0°C), and the supernatant was diluted by the phosphate buffer so that its optical density at 280 nm was 1.0. The resulting solution was immediately used for kinetic studies of the chromophore maturation, the incubation of the lysate being started under aerobic conditions at various temperatures in the range 4-80°C. After different time of incubation, samples were taken from the incubation mixture. The fluorescence of the samples was measured after 5 min of incubation of the samples directly in a spectrophotometric cuvette in a thermostatted (25°C) cell of a fluorimeter. The optical density of the lysate at 650 nm did not exceed 0.05 during the experiment. The time between the beginning of the cell lysis and the beginning of fluorescence studies did not exceed 30 min at 0°C.
A Shimadzu UV-1601 spectrophotometer, Perkin-Elmer LS50B spectrofluorimeter, and Jasco-720 spectropolarimeter were used to study the absorption, fluorescence, and CD, respectively. A Beckman XL-1 analytical centrifuge was used for equilibrium centrifugation of the samples at 15,000 rpm for 48 h. All the spectroscopic studies and centrifugation of the samples were performed at 25 and 20°C, respectively. Experimental data were analyzed using Microcal Origin v. 5.0 (Microcal Software) and MatLab v. 5.3 (MathWorks) programs.
RESULTS AND DISCUSSION
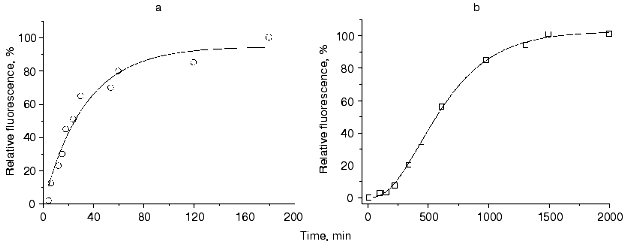
The kinetics of maturation of the wild type red fluorescent protein (DsRed) detected by the change in its fluorescence intensity was studied over a wide range of temperatures, from 4 to 80°C. For all the temperatures, the kinetic curves of maturation of DsRed exhibited a lag-period with subsequent acceleration, an inflection point, deceleration, and a plateau. Figure 1 illustrates kinetic curves of maturation of the mutant green fluorescent protein (EGFP) (Fig. 1a) and DsRed (Fig. 1b) at 37°C. The kinetics of maturation of EGFP is a single exponential curve and can be adequately described by a single-step model according to which a nonfluorescent precursor A transforms into a fluorescent product B in the course of a first order reaction with a rate constant k1:
If at zero time all the protein exists in the formA (i.e., at t = 0, [A] = A0 and [B] = 0), the kinetics of concentration changes for A and B is described by the equations:
where t is the time. The solid line in Fig. 1a corresponds to Eq. (3), where k1 = 5.01*10-4 sec-1.
The kinetics of maturation of DsRed cannot be described by either the single-step model or by a two-step model. Approximation of the experimental data by the single- or two-step models results in the appearance of systematic deviations between the experimental data and the theoretical values (data not shown). An adequate approximation can be achieved by using a three-step model:Fig. 1. a, b) Kinetics of maturation of recombinant green fluorescent protein EGFP (open circles) and the recombinant red fluorescent protein DsRed (open squares), respectively. The reaction was detected by the change in the fluorescence intensity of purified bacterial lysate at 37°C. The experimental data were approximated by Eq. (3) in the case of EGFP (solid line) and by Eq. (8) in the case of DsRed (solid line).
The model suggests transformation of nonfluorescent precursors A, B, and C into a fluorescent product D in the course of three consecutive first-order reactions with rate constants k1, k2, and k3, respectively. If at zero time all the protein exists in the formA (i.e., at t = 0, [A] = A0, [B] = 0, [C] = 0, and [D] = 0), kinetics of the changes in concentrations of A, B, C and D is described by the equations:
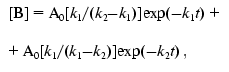 (6)
(6)
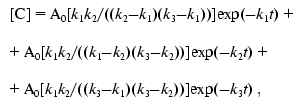 (7)
(7)
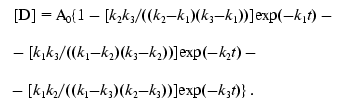 (8)
(8)The solid line in Fig. 1b corresponds to Eq. (8), where k1 = 7.6411*10-5 sec-1, k2 = 7.6416*10-5 sec-1, and k3 = 7.6408*10-5 sec-1.
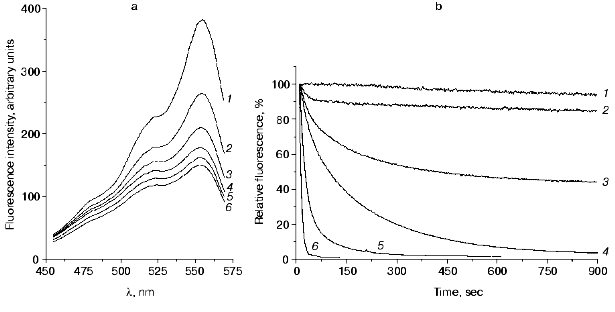
The fluorescence of DsRed little depends on pH of the medium in the range 5.0-12.0, but decreases outside this range [10, 11]. We demonstrated that the decrease in fluorescence observed in acid medium was a partially reversible process [10]. Figure 2a presents excitation spectra of the red fluorescent protein recorded after different times of incubation at pH 4.3. The fluorescence intensity of DsRed decreases with time at 450-570 nm excitation. The kinetics of the decrease in the fluorescence at a certain pH value (Fig. 2b) can be described by a double-exponential curve:
where IA is the fluorescence intensity, A0 is the residual fluorescence intensity at infinite time, A1, A2, t1A, and t2A are kinetic parameters depending on pH. It should be noted that the experimental data can be adequately described by only a function containing at least two exponential curves.
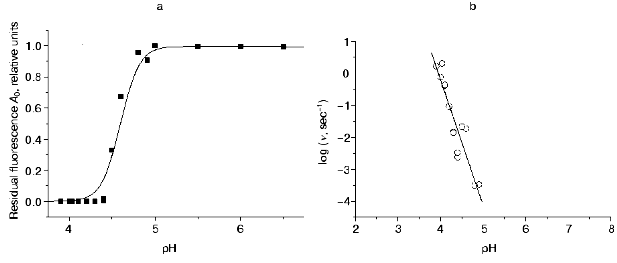
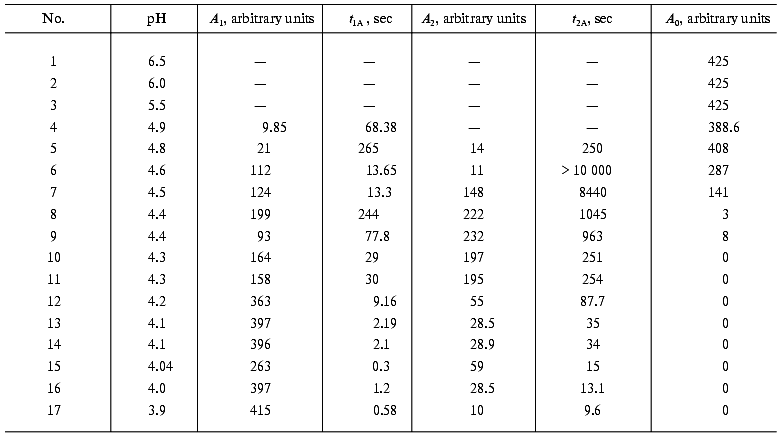
Decrease in pH of the medium results in a significant increase in the A1 values and decrease in the A0, t1A, and t2A values, while the A2 exhibits a bell-shaped dependence with a maximum at pH 4.4 (Table 1). At pH 4.55, the residual fluorescence A0 of DsRed is a half of its initial value; however, at pH 4.4, the fluorescence of DsRed almost completely disappears (A0 = 0) (Fig. 3a). The rate of the decrease in the fluorescence also significantly depends on pH. The relative initial rate of the fluorescence decrease (nu) is presented in Fig. 3b as a function of proton concentration on a logarithmic scale. This dependence can be presented as a straight line with a slope of -4.0 (p < 0.001). Such a sharp drop in the fluorescence of DsRed on decreasing pH is in great contrast to the behavior of the wild-type green fluorescent proteins, which exhibit a relatively slow loss of the fluorescence on pH decrease from 6.5 to 4.5 in the case of Aequorea GFP [12] and from 6.0 to 2.5 in the case of Renilla GFP [13].Fig. 2. a) Excitation spectra of DsRed: 1) at pH 5.0; 2-6) after incubation at pH 4.3 for 29, 62, 104, 244, and 495 sec, respectively. No significant changes in the spectrum were observed at pH 5.0 for the indicated time intervals (data not shown). b) Kinetics of the decrease in DsRed fluorescence in acid medium: 1-6) pH 4.8, 4.5, 4.4, 4.3, 4.2, and 3.9, respectively.
Table 1. Values of the parameters
A0, A1, A2,
t1A, and t2A of Eq. (9) used for the approximation of acidic inactivation
of DsRed at different pH of the solution. The conditions are given in
the legend to Fig. 2b
The pH dependencies of the green fluorescence proteins described in [12, 14, 15] can be approximately described using a simple fast equilibrium model suggesting protonation of a single ionogenic group of the protein:Fig. 3. a) The pH dependence of normalized values of residual fluorescence of DsRed (A0, filled squares). The A0 values were calculated from the experimental data presented in Fig. 2b using Eq. (9). The A0 value at pH 5.0 was taken as 1.0. The solid line is the approximation of the experimental data according to Eq. (18), where Ka = 0.0025 and Kd = 108. b) Dependence of the relative initial rate of the fluorescence decrease (nu) on proton concentration in a logarithmic scale. The nu values were calculated from the slope of the kinetic curves (Fig. 2b) at zero time using Eq. (9). Solid line is a linear approximation of the experimental data.
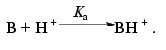 (10)
(10)Assuming that only the deprotonated form is capable of emitting, the theoretical dependencies of the normalized concentration of the deprotonated form [B] on proton concentration can be described for Scheme (10) using Eq. (11):
This dependence is in a good agreement with pH dependencies of the normalized fluorescence of a number of GFP mutants [14, 15]. Analysis of the data presented in [14] shows that in the case of ECFP (a cyan fluorescent mutant of GFP), the theoretical dependence fits the experimental data only assuming that both the deprotonated form B and the protonated form BH+ (see Scheme (10)) are capable of emitting, the contributions of the protonated and the deprotonated forms being 0.5 and 1.0, respectively. Consequently, the dependence of the normalized fluorescence (F) of this mutant GFP form is described by the equation:
Thus, in spite of some differences in mechanism, the pH dependencies of the fluorescence of all GFP mutants can be described by the fast equilibrium protonation model (Eq. (10)), differing greatly from those of the red fluorescent protein.
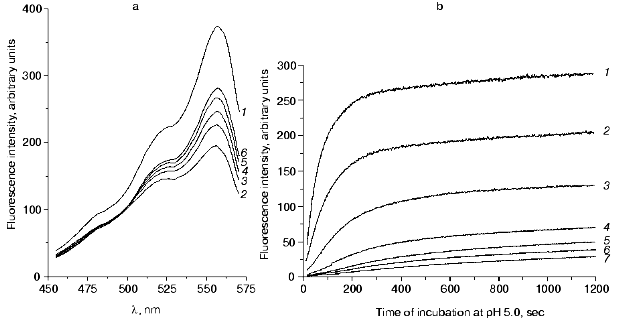
We investigated the recovery of the fluorescence of DsRed after incubation of the protein at pH values below 4.8, the pH value of the solution being then adjusted back to 5.0. The alkalization of the incubation medium to pH 5.0 resulted in a significant time-dependent recovery of the fluorescence, although its level was lower than the initial one. The fluorescence intensity increased greatly under 510-570 nm excitation (Fig. 4a).
Figure 4b illustrates the kinetics of the fluorescence recovery for the samples of DsRed incubated at pH 4.1 for different times. After the incubation of the protein at pH 4.1 for the times indicated in Fig. 4b, no residual fluorescence was observed (A0 = 0). Under 556-nm excitation, the kinetics of the fluorescence recovery after alkalization of the solution to pH 5.0 could be described by a function containing at least three exponential components:Fig. 4. a) Excitation spectra of DsRed at pH 5.0: 1) initial spectrum; 2-6) the protein was incubated at pH 4.4 for 90 sec, then the pH of the solution was adjusted to 5.0 and the protein was incubated at pH 5.0 for 68 (2), 134 (3), 200 (4),355 (5), and 839 sec (6). b) Recovery of the fluorescence of DsRed at pH 5.0. The protein was incubated at pH 4.1 for 30 (1), 50 (2), 99 (3), 200 (4), 372 (5), 435 (6), and 710 sec (7), then the solution was adjusted to pH 5.0.
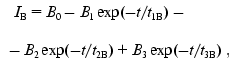 (13)
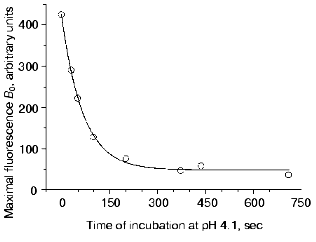
(13)where IB is the fluorescence intensity, B0 is the maximal fluorescence at infinite time, B1, B2, B3, t1B, t2B, and t3B are kinetic parameters depending on the time of incubation of DsRed at pH 4.1. An increase in the incubation time at pH 4.1 results in a decrease in the values of B0, B1, B2, and B3 and in an increase in the parameters t1B, t2B, and t3B (Table 2). Such a decrease in the maximal fluorescence (B0) is related to an irreversible inactivation, i.e., the inactivation that cannot be reversed by alkalization of the medium to pH 5.0. The rate constant of the irreversible inactivation of DsRed at pH 4.1 can be calculated from the dependence of B0 on incubation time (Fig. 5).
Table 2. Values of the parameters
B1, t1B, B2,
t2B, B3, t3B, and
B0 of Eq. (13) used for the
approximation of the recovery of DsRed fluorescence after different
times of incubation at pH 4.1 (tac). The
conditions are given in the legend to Fig. 4b
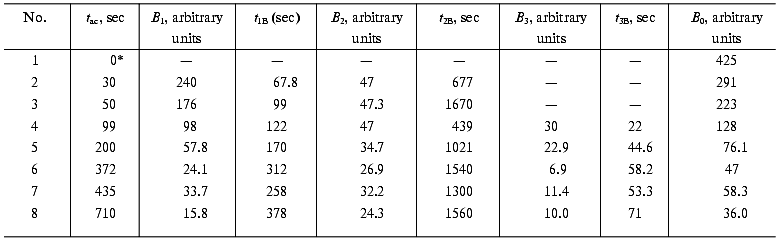
*Sample was not incubated in acid medium.
As seen from the data presented in Fig. 4b, the recovery of the fluorescence is described by a triple-exponential curve. The simplest kinetic model of this process includes three consecutive monomolecular reactions resulting in the formation of a completely fluorescent product from a nonfluorescent form of DsRed. In the simplest case, the mechanism of DsRed inactivation in acid medium must include the same intermediate steps as those after returning the pH value to the initial level. Considering that some form of acid inactivated DsRed cannot be recovered upon alkalization (Fig. 5), the kinetic scheme for the inactivation-reactivation of DsRed is as follows:Fig. 5. Dependence of the maximal value of the fluorescence at infinite time of reactivation at pH 5.0 (B0) on the time of DsRed incubation at pH 4.1. Experimental data are shown by open circles; the theoretical curve B0 = 377exp(-1.51*10-2 t) + 48.3 is shown by a solid line.
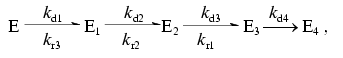 (14)
(14)where E is the fluorescent form of DsRed subjected to acidic inactivation; E1, E2, E3, and E4 are the products of the acidic inactivation of DsRed; kd1, kd2, kd3, and kd4 are the inactivation rate constants for the acidic inactivation of DsRed; kr1, kr2, kr3 are the reactivation rate constants for the reactivation of DsRed after alkalization of the medium. It is assumed that E4 is the irreversibly inactivated protein form that and cannot be reactivated. Experiments on kinetic of inactivation of DsRed (Fig. 2b) can be described by the Scheme (14) assuming that both E and E1 forms are capable of emitting.
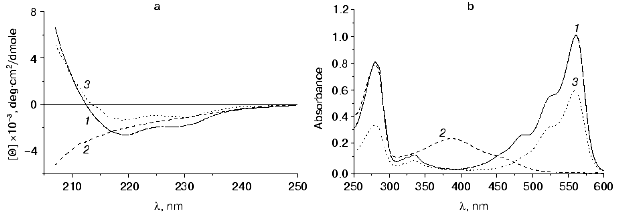
To study the molecular mechanism of the disappearance and recovery of the fluorescence of DsRed, we investigated the absorption and CD spectra of the protein. Figure 6a illustrates the effect of the acid medium (pH 4.1) on the secondary structure of DsRed. The far-UV CD spectrum of native DsRed at pH 5.0 (Fig. 6a, curve 1) has a pronounced minimum at 219 nm and a local minimum at 230 nm, this indicating a significant contribution of beta-structure to the spectrum of the protein.
This secondary structure breaks down at pH 4.1; as a result, the CD spectrum of the protein (curve 2 in Fig. 6a) becomes similar to the spectrum of a chaotic globule. Further incubation of the protein at pH 4.1 for 48 h results in no additional changes in the CD spectrum. These data are in agreement with previous studies of the CD of the green fluorescent protein denatured in the presence of guanidine hydrochloride [16]. The absorption spectrum of DsRed after the incubation at pH 4.1 (curve 2 in Fig. 6b) differs greatly from that of the native DsRed (curve 1 in Fig. 6b). Instead of the three unresolved bands in the visible region, which are characteristic for the native DsRed, the protein after incubation in acid medium has a broad absorption band with a maximum at 387 nm, this suggesting a significant changes in the microenvironment of the chromophore. This value is close to that of the absorption maximum of GFP under strongly acidic conditions (383 nm) [1], suggesting that the protonated chromophores of GFP and DsRed have similar structures [7].Fig. 6. a) CD spectra of DsRed: 1) at pH 5.0; 2) after 480 sec of incubation at pH 4.1; 3) the protein was incubated at pH 4.1 for 60 sec, then the pH of the sample was adjusted to 5.0. Samples 1 and 2 contained 0.24 mg/ml DsRed in 10 mM citrate-phosphate buffer. Curve 3 is the spectrum of pooled fluorescent fractions after alkalization of the sample from the pH 4.1 to 5.0. DsRed was incubated at pH 5.0 for 1 h, then the fluorescent fractions were isolated by gel filtration (see “Materials and Methods”). The final volume of the sample was equal to the initial volume. [Theta] is the molar ellipticity calculated per average residue. A spectrum similar to spectrum 2 was obtained after incubation of the protein at pH 4.1 for 60 sec (data not shown). b) Absorption spectra of DsRed: 1) at pH 5.0; 2) after 480 sec of incubation at pH 4.1; 3) the protein was incubated at 4.1 for 60 sec, then the pH of the sample was adjusted to 5.0. Samples 1 and 2 contained 0.6 mg/ml DsRed in 100 mM citrate-phosphate buffer. The sample 3 was prepared as indicated in the legend to Fig. 6a.
Incubation of DsRed at pH 4.1 for 60 sec with subsequent alkalization to pH 5.0 results in renaturation of DsRed, which can be detected by the changes in the CD spectrum. The CD spectrum of the fluorescent fraction of DsRed obtained by gel filtration (curve 3, Fig. 6a) is similar to that of the original sample of DsRed (curve 1, Fig. 6a). The absorption spectrum of this fraction (curve 3, Fig. 6b) is also similar to that of the native DsRed (curve 1, Fig. 6b). These data together with the data presented above indicate that the disappearance of the fluorescence and its recovery are due to the denaturation of DsRed at the pH values below 5.0 and its renaturation at pH 5.0, respectively.
Recently it has been shown that the chromophore of DsRed exists in two different states: immature green (475 nm excitation and 500 nm emission) and mature red (558 and 583 nm, respectively) [11]. The ratio of the immature protein form to the mature one in solution is approximately 1 : 1. However, fluorescence of the green form does not exceed 1% of the maximal red fluorescence, because the extinction coefficient at 475 nm as well as the quantum yield of the green fluorescence (500 nm) are significantly lower than the corresponding values of the red form of the chromophore. Also, the emissionless transition of the energy from the green to the red chromophore can proceed [11]. As a result, the absorption and excitation fluorescence spectra of the protein solution have a well detectable component of the green chromophore: the left shoulder with a maximum at 487 nm in the complex structure of DsRed spectrum (Figs. 4a and 6b, and [6]). It is noteworthy that the worst recovery of both the excitation spectrum (curves 2-6 in Fig. 4a) and the absorption spectrum (curve 3, Fig. 6b) after the incubation in acid medium is observed in the range of 470-500 nm, which corresponds to the green state of the chromophore. This suggests that the acidic denaturation results in either irreversible denaturation of the green state of the DsRed chromophore or its transition into the red state after renaturation at pH 5.0.
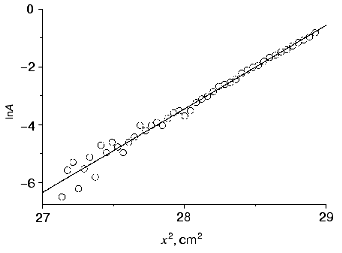
It is now well known that DsRed associates in solutions yielding stable tetramers [10, 11]. Also, the pH dependence of the initial rate of the fluorescence decrease is linearized in a logarithm scale with a slope of -4 (Fig. 3b). This suggests that four protons are involved in the inactivation process. Figure 7 presents the results of the sedimentation equilibrium analysis of a DsRed sample at pH 5.0. Concentration profiles of DsRed in a centrifugal cell after 48 h of centrifugation can be linearized on a logarithmic scale yielding a straight line with a slope of 2.9 (p < 0.001). Longer centrifugation did not alter the concentration profiles, suggesting that a thermodynamic equilibrium with a single type of DsRed particles was achieved. The molecular weight of the particles was calculated from the slope of the mentioned straight line as described in [17]. The value of the molecular weight was 115.2 kD, this corresponding to tetramers of DsRed. Tetramer formation was observed at protein concentrations in the range 0.15-1.0 mg/ml protein and at pH values in the range 5.0-8.0 (data not shown). This indicates that DsRed associates in solution yielding stable non-dissociating tetramers.
Our experiments on analytical ultracentrifugation have show that other GFP-like proteins zFP538 and zFP506 from nonbioluminescent coral Zonatus sp. [6] as well as amFP486 from Anemonia majano [6] form stable complexes even at low protein concentrations (data not shown). Fluorescent proteins asFP499 and asFP595 (homologous to GFP) isolated from the nonbioluminescent organism Anemonia sulcata also exist as stable dimers and tetramers [18]. There is also some information that GFP-like proteins of some bioluminescent representatives of Hydrozoa, including Aequrea GFP, at high concentrations close to physiological (more than 10 mg/ml) are capable of forming Ca2+-sensitive heterodimers with aequorin (GFP2-aequorin2)[19]. However, at low concentrations these proteins exist as nondissociating dimers, which can only be broken down by irreversible denaturation (except for Aequrea GFP, which exists mostly in monomeric state at concentrations lower than 0.5 mg/ml) [16]. These data, as well as X-ray studies [4, 8, 20] indicate that the beta-barrel of a monomer of most fluorescent proteins has two different sites involved in interactions with other subunits. One of these sites is usually involved in the formation of homodimers; functions of the other site have diverged in the course of evolution: in the case of bioluminescent species Anthozoa and Hydrozoa, it binds aequorin, while in the case of nonbioluminescent organisms, it binds the second dimer of the fluorescent protein.Fig. 7. Dependence of DsRed sample absorbance at 280 nm (A) on centrifugal cell path (x) after equilibrium centrifugation in the indicated scale. The sample contained 0.15 mg/ml DsRed in 100 mM citrate-phosphate buffer (pH 5.0). Experimental data (open circles) and linear approximation (solid line) are shown.
The fact that the denaturation of DsRed, which is detected by decrease in its fluorescence, is observed in a rather narrow range of pH (Fig. 3a), and also the fact that the initial velocity of the decrease in the fluorescence is proportional to the forth power of proton concentration (Fig. 3b) suggest that the denaturation results from simultaneous protonation of at least four ionogenic groups of DsRed. It remains unclear, whether the pKa values of these groups differ. Considering that DsRed is a tetramer, these ionogenic groups can be either the groups of a single subunit, or the groups of different subunits within the tetramer. If all monomers of DsRed are independently inactivated, simultaneous protonation of four ionogenic groups of a monomer is necessary to denature this monomer. If the whole tetramer is inactivated, protonation of a single ionogenic group in each monomer is necessary to denature the tetramer. For both cases, the pKa value estimated for these ionogenic groups (assuming that they have equal values) is in the range pKa < 4.0 [21]. Thus, a relation between the completely mature red fluorescent protein (D in Scheme (4)) and the protein form subjected to acidic denaturation (E in Scheme (14)) can be described by the scheme:
This scheme considers only the completely deprotonated form (D) and the completely protonated form (E) and reflects a sequential process of independent protonation/deprotonation of four ionogenic groups D with dissociation constant value of Ka. It should be noted that, unlike the green fluorescent proteins, protonation of the red fluorescent protein (yielding the E form in Scheme (15)) does not result in any detectable changes in its fluorescence, but triggers further transformations (Scheme (14)) leading to denaturation of the protein and disappearance of its fluorescence. According to Schemes (14) and (15), the relative initial velocity of the fluorescence decrease (nu) can be expressed as a function of proton concentration as follows:
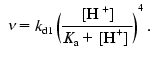 (16)
(16)The linear dependence nu([H+]) on a logarithmic scale (Fig. 3b) indicates that the studies were performed under conditions when [H+] << Ka and, consequently, pKa < 4. The experimental value A0 (Fig. 3a) reflects equilibrium between the fluorescent and nonfluorescent forms of DsRed. In the case of the equilibrium between the nonprotonated form D, its partially protonated forms Dx, the completely protonated form E, and the nonfluorescent form NF
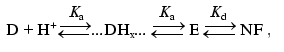 (17)
(17)the normalized fluorescence intensity (I) is evaluated as follows:
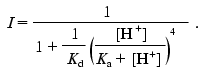 (18)
(18)As seen from Fig. 3a, Eq. (18) fits the experimental data reasonably well.
Thus, maturation of the red fluorescent protein and its renaturation after acidic denaturation are very similar in terms of kinetics, being sequential processes. Protonation of the tetramer of DsRed with four protons (probably, each monomer of the tetramer is protonated) results in partial and reversible denaturation of the protein with subsequent disappearance of its fluorescence. Further denaturation results in irreversible changes, the intensity of the changes depending on the time of the protein incubation in acid medium. The data available at the present time concerning the chemical nature of maturation of the red fluorescent protein [7] suggest that two of three kinetically significant reversible steps in the process of acidic denaturation of DsRed (Scheme (14)) can reflect unfolding of the protein chain and hydration of acylimine yielding carbinolamide. The irreversible step of E4 formation (Scheme (14)) can correspond to the process of carbinolamide hydrolysis with subsequent cleavage of the polypeptide chain of DsRed between Phe65 and oxidized Gln66, yielding two fragments [7]. Three steps of the model describing maturation of DsRed (Scheme (4)) can correspond to the processes of the protein folding, cyclization, and oxidation of the Gln66-Tyr67-Gly68 polypeptide fragment and oxidation of the Gln66 residue by oxygen [7].
In the present work an effect of the sharp dependence of DsRed fluorescence on pH in the range pH < 5.0 was investigated. It was shown that slight changes in pH resulted in complete disappearance (or appearance) of the fluorescence according to the “all or none” principle, which is extremely rarely realized in protein systems. Such a unique pH sensitivity allows using of this protein as an indicator in inner cell structures having low pH values (lysosomes, endosomes, Golgi apparatus), as well as in biological media of an organism during the expression of the protein.
V. V. Verkhusha thanks S. Tsukita and JST Corporation for the possibility of doing part of this work in the framework of the Tsukita Cell Axis project (Japan). The work was supported in part by the Russian Foundation for Basic Research (grant 00-04-48777).
REFERENCES
1.Tsien, R. Y. (1998) Annu. Rev. Biochem.,
67, 509-544.
2.Cody, C. W., Prasher, D. C., Westler, W. M.,
Prendergast, F. G., and Ward, W. W. (1993) Biochemistry,
32, 1212-1218.
3.Niwa, G., Inouye, S., Hirano, T., Matsuno, T.,
Kojima, S., Massayuki, K., Ohashi, M., and Tsuji, F. I. (1996) Proc.
Natl. Acad. Sci. USA, 93, 13617-13622.
4.Yang, F., Moss, L. G., and Phillips, G. N. (1996)
Nature Biotechnol., 14, 1246-1251.
5.Phillips, G. N., Jr. (1998) in Green Fluorescent
Protein: Properties, Applications, and Protocols
(Chalfie, M., and Kain, R., eds.) Wiley-Liss, New York, pp.
77-96.
6.Matz, M. V., Fradkov, A. F., Labas, Y. A.,
Savitsky, A. P., Zaraisky, A. G., Markelov, M. L., and Lukyanov, S. A.
(1999) Nat. Biotechnol., 17, 969-973.
7.Gross, L. A., Baird, G. S., Hoffman, R. C.,
Baldridge, K. K., and Tsien, R. Y. (2000) Proc. Natl Acad. Sci.
USA, 97, 11990-11995.
8.Wall, M. A., Socolich, M., and Ranganathan, R.
(2000) Nature Struct. Biol., 7, 1133-1138.
9.Reid, B. G., and Flynn, G. C. (1997)
Biochemistry, 36, 6789-6791.
10.Vrzheshch, P. V., Akovbian, N. A., Varfolomeyev,
S. D., and Verkhusha, V. V. (2000) FEBS Lett., 487,
203-208.
11.Baird, S. G., Zacharias, D. A., and Tsien, R. Y.
(2000) Proc. Natl. Acad. Sci. USA, 97, 11984-11989.
12.Bokman, S. H., and Ward, W. W. (1981) Biochem.
Biophys. Res. Commun., 101, 1372-1380.
13.Ward, W. W. (1981) in Bioluminescence and
Chemiluminescence: Basic Chemistry and Analytical Applications
(DeLuca, M., and McElroy, D. W., eds.) Academic Press, New York, pp.
235-242.
14.Llopis, J., McCaffery, J. M., Miyawaki, A.,
Farquhar, M. G., and Tsien, R. Y. (1998) Proc. Natl. Acad. Sci.
USA, 95, 6803-6808.
15.Elsliger, M., Wachter, R. M., Hanson, G. T.,
Kallio, K., and Remington, S. J. (1999) Biochemistry, 38,
5296-5301.
16.Ward, W. W. (1998) in Green Fluorescent
Protein: Properties, Applications, and Protocols
(Chalfie, M., and Kain, R., eds.) Wiley-Liss, New York, pp. 45-75.
17.Freifelder, D. (1976) Physical
Biochemistry, W. H. Freeman and Company, San Francisco.
18.Wiedenmann, J., Elke, C., Spindler, K.-D., and
Funke, W. (2000) Proc. Natl. Acad. Sci. USA, 97,
14091-14096.
19.Culter, M. W., and Ward, W. W. (1997) in
Bioluminescence and Chemiluminescence: Molecular Reporting with
Photons (Hastings, J. W., Kricka, L. J., and Stanley, P. E., eds.)
Wiley, New York, pp. 403-406.
20.Yarbrough, D., Wachter, R. M., Kallio, K., Matz,
M. V., and Remington, S. J. (2001) Proc. Natl. Acad. Sci. USA,
98, 462-467.
21.Varfolomeyev, S. D., and Gurevich, K. G. (1999)
in Biokinetics: Practical Course [in Russian], FAIR-PRESS,
Moscow.