Effect of Hinge Region State on Interaction of Human IgG3 with the Complement System
V. M. Tischenko1,2
1Institute of Biological Instrumentation, Russian Academy of Sciences, Pushchino, Moscow Region, 142292 Russia2Institute of Immunological Engineering, Lyubuchany, Moscow Region, 142380 Russia; fax: (095) 924-0493; E-mail: tischen@vega.protres.ru
Received June 4, 2001; Revision received September 3, 2001
Covalently cross-linked rod-like dimers of human myeloma IgG3 are more efficient in the inhibition of the complement system reaction than compact dimers. This correlates with an increase in the stability of CH2 domains of the immunoglobulin in the state with the rod-like hinge region.
KEY WORDS: third subclass immunoglobulins, complement system, dimers, hinge region, compact and rod-like forms
Abbreviations: SPDP) N-succinimidyl 3-(2-pyridyldithio)propionate; DNS) 1-dimethylaminonaphthalene-5-sulfonyl; DTT) dithiothreitol.
The dimensions of the human IgG3 hinge region are significantly greater
than the dimensions of a similar structure of any other immunoglobulin
[1-3]. Such a unique structure
of IgG3 is suggested to appear as a result of quadruplicating of the
exon encoding the hinge region of IgG1 [3, 4]. Most current models of human IgG3 suggest this
region to be rod-like [5-9]. An
alternative model was suggested upon studies on human myeloma IgG3 Kuc
[10]. Electron microscopy shows a compact
structure of the hinge region, and only this structure of human IgG3 in
solution under physiological conditions does not contradict
hydrodynamic and calorimetric data. By all these methods, the compact
hinge region transition into the rod-like state was shown under
nondenaturing changes in pH and temperature [11,
12]. Such a significant transformation resulting
in changes of various physical characteristics of IgG3 molecules can be
also important for their biological activities, which include the
so-called effector functions most of which are determined by the
Fc-subunit. In the present work, the effect of the hinge region
transition from one state into the other was studied based on the
interaction of myeloma IgG3 Kuc with the complement system.
MATERIALS AND METHODS
The earlier described [10, 13] human myeloma IgG3 Kuc was studied. The protein is an immunoglobulin of G class; its molecular weight is 170 kD. It interacts with an antiserum to the third subclass and fails to interact with the A protein of Staphylococcus aureus. Therefore, this protein was assigned to thethird class. The Fab- and Fc-fragments were prepared and purified as described in [5, 14]. The preparation and characterization of human myeloma IgG4 are described in [10]. The homogeneity of all proteins and of their fragments was monitored by SDS electrophoresis in 10% polyacrylamide gel in both the presence and absence of reducing agents by the method of Weber and Osborn [15]. The homoconjugation of IgG3 Kuc molecules was performed within 45 min at 25°C at the protein concentration of 30 mg/ml and at the protein/SPDP molar ratio of 1 : 1 [16].
The molecular weight was determined by two equilibrium centrifugation methods [17, 18] using MOM (Hungary) and Beckman (USA) centrifuges and by interference optics. Sedimentation experiments were performed with a Beckman centrifuge in the protein concentration range from 2 to 8 mg/ml (Schlieren optics) and of 0.2-0.25 mg/ml.
The interaction of the immunoglobulin with complement system proteins was determined by the method described in [19]. This method is based on determination of the residual activity of the complement system C1 factor [20]. The activity was associated with the C1 fraction that failed to bind to the immunoglobulin under study, and, consequently, could induce erythrocyte lysis [21].
DNS-IgG3 Kuc was prepared as described in [22, 23]. The protein fraction containing CH2 domains modified with a fluorescent label was prepared as described in [12]. The fluorescence was determined using an MPF-44A Perkin Elmer spectrofluorimeter (USA). The exciting light wavelength was 280 nm.
RESULTS AND DISCUSSION
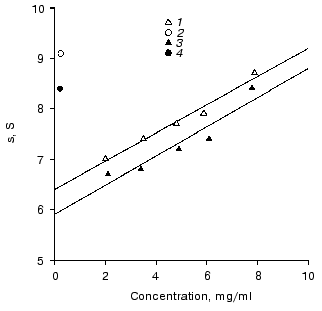
Description of two monomeric forms of IgG3 Kuc. The sedimentation coefficient so20 of the resulting IgG3 Kuc was 6.4 S (Fig. 1), this being significantly higher than the sedimentation coefficients of the polyclonal and of standard polyclonal human myeloma IgG3 [24, 25]. The molecular weight of this protein is 171 ± 8 and 176 ± 12 kD by data of high-speed and low-speed ultracentrifugation, respectively. This state will be further denoted as compact.
Heating in 50 mM phosphate buffer (pH 7.0) to 55°C resulted in partial precipitation of the protein. This procedure was performed in thin capillaries to decrease the fraction of the protein precipitated. This fraction was removed by sequential centrifugation at 50,000 rpm for 3 h with subsequent gel filtration on a column with AcA-34 equilibrated with 50 mM phosphate buffer. As a result, a protein was obtained with sedimentation coefficient so20 of 5.7 S that was somewhat lower than the standard value for the third subclass immunoglobulins. This state will be further denoted as elongated. The molecular weight of IgG3 obtained after the thermal treatment was 173 ± 8 and 169 ± 12 kD by data of high-speed and low-speed ultracentrifugation, respectively. In the absence of reducing agents, IgG3 Kuc gave a single band on SDS electrophoresis, and upon addition of DTT two standard bands appeared which corresponded to heavy and light chains. These findings suggested two conclusions. First, both protein forms should be monomers. Second, the transition of the compact protein into the elongated state should be a purely intramolecular process not associated, in particular, with releasing of various low-molecular-weight factors that could be present in immunoglobulins [26-28].Fig. 1. Determination of sedimentation coefficients so20 of IgG3 Kuc monomers (1, 3) and dimers (2, 4) with the compact and elongated hinge regions (light and dark symbols, respectively). Conditions: 20 mM phosphate buffer, 150 mM NaCl (pH 7.0).
Such a soft thermal treatment failed to markedly change the structure of the Fab- and Fc-subunits because their thermostability is significantly higher [12, 13]. This treatment did not also cause the loss of their functional activities (the interactions with the staphylococcal A protein, with the streptococcal G protein, and with the complement system proteins). This was concluded because the Fab- and Fc-fragments prepared by proteolysis from two forms of the protein were capable of these interactions. However, the hinge region on the moderate heating changed its compact state for the rod-like one in both the intact protein [11] and the isolated pFh-fragment [29]. These findings suggested that changes in the hydrodynamics parameters and in the IgG3 Kuc molecule form should be determined by the conformational changes only in the hinge region of the protein. Certainly, this is not to say that no other conformational changes occur, especially in the areas of the molecule immediately adjacent to the hinge region.
Preparation of IgG3 Kuc dimers using a bifunctional reagent. According to high-speed centrifugation in 10 mM phosphate buffer (pH 7.0) supplemented with 150 mM NaCl, IgG3 Kuc was a monomer. However, on increasing the pH to 7.5 and the ionic strength to 300 mM a tendency for dimer formation was observed. But the association constant was low and the equilibrium in the range of concentrations (0.06-1.5 mg/ml, 4·10-7-10-5 M) during centrifugation experiments was shifted to the monomers. The dimerization was provided by the Fab-subunits because this tendency was found for the F(ab)2-fragments but not for the Fc-fragments. When the heterobifunctional reagent SPDP was used at high concentrations of IgG3 Kuc, 6% of the immunoglobulins produced covalently cross-linked dimers. These dimers were purified by gel filtration on two columns in series. The first column contained Sephadex G-200 and the second column contained Ultragel AcA-22. These columns were equilibrated with 10 mM phosphate buffer (pH 7.0).
Papain or pepsin hydrolysis resulted in the cross-linking either of the F(ab)2- or of the Fab-fragments. This suggested that the dimerization should be a result of modification of groups located in the Fab-fragments. Therefore, this modification seemed to have no effect on the state of other regions of the molecule.
The IgG3 Kuc dimers produced from the elongated monomers were characterized by sedimentation coefficient of 8.4 S. A similar value was obtained for dimers of the recombinant IgG3 produced during the interaction with a bivalent hapten [30]. The compact dimer was characterized by sedimentation coefficient of 9.1 S (Fig. 2). These clear distinctions in characteristics of the two dimers and also the IgG3 transition in the dimer from the compact to the elongated state upon heating to 55°C unambiguously indicated that the hinge region state did not change during the dimerization, as was suggested.
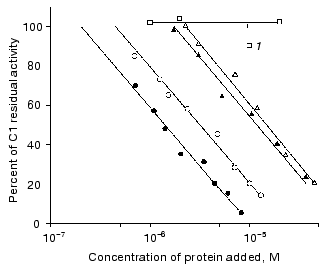
Interaction of immunoglobulin with complement. Figure 2 shows that the monomeric compact IgG3 Kuc at concentration of about 10-5 M binds 50% of the complement activity. The same immunoglobulin (also in the monomeric state) with the rod-like hinge region provided a similar effect at a somewhat lower concentration. The distinctions observed were not significant.Fig. 2. Effect of immunoglobulins on the activity of the complement system C1 factor: 1) IgG4; the other symbols are the same as in Fig. 1.
The picture was clearer when IgG3 Kuc dimers were used for the complement fixation. First, the dimers bound 50% of the complement activity at significantly lower concentrations of the protein (Fig. 2). Second, the activities of the two IgG3 Kuc forms were clearly different. The elongated form was more efficient in the interaction with the complement system proteins. Thermal aggregation of the two dimer types additionally increased their complement-binding ability, and the activities of both protein forms were similar. This was also observed in the case of aggregations produced during the heating of the compact and elongated forms when the latter form was initially monomeric.
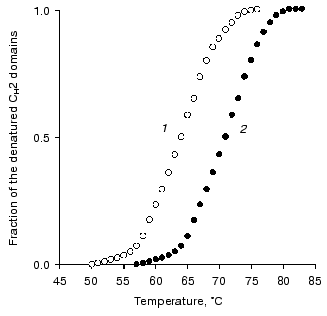
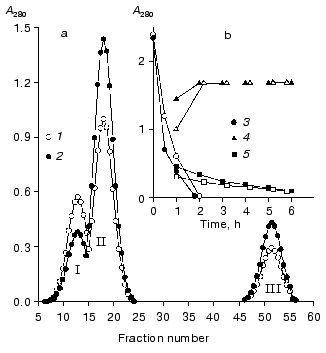
Studies on the state of CH2 domains of the two IgG3 Kuc forms. It is known that during the development of the complement system reactions by the classic pathway immunoglobulins first interact with the C1q factor [31]. This interaction is provided by CH2 domains [32] where amino acid residues forming the active site are located [33]. Therefore, the stability of these domains was studied inside IgG3 Kuc, in both the compact and elongated forms of the latter. Figure 3 presents the findings with the fluorescent label that has selectively modified the CH2 domains. The thermostability of these domains was markedly decreased in the compact IgG3 Kuc. This destabilization of the CH2 domains clearly correlated with a decrease in their resistance to pepsin (Fig. 4) even under conditions (pH 4.5) of destabilization of the compact structure of the hinge region itself [29]. This effect was suggested to be due to the decreased value of free energy of stabilization of the domain native structure [35]. If the hinge region stability was increased (as performed in [36, 37]) by the disulfide bond between its subdomains [29, 38], the effect on the CH2 domains would be increased more.
Fig. 3. Temperature-dependent changes in the fraction of the denatured CH2 domains determined using the fluorescent label for the compact (1) and elongated (2) forms of IgG3 Kuc.
At present, it is impossible to give an exhaustive structural and thermodynamic interpretation of the CH2 domain destabilization found. Electron microscopy clearly shows the existence of a large contact zone between the CH2 domains and the hinge region in the compact IgG3 Kuc [11]. On the other hand, according to X-ray diffraction data a pair of the CH2 and CH3 domains interacts with one another by surfaces of about 400 Å2 [39]. Because interdomain interactions can result in either domain stabilization and just the opposite effect [40], the hinge region is suggested to immediately destabilize the CH2 domains. Such a viewpoint is in agreement with the character of interdomain interactions in the Fc-fragment [12]. The possibility of weakening of the CH2 domain interaction with the CH3 domains must also not be ruled out as the main effect of the hinge region on the CH2 domains. To check these hypotheses, it is necessary to obtain such a state when the CH2 and CH3 domain interactions would be either absent (as in the Facb-fragment [41]), or be strongly weakened (as with the metastable state of the the Fc-subunit [42]).Fig. 4. Effect of the hinge region state on IgG3 Kuc hydrolysis rate. a) Results of gel filtration on the AcA-34 column of the 1-h hydrolysis products of the compact (1) and elongated (2) forms of the protein. b) Time-dependent changes in the ratio between the intact protein (3), F(ab)2 (4), and pFc fragments (5). The compact and elongated forms are indicated by light and dark symbols, respectively. The proteolysis was performed at 37°C in 100 mM acetate buffer (pH 4.5), at enzyme/substrate ratio of 1 : 100 [34].
This work was supported by the International Science Foundation (project RL-1300), the International Science and Technology Center (project No. 091-94), the Russian Foundation for Basic Research (project No. 99-04-48329), and the Russian Foundation for Basic Research - Science Towns near Moscow (project No. 01-04-97010).
REFERENCES
1.Michaelsen, T. E., Frangione, B., and Franklin, E.
C. (1977) J. Biol. Chem., 252, 883-889.
2.Michaelsen, T. E., and Natvig, J. B. (1972) FEBS
Lett., 28, 121-124.
3.Michaelsen, T. E., and Natvig, J. B. (1974) J.
Biol. Chem., 249, 2778-2785.
4.Huck, S., Fort, P., Crawford, D. H., Lefranc,
M.-P., and Lefranc, G. (1986) Nucleic Acids Res., 14,
1779-1789.
5.Soberg, B., Rosenquist, E., Michaelsen, T., Pap,
S., and Osterberg, R. (1980) Biochim. Biophys. Acta, 625,
10-17.
6.Gregory, L., Davis, K. G., Sheth, B., Boyd, J.,
Jefferis, R., Naves, C., and Burton, D. R. (1987) Mol. Immunol.,
24, 821-829.
7.Pumphrey, R. (1986) Immunol. Today,
7, 174-178.
8.Phillips, M. L., Tao, M.-H., Morrison, S. L., and
Schumaker, V. N. (1994) Mol. Immunol., 31, 1201-1210.
9.Roux, K. H., Strelets, L., and Michaelsen, T. E.
(1997) J. Immunol., 159, 3372-3382.
10.Burton, D. R., and Woof, J. M. (1992) Adv.
Immunol., 55, 1-84.
11.Ryazantsev, S., Tishchenko, V., Vasiliev, V.,
Zavyalov, V., and Abramov, V. (1990) Eur. J. Biochem.,
190, 393-399.
12.Tischenko, V. M., Abramov, V. P., and Zav'yalov,
V. P. (1998) Biochemistry, 37, 5576-5581.
13.Denesyuk, A. I., Tischenko, V. M., Abramov, V.
P., and Zav'yalov, V. P. (1983) Mol. Biol. (Moscow),
17, 1262-1271.
14.Porter, R. R. (1959) Biochem. J.,
73, 119-126.
15.Weber, K., and Osborn, M. (1969) J. Biol.
Chem., 244, 4406-4412.
16.Carlsson, J., Drevin, H., and Axen, R. (1978)
Biochem. J., 173, 723-730.
17.Van Holde, K. E., and Baldwin, R. L. (1958) J.
Phys. Chem., 62, 734-743.
18.Yphantis, D. A. (1964) Biochemistry,
3, 297-317.
19.Augener, W., Grey, H. M., Cooper, N. R., and
Muller-Eberhard, H. J. (1971) Immunochemistry, 8,
1011-1019.
20.Cabat, E. A., and Mayer, M. M. (1964)
Experimental Immunochemistry [Russian translation], Meditsina,
Moscow.
21.Yasmeen, D., Ellerson, J. R., Dorrington, K. J.,
and Painter, R. H. (1976) J. Immunol., 116,
518-526.
22.Steiner, R. F., and McAlister, A. J. (1957) J.
Polym. Sci., 24, 105-123.
23.Zagyansky, Yu. A., Nezlin, R. S., and Tumerman,
L. A. (1969) Immunochemistry, 6, 787-800.
24.Schmidt-Kessen, A., and Lustig, A. (1981)
Hoppe-Seyler's Z. Physiol. Chem., 362, 18-19.
25.Kilar, F., Simon, I., Lakatos, S., Vonderviszt,
F., Medgyesi, G. A., and Zavodszky, P. (1985) Eur. J. Biochem.,
147, 17-25.
26.Carson, S. D., and Bowman, B. H. (1981)
Biochim. Biophys. Acta, 667, 23-34.
27.Hautanen, A., Sarnesto, A., Keski-Oja, J., and
Seppala, I. (1982) Immunol. Lett., 5, 227-232.
28.Dudich, I. V., Dudich, E. I., and Timofeev, V. P.
(1983) Mol. Immunol., 20, 1273-1276.
29.Tishchenko, V. M. (2000) Biochemistry
(Moscow), 65, 1227-1230.
30.Phillips, M., Tao, M.-H., Morrison, S. L., and
Shumaker, V. N. (1994) Mol. Immunol., 31, 1201-1210.
31.Porter, R. R., and Reid, K. B. M. (1979) Adv.
Protein Chem., 33, 1-71.
32.Kehoe, J. M., and Fougereau, M. (1969)
Nature, 224, 1212-1213.
33.Dunkan, A. R., and Winter, C. (1988)
Nature, 332, 738-740.
34.Turner, M. W., Bennich, H. H., and Natvig, J. B.
(1970) Clin. Exp. Immunol., 7, 603-625.
35.Wang, L., and Kallenbach, N. R. (1998) Protein
Sci., 7, 2460-2464.
36.Tishchenko, V., Lund, J., Goodall, M., and
Jeffeis, R. (1996) in The First Int. Conf. on the Applications of
Biocalorimetry, Oxford, UK, Plenum Press, p. R-04.
37.Tischenko, V. M. (1999) in New Trend in
Calorimetry and Its Application, Moscow, p. IV-54.
38.Tischenko, V. M. (2000) Mol. Biol.
(Moscow), 34, 110-115.
39.Deisenhofer, J. (1981) Biochemistry,
20, 2361-2370.
40.Privalov, P. L., and Potekhin, S. A. (1986)
Meth. Enzymol., 131, 4-51.
41.Colomb, M., and Porter, R. R. (1975) Biochem.
J., 145, 177-183.
42.Tischenko, V. M. (2000) J. Therm. Anal.
Cal., 62, 63-68.