X-Ray Analysis of Ribosomes: the Static of the Dynamic
A. M. Kopylov
School of Chemistry and Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-3181; E-mail: kopylov@rnp-group.genebee.msu.su
Received September 17, 2001; Revision received October 15, 2001
This review considers a brief history, comments, and consequences of recent remarkable achievements: X-ray analysis on the level of atomic resolution of structures of bacterial ribosomes, their subunits, and functional complexes.
KEY WORDS: bacteria, ribosome, structure, function, X-ray analysis, Thermus thermophilus, Haloarcula marismortui
The end of the millennium was marked by an outstanding event, the X-ray analysis (XRA) of a fine structure of bacterial ribosomes on the level of atomic resolution. No wonder that the reports about it were placed on the covers of Nature [1, 2]and Science [3] and were accompanied by a flurry of comments [4-6].
The traditional Conference on Ribosomes, near the famous Helsingor castle of Hamlet (Denmark, June 13-17, 1999), was the place where the first high-resolution XRA data on extremophilic ribosomes were reported [7-9]. Thus, June 1999 can be conventionally called the date of promulgation of these remarkable results and also the date of the beginning of the race of the mightiest project in the field of XRA of biological macromolecules and their complexes [Footnote: Many researchers suggest that the ribosome should be the most complicated biological object that in principle can be crystallized (but it is not the largest object because virus crystals now exist which are larger than ribosome crystals)].
Then the events developed as follows. The article of Venki Ramakrishnan and Brian Wimberly with colleagues describing the structure of a small ribosomal subunit from the bacterium Thermus thermophilus at the resolution of 5.5 A was submitted to Nature on June 14, 1999 and was published in this journal on August 26 of the same year [1]. The article by Thomas Steitz and Peter Moore with colleagues about the structure of a large subunit of bacterial ribosomes from the extreme halophile archaeon Haloarcula marismortui at the resolution of 5 A was received two weeks later, July 2, and was published in the same issue of the journal [2]. The article of Harry Noller with colleagues on the structure of bacterial 70S ribosomes from T. thermophilus and of ribosomal functional complexes at the resolution of 7.8 A was received two months later, August 24, and was published September 24, 1999 in Science [3], i.e., only a month after the publication about the structure of individual subunits.
Exactly a year later, in summer 2000, results of the next stage of the race were summarized. At the annual RNA-2000 Conference of the RNA Society (University of Wisconsin, Madison, USA, May 30-June 4, 2000) there were presented reports of Ada Yonath with colleagues about the structure of the small subunit at the resolution of 3.6 A [10] and of Steitz and Moore with colleagues about the structure of the large subunit at the resolution of 2.7 A [11]. The teams of Ramakrishnan and of Noller presented no data. Immediately afterwards, at the most prestigious XVIII International Congress on Biochemistry and Molecular Biology (July 16-20, 2000, Birmingham, UK) everybody made reports [12-14] except Noller.
Let us regard again the publication chronicle. Steitz and Moore with colleagues shot up two weeks ahead with their data on the structure of the large ribosomal subunit at the resolution of 2.4 A (the article was received July 24, 2000, and was published in the Science issue of August 11, 2000 [15]). In addition, Steitz and colleagues in the concomitant article published a version of the catalytic mechanism for the peptide bond generation, i.e., the first functional interpretation of findings based on the structure of the large ribosomal subunit [16]. Finally, an article by Yonath, who is a pioneer in XRA of ribosomes, appeared which presents data on the structure of the small ribosomal particle at the resolution of 3.3 A. The article was received June 23 and published in the Cell issue ofSeptember 1, 2000 [17]. Ramakrishnan and colleages were a little late: their data at the resolution of 3 A appeared three weeks later but the resolution was better than that obtained by Yonath. Their article was sent July 14 and published in the Nature of September 21, 2000 [18]. And in the concomitant article Ramakrishnan and colleagues presented a version of the action mechanism of some translational antibiotics, i.e., the first functional interpretation of the data based on the structure of the small ribosomal subunit [19]. Noller's team kept silence with their data on the 70S ribosome.
And, finally, the data were analyzed in detail at the 66th Quantitative Biology Symposium (May 31-June 5, 2001, Cold Spring Harbor, USA), which was entirely dedicated to ribosomes.
But who could imagine that the time would be counted in days when the race was for such an unbelievably complicated structure as the ribosome? [Footnote: There are three components to this discovery: the historical aspect [5], current working conclusions, and predictions. Even in this short publication it is necessary to remark on an outstanding contribution of two leading ribosomologists who, ahead of time, were bold enough to set the task long before the elaboration of approaches for its solution: these are A. S. Spirin in Russia and H. G. Wittmann in Germany.]
The translation of genetic information encoded by the mRNA template into the protein polypeptide chain is a unique process that binds the genotype with the phenotype. The translation is executed by the ribosome, the most ancient ribonucleoprotein particle, and its main structure and general functional mechanism are conservative for all forms of life. The ribosome is the most important particle of each cell because it provides both a workshop and tools for synthesis of all proteins in the cell. [Footnote: A. S. Spirin suggests that the ribosome and the translation itself should be the basis for all forms of life and also the starting point for the origin of life.]
Amino acids produced as a result of catabolism of various protein products (and also of de novo synthesis) are bound to tRNA and again introduced into the protein building on the ribosome. And although the ribosome-catalyzed chemical reaction itself is simply the binding of amino acids by an amide (peptide) bond, the ribosome is responsible for the most difficult problem of choosing the tRNA that carries the appropriate amino acid to be added to the growing polypeptide chain in accordance with the codon text of mRNA.
Among all ribosomes, bacterial ribosomes are relatively small and are the best studied. Their molecular weight is ~2.5 MD, they can dissociate to small and large subunits (30 and 50S, respectively), which are denoted by their apparent sedimentation coefficient in Svedberg units (S). The 30S subunit consists of 16S rRNA of about 1500 nucleotides in length and of approximately 20 various proteins of medium size; the 50S subunit contains 23S rRNA of about 2900 nucleotides in length, 5S rRNA of 120 nucleotides in length, and more than 30 various proteins of medium size. Thus, the whole 70S ribosome contains about 4500 nucleotides and more than 50 various proteins. Such a highly complicated structure of the ribosome reasonably corresponds to the intricacy of its biological functions and to the size of its “substrates” which it interacts with, such as mRNA and tRNA.
Protein synthesis starts on the 30S subunit by the mRNA binding to it, and in many cases it is accompanied by an unfolding of the macromolecular structure of mRNA. The molecule of fMet-tRNAfMet carrying on the 3´- terminus the first amino acid for a new protein binds to the binary complex of the 30S subunit with mRNA. This occurs concurrently with a specific interaction of heterocyclic bases of the tRNA anticodon with the codon triplet of mRNA. And this process essentially determines what amino acid will be incorporated into the protein chain.
Afterwards, the 50S subunit binds to the triple complex of the 30S subunit, and the aminoacylated 3´-terminus of tRNA enters the region of the active center of the large subunit. Another aminoacylated tRNA binds to the neighboring codon on the small subunit, and its amino acid also enters the region of the active center of the large subunit that was succeeded by the generation of a peptide bond connecting two amino acids. Just this was the reason for Moore to call the ribosome a polypeptide polymerase enzyme [20]. On completion of the polypeptide synthesis, a special termination factor recognizes the stop codon and promotes the newly synthesized protein releasing through a channel in the 50S subunit. Then the two ribosomal subunits dissociate.
Thus, the ribosome substrates are aminoacylated tRNA molecules for which the ribosome has three different binding sites: A- (aminoacyl-), P- (peptidyl-), and E- (exit) sites; before the dissociation from the ribosome, the deacylated tRNA binds to the last of the sites. Each site is distributed between two subunits that, according to Noller, can result in the existence of at least six hybrid sites of the tRNA interaction with the ribosome. Naturally, tRNA should be located on the interface of the subunits.
The elongation cycle of translation is determined by three fundamental processes on the ribosome: the first is the selection of aminoacylated tRNA, the second is the generation of a peptide bond, and the third is the translocation of tRNA from one site into the next one. Each stage of the translation requires an involvement of protein factors that bind to tRNA, mRNA, and ribosomal subunits and, thus, are responsible for the rate, faithfulness, and movement of tRNA and mRNA relatively to the ribosome. The in vivo stages of the tRNA selection and translocation on the ribosome need an involvement of elongation factors EF-Tu and EF-G, respectively, and also the degradation of GTP to GDP. Under certain in vitro conditions, the two stages can be executed by the ribosome itself without the involvement of the factors. Thus, all three main steps in the elongation cycle of translation should be based on properties of the ribosome itself and, most likely, on properties of its rRNA. [Footnote: The structure of ribosomes and the translation are described in more detail in a book by A. S. Spirin [21] and in his recent reviews [22, 23].]
Molecular mechanisms responsible for these functions of the ribosome in many respects remain a mystery. And although the knowledge of the ribosome fine structure is not expected to immediately explain all specific features of the translation, it is clear that the deeper concept on the ribosome work will be based just on this knowledge. [Footnote: A comment of Anders Liljas, the known specialist in the structure of ribosomes, published in Science [6] was entitled as: “Function is a structure”. This popular fallacy established during the first stages of development of molecular biology has been for long disproved by enzymologists. The understanding of the operating mechanism is contributed to not only by the knowledge of structures of the interacting molecules but also by studies on thermodynamics of their interaction. And although studies on the structure are still prevailing now, the advantage of the most harmonious combination of both approaches is brilliantly demonstrated by the book by Alan Fersht, the second edition of which appeared quite recently [24].]
Studies on individual components of the ribosome have required immense efforts that needed many new approaches to be developed and used. The most demonstrative are the results of the so-called “phylogenetic approach” in the prediction of double-stranded regions of elongated rRNA molecules, which have been brilliantly confirmed by XRA of the ribosome. Moreover, the comparison of nucleotide sequences of rRNA from various cells for the first time resulted in the construction of an adequate evolutionary tree and, as a consequence, in the discovery in 1977 of the third kingdom of living matter, the Archaea.
According to the opinion of a leading ribosomologist, Roger Garrett [4], the progress in the comprehension of the ribosome structure and function was for a long time restrained by two conceptual circumstances. The first circumstance could be overcome, whereas the other was insuperable. “Avoidable was the tradition of viewing the ribosome as two pieces of rock containing two (later three) shallow cavities where transfer RNAs carrying amino acids bind and interact. This produced the erroneous view that the tRNAs, the messenger RNA (mRNA) template, and protein factors are located at one cavity or another, and that the ribosome itself is more or less irrelevant to the process of protein production. This view negated the concept of ribosomal movements. [Footnote: Justice demands that it be noted that A. S. Spirin even at the dawn of ribosomology had formulated the concept on the ribosome dynamic functioning in protein biosynthesis [25].]
But we now know, from studies of cells containing mutated ribosomes with altered properties, and from investigations into how antibiotics block different steps of protein synthesis, that the ribosome is a dynamic machine. This evidence is strongly reinforced by physical measurements of the differences in shape observed for ribosomes in different functional states.
An unavoidable block to progress in solving the ribosome's structure was the fact that, at a very early stage in evolution, the ribosome became highly complex (fossilized deposits of cyanobacteria and other bacteria and archaea containing ribosomes date back up to three billion years). Efficient proofreading (to ensure that the mRNA template was followed correctly) and a fine balance between the speed and accuracy of peptide elongation developed. During this stage, the ribosome also defined the size of evolving genes, because the capacity to translate larger regions of mRNA more accurately allowed larger functional proteins to be produced. So, there is no simpler ribosome structure than the bacterial one (except for some highly degenerate ribosomes within the cell's respiration center, the mitochondria)”. [Footnote: A rather common error should be avoided when the evolution of ribosomes (and also of cells as they are) from bacteria to eukaryote is under consideration. The standpoint also supported by the author of the present publication considers bacteria to be a similarly advanced branch of evolution as eukaryote and they both independently descend from a progenote. During the evolution, bacteria have refined the structure of their ribosomes to provide the fastest biosynthesis of proteins that could be also under an effective and dynamic regulation.]
Studies by electron microscopy on the ribosome morphology was one of the first steps in the establishing of its structure. Later, immunoelectron microscopy was developed, and this allowed epitopes of ribosomal proteins and individual regions of rRNA to be mapped. Using neutron diffraction, a three-dimensional distribution of weight centers of ribosomal proteins was mapped. Significant progress was recently achieved by cryo-electron microscopy in the elucidation of structures of ribosomal subunits, of the ribosome itself, and of its functional complexes. Based on abundant biochemical data for most proteins and for rRNA and on the results of cryo-electron microscopy, every subunit was modeled at the resolution of ~10-20 A. [Footnote: Computerized models based on data of cryo-electron microscopy at the resolution of 15-20 A had, in fact, a conventional resolution about that at the atomic level. Within several years, these models were not only extremely useful, but in many respects adequate, e.g., for 5S rRNA, the mRNA-binding center. Moreover, cryo-electron microscopy seems promising as an approach for detection of significant conformational changes associated with the ribosome functioning [26], i.e., it is expected to play the decisive role in the transition from studies on statics of ribosomes to studies on their dynamics.]
However, up to the last two years, data of X-ray diffraction were insufficiently good for exact modeling of ribosomes, and their large dimensions and asymmetry in no case simplified this problem.
The atomic-level resolution became available due to elaboration of high-capacity sources of synchrotron radiation, to wide-area detectors, to an immense improvement in computerized calculations, and to development of cryo-crystallography. Pioneer efforts in the crystallization of ribosomes from a T. thermophilus bacterium were made in the 1980s by A. S. Spirin with colleagues in Russia [27] and by H. G. Wittmann and A. Yonath with colleagues in Germany [28]. The slow development of events in this field instilled a certain hope that some day a result would be obtained, but nobody except the authors of these works would risk forecasting such an impetuous development of XRA of ribosomes.
According to the reasonable remark of Masayasu Nomura [29]: “There can be two directions in ribosome research in this respect. One is to simplify the complexity of ribosomes and to identify key elements responsible for carrying out basic biochemical reactions involved in ribosome function. The other direction is to understand the significance of complexity of ribosome structure”. However, the real studies on ribosomes were developing according to quite another logic: individual components were fully characterized. Structures of ribosomal proteins and rRNA fragments were determined by XRA and NMR spectroscopy. And, finally, the publications cited in the beginning appeared. Many experts are agreed on the opinion that these achievements are the final chapter in the static structural studies on ribosomes that have been performed for more than four decades.
And what new the crystallography data have told us about the ribosome? First of all, they provided a strong basis for the structural interpretation of a huge set of biochemical data accumulated on the ribosome structure and functions.
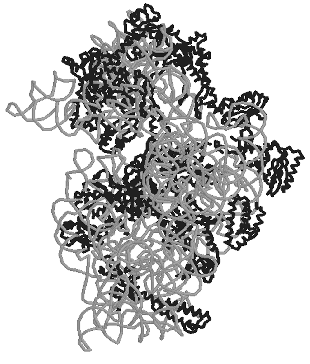
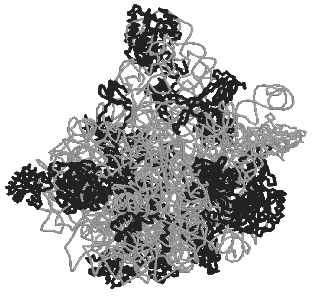
The small ribosomal subunit. Structures of all ordered regions of 16S rRNA and of 20 proteins that constitute 99% of all 16S rRNA and 95% of all proteins were presented in the work of Ramakrishnan with colleagues [18] about the structure of the 30S subunit at the resolution of 3 A: only locations of the terminal regions of the molecules were not determined. The general shape of the 30S subunit was found to correspond to its image based on data of electron microscopy and, later, of cryo-electron microscopy. Moreover, its shape was found to be more like the subunit structure paired with the 50S subunit inside 70S ribosomes than its own structure in the free state. The shape of the 30S subunit is determined by the structure of 16S rRNA, and neither essential morphological element of the structure is built only of ribosomal proteins. In a traditional frontal projection as viewed from the 50S subunit side, the small subunit appearance is described by the following morphological elements (Fig. 1): a “head” with a “beak” (directed leftward), a “body” with a “shoulder” (at the top left) and a “spur” (on the lower left), and also with a “platform” (at the top right).
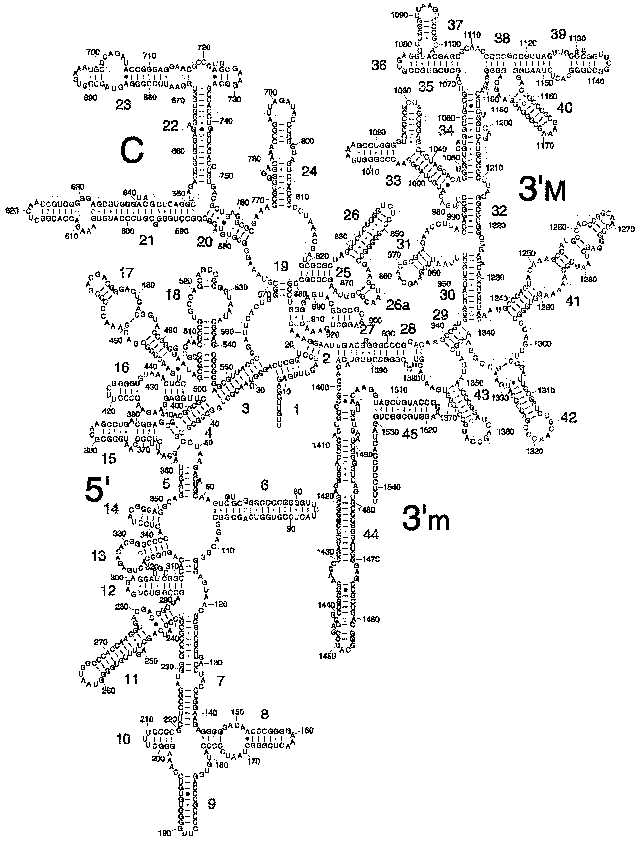
The secondary structure of 16S rRNA (Fig. 2) is formed by more than 50 regular double-stranded regions (which are grouped and denoted as H1-H45) connected to one another by irregular single-stranded loops. The crystal structure of most of these formally single-stranded loops is really more like slightly distorted extensions of the neighboring regular helices. Thus, most of the 16S rRNA structure elements can be described as helical or nearly helical. The helical elements interact with each other by butt-ends (a coaxial stacking) and also horizontally (Fig. 3c), e.g., the “side-by-side” packing of the helices with small grooves. The coaxial stacking occurs rather frequently: most of 16S rRNA helices are organized into 13 similar groups, and only 23 helices lack such stacking. As a result, there are 36 helical elements. Just the packing of these elements mainly determines the general packing of each of four 16S rRNA domains which, in turn, form large morphological elements of the subunit, as it was supposed in the early models by Noller with colleagues [3] and by Brimacombe [30]. By distant interactions, short single-stranded regions stabilize the dense packing of the helices. Binding to several helices, proteins are also involved in the stabilization of the tertiary structure of rRNA.Fig. 1. The spatial structure of the 30S small subunit of ribosomes from the hyperthermophilic bacterium T. thermophilus based on data of XRA (PDB 1FJF) [18]. The traditional frontal projection as viewed from the 50S subunit side is described by the morphological elements as follows: a “head” with a “beak” (directed leftward), a “body” with a “shoulder” (at the top left) and a “spur” (on the lower left), and also with a “platform” (at the top right). Proteins and 16S rRNA are shown with the dark and light bands, respectively.
Fig. 2. Model of secondary structure of 16S rRNA from E. coli with a standard indexing of the double-stranded regions of H1-H45 [35].
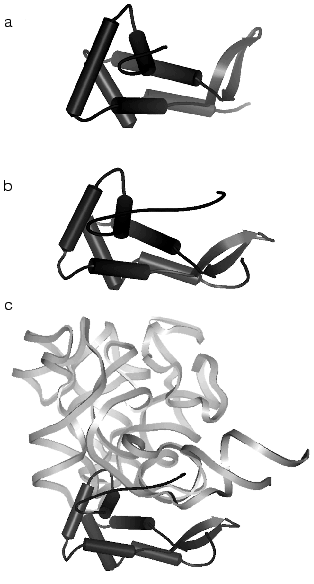
The 5´-domain of 16S rRNA forms the “body” and consists of 19 helices. The terminal “spur” of the domain is formed by the H6 helix, which varies in size in ribosomes from various cells. The central domain forms the “platform” and consists of nine helical elements. The minor 3´-domain is a single long helix which forms a part of the “body” in the region of the subunit contact.Fig. 3. Spatial structure of the ribosomal S7 protein isolated from T. thermophilus by data of XRA (PDB 1RSS) [44] (a), of the ribosomal S7 protein obtained by subtraction from the structure of the 30S subunit from T. thermophilus (PDB 1FJF) [18] (b), and of the complex of a fragment of the 3´-terminal domain D3LH (H28, H29, H41-H43) of 16S rRNA [45, 46] with the S7 protein obtained by subtraction from the structure of the 30S subunit from T. thermophilus (PDB 1FJF) [18] (c) (the complex orientation does not correspond to the 30S subunit projection in Fig. 1). The rRNA fragment and the S7 protein are shown with light bands and with dark color, respectively.
The major 3´-domain forms the subunit “head” and consists of five helical elements. The “head” appearance of the “left hand” type narrows leftwards to the “beak” formed by rRNA. Unlike the 5´- and the central domains, most helices of this domain fail to form the coaxial stacking, except H35-H36-H38-H39. It is interesting that the functionally important helices H31 and H34 have a very irregular structure and are packed rather loosely compared to other helices of rRNA.
All four domains of 16S rRNA are joined especially tightly in the central region of the subunit “neck”, which is the most functionally important.
The structures of all 20 proteins of the small subunit of T. thermophilus ribosomes are well detectable at the resolution of 3 A. As a rule, the protein structure has one or several globular domains. Different proteins can have the same folding type, i.e., S12 and S17 present a beta-barrel, whereas S3, S6, S10, and S11 present an alpha-helix/beta-layer. In addition to globular domains, nearly all proteins have strongly elongated structural elements, which are often called “hands” or “tails”. Some of these elements are helical (the S2, the C-terminal of S13), or long beta-hairpins, loopings (S10, S17), or elongated N- or C-terminal “tails” (Figs. 3b and 4) (S4, S7, S9, S11, S12, S13, S19). These elongated elements form close contacts with rRNA (Fig. 3c) and usually are not revealed at the analysis of isolated proteins (compare Figs. 3a and 3b) in the absence of rRNA when their structure is unordered.
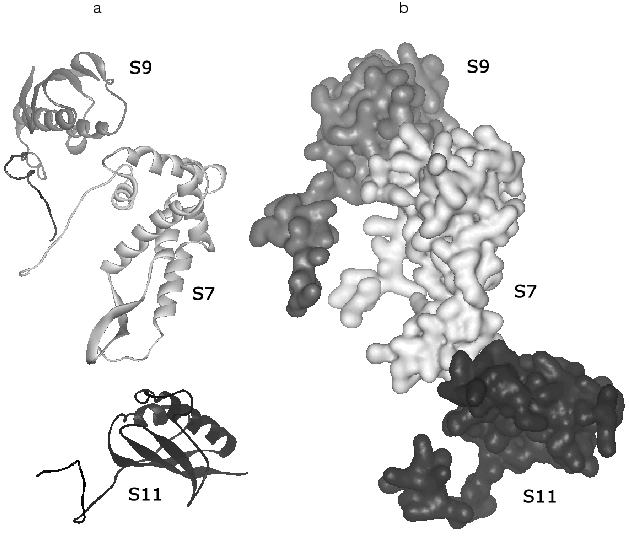
Consider briefly the interaction of ribosomal proteins with 16S rRNA in the small subunit. First of all, note a very interesting feature that is likely to be very essential for elucidation of mechanisms of the ribosome biogenesis: the folding elements common for different proteins interact differently with rRNA. Thus, in the protein group of S3, S10, S6, and S11 the beta-layer of the S11 protein is located flatly relatively to the small groove of RNA, the beta-layer edge of S6 interacts with RNA, whereas the alpha/beta-domain of S3 has no contact with RNA. “Tails” of ribosomal proteins can penetrate far into the environment of 16S rRNA and provide the contact of a single protein with several sites of rRNA that seems important for stabilization of its tertiary structure. The elongated structure of these “tails” fits this task especially well. Moreover, basic amino acids in their structure can neutralize the effect of the charge of the rRNA skeleton. [Footnote: It is suggested that the anchor role of these elements of protein structure in the RNA-protein interactions should be more informative.]Fig. 4. Spatial structure of a protein trimer S9-S7-S11 that is responsible for the “head” contact with the “platform” obtained by subtraction from the structure of the 30S subunit from T. thermophilus (PDB 1FJF) [18]. The orientation of the 30S subunit is the same as in Fig. 1. a) The structure of the main polypeptide chain; b) a complete volume model. In this projection the “tails” of all three proteins are well shown: the C-terminals of S9 and S11 and the N-terminal of S7.
A number of proteins bind to the joining point of several helices of 16S rRNA. S4 binds to the joining point of five helices (H3, H4, H16, H17, and H18) in the 5´-domain and S7 binds to the joining point of four helices (H29, H30, H41, and H42) (Fig. 3c) in the main 3´-domain. It seems not accidental that these proteins are very important for the early assembly stages of the “head” and “body”, respectively, of the 30S subunit. [Footnote: The binding site of the S7 protein is also created by the neighboring fifth helix H43.]
Similarly, the S8, S15, and S17 proteins bind to the joining point of three helices (H20, H21, and H22) that determines the assembly of the central domain of the subunit. Such an interaction can be a paradigm: the binding of proteins just to the joining point of the helices determines the initiation of folding of a correct tertiary structure of 16S rRNA.
A number of proteins have contacts with the sites of 16S rRNA from different domains and thus help in the organization of the general structure of the 30S subunit. Thus, S17 contacts with H7 and H11 of the 5´-domain and with H21 of the central domain, whereas S20 promotes the contact between H44 of the minor 3´-domain and the 5´-domain in the base of the “body”. The S7 protein of the major 3´-domain contacts with the H23 hairpin of the central domain. Rather many contacts between the subunit “head” and “body” are mediated by such proteins as S2 and S5.
In general, the structure of the 16S rRNA-protein contacts is in good agreement with biochemical findings of Nomura and colleagues and of Noller and colleagues (references in [3]) for the assembly of the subunit, although some discrepancies can also occur. Thus, S20 binding in the lower part of the 30S subunit is unlikely to affect S13 binding in the “head”. A good agreement is also observed for other biochemical data: the hydroxyl-radical foot-printing and the UV-induced cross-linking.
In addition to close contacts with rRNA, XRA reveals a number of protein-protein contacts. The S3-S10-S14 proteins produce a dense hydrophobic cluster that is wedged into a V-shaped “gap” between two 16S rRNA subdomains in the “head” and thus promotes the stabilization of the whole domain structure. By contrast, other protein clusters (S4-S5-S8) are mainly produced at the cost of electrostatic interactions and hydrogen bonds. A protein trimer S9-S7-S11 (Fig. 4) is responsible for the “head” contact with the “shoulder” and thus can be involved in their positioning. It is interesting that contacts of the S4-S5 dimer can be rearranged during functioning of the 30S subunit.
To a globular S12 domain on the interphase side of the 30S subunit, a long element is attached which is “snaking” through 16S rRNA of the “body” and then coils into a short helix contacting S8 and S17 on the back side of the subunit. Similar, although less pronounced, interactions are observed between the C-terminal “tail” of S9 and the S14 and S10 proteins on the backside of the “head”. Such interactions of proteins located on the opposite sides of a subunit are likely to contribute to the general structure of 16S rRNA. The proteins and 16S rRNA are distributed in the 30S subunit asymmetrically, as supposed earlier based on the results of neutron scattering for the S13, S14, S16, and S19 proteins, and only the S20 location is different. The proteins are mainly concentrated on the top, sides, and “back”. Neither protein is incorporated inside any 16S rRNA domain, only the S20 protein is located between the 5´- and 3´-minor domains.
The region of contact with the 50S subunit is mainly free of proteins except S12, which is located near the mRNA-binding site on the top of the long H44 hairpin that is oriented from the top downwards in the region of the subunit contact [31]. Other proteins are located along the periphery of the region of the subunit contact and, probably, also have contacts with the 50S subunit.
The first conclusions concerning the possible operating mechanisms of the small subunit were presented in the concomitant article about the structural bases of the action mechanism of some translational antibiotics [19]; this subject deserves separate comment.
The large ribosomal subunit. The first description in detail of the large ribosomal subunit structure has been recently published by Richard Brimacombe [30], and this allows us to give only short comments in the present article. The data of Steitz and Moore with colleagues [15] on the structure of the large ribosomal subunit at atomic resolution became for the first time an experimental basis for the hypothesis that just 23S rRNA can catalyze the key peptidyltransferase reaction executed by the ribosome [16]. Thus, 23S rRNA is not a simple scaffold for the structural organization of proteins that would provide a traditional protein catalysis. The proteins should be considered instead as structural components that help in the organization of a key ribosome catalytic center of rRNA, and this is just the idea that for a long time has been promoted by Harry Noller, Carl Woese, and others.
At the resolution of 2.4 A nearly the whole chain of 23S rRNA and also of 5S rRNA (altogether 3043 nucleotides) fits into the electron density map of the large ribosomal subunit (Fig. 5). The secondary structure of 23S rRNA proposed earlier based on theoretical comparative phylogenetic analysis [32] is consistent with the XRA data. Moreover, many tertiary interactions have been found in the structure of rRNA. As a result, 23S rRNA presents a whole giant bulk of tightly packed helices rather than six separate discrete domains joined by flexible linkers, as could be assumed from the scheme of its secondary structure.
But where are all the ribosomal proteins located and what are their functions? Globular domains of 26 proteins are mainly found on the external surface of the subunit. Twelve of these proteins have unusual “snake-like” elements that lack a tertiary structure (and sometimes they even lack a regular secondary structure), and one protein is fully unfold. These specific features of the protein structure determine their interaction with 23S rRNA. The comparison of such structures with data available for protein cofactors that promote functioning of other ribozymes induces the following most likely conclusion: the ribosomal proteins form, maintain, and stabilize the flexible structure of 23S rRNA and thus provide its specific and functionally active conformation.Fig. 5. Appearance of the 50S large subunit of bacterial ribosomes of the extreme halophile archaeon H. marismortui (PDB 1FFK) [15]. The conventional frontal projection as viewed from the side of the 30S subunit is presented. Proteins are shown with dark bands, 23S and 5S rRNAs are shown with light bands.
The least number of proteins was found in the region of the 50S subunit active center that is responsible for the peptidyltransferase reaction. This reaction is suggested to occur not only due to the entropy factor due to a correct orientation of 3´-terminus of two tRNA molecules. It is reasonably suggested to be associated with a typical acid-base catalysis.
Certainly, this catalysis could be also provided by the active site of any ribosomal protein. Therefore, a question arises inevitably: why does Nature use the RNA-provided catalysis for protein synthesis [33]? A possible answer can be sought among concepts on the evolution. If initially an “RNA World” really existed [34] where RNA molecules not only determine the genetic information but also provided various catalytic functions, it is reasonable to suggest that protein synthesis should be initially catalyzed by the RNA molecule. Afterwards, this ribosomal ribozyme could acquire an additional protein component. And because the ribosomal RNA continued its functioning sufficiently well, it was not later replaced by a protein catalyst.
In addition to the evolutionary explanation, a rather convincing chemical explanation can be proposed. Substrates of the ribosome are RNA molecules, aminoacylated tRNA and mRNA, and just the RNA molecule is well adapted to be specifically recognized by other RNAs via generation of complementary pairs and other interactions. Moreover, the RNA molecule is also very well adapted for large scale conformational changes required for the protein synthesis.
The 50S subunit includes two main regions which are responsible for the work of at least one protein factor, the elongation factor EF-G, which promotes the movement of the tRNA-mRNA complex along the ribosome after the next peptide bond has been generated. One of these regions is represented by a small segment of 23S rRNA and another region is the rRNA-protein complex. Both these regions were earlier isolated and analyzed using NMR and XRA. Conformational transitions in these regions are inhibited by antibiotics and toxins resulting in a decrease in the rate of cell growth or even in cell death. Thus, the function of the rRNA-protein complex is inhibited by the antibiotics thiostrepton and micrococin. On the other hand, the modification of the 23S rRNA segment by some toxins including ricin and alpha-sarcin also results in the inactivation of ribosomes (references in [4]).
The 70S ribosome. The work of H. Noller and colleagues was another essential step in the structural analysis of ribosomes and presented an analysis of the 70S ribosome operating, because the authors succeeded in crystallization of some functional complexes of the ribosome [3]. The structure of functional complexes of the 70S ribosome [Footnote: The ribosomes were purified and crystallized as described in earlier works by A. S. Spirin and colleagues (references in [3]) with some modifications. A fragment of mRNA of the gene 32 of T4 phage had 36 nucleotides in length and had changes in the Shine-Dalgarno sequence and in the first three codons for Met, Phe, and Lys. ASL of tRNAPhe, a fragment of tRNA of 19 nucleotides in length, had six pairs of the anticodon stem-and-loop in the double-stranded region. The first complex was prepared with mRNA and ASLPhe in the P-site of the ribosome, the second complex was prepared on addition of deacylated tRNALys to the A-site of the ribosome. The third complex was prepared with mRNA and a full-sized tRNAfMet at the P-site. The complex with ASL at the P-site and tRNALys in the A-site of the ribosome was prepared by soaking the crystals of the triple complex with ASL in tRNALys solution.] which contained a fragment of mRNA and tRNA (and/or its analog) was established for ribosomes from T. thermophilus at the resolution of 7.8 A. [Footnote: After this manuscript had been written, a publication by Noller with colleagues appeared on the structure of the ribosome and its functional complexes at the resolution of 5.5 A [35].]
Even such a low resolution allowed us to recognize elements of the protein structure and the packing of rRNA helices, including those on the interphase of the subunits. A complicated network of functional interactions is concentrated around the long H44 hairpin 16S rRNA on the subunit interphase, including the switching hairpin and the mRNA-binding site of 16S rRNA and also RNA bridges with the 50S subunit [31]. Moreover, rather many details in the tRNA interactions with the ribosome became recognizable. A number of contacts between the 30S subunit and the anticodon hairpin of tRNA in the P-site was analyzed, and the anticodon region of tRNA in the A-site was shown better. The tRNA interactions with the ribosome in all three sites (A, P, and E) are comprehensively visible, assuring new data for the description of the translation mechanism in detail.
And, finally, we present some comments from the field of predictions: R. Garrett writes [4]: “When I first saw these X-ray structures I thought that this is how it must have felt, for those involved, to see the final stones being placed on the first pyramid. But on reflection, although this analogy may be appropriate for the amount of collective effort that has been expended, it would also repeat the conceptual mistake that has plagued the ribosome field (more pieces of rock).”
Garret writes further that the ribosome with all its accessories is the most elaborate molecular machine that has ever been created. All its components are active and move releasing at each step only GDP and phosphate that occurs in a complete harmony with the intracellular medium. The XRA achievements in the modeling of ribosomes should be first of all carefully “relished”. [Footnote: This seems to relate to a laborious work on searching for correlations between the earlier biochemical data with XRA results. Such work is already started, and this is partially the reason for the delayed appearance of the present publication. In addition to comments in the articles with XRA data, the works began to appear that entirely deal with analysis of these correlations [36, 37], let alone all subsequent experimental works simply must include such an analysis.]
And then it will be evident that the time has come for new thinking, for development of new approaches, and also for an active attraction of young people. Static models obtained by cryo-electron microscopy and XRA will be a retiring stage because the next decade will deal with studies on the dynamics of the ribosomal machine. [Footnote: Not without reason the next conference on ribosomes (from January 27 to February 1, 2002, in Queenstown, New Zealand) is entitled Dynamics of Structure and Functions of Ribosomes.]
Thomas Cech [33] comes to a similar conclusion: “These most recent contributions of Steitz, Moore, and colleagues provide a milestone, but not the finish line. This one structure contains more RNA-RNA and RNA-protein interactions than all previous atomic-level structures combined, so ribophils can look forward to years of additional analysis.
The whole 70S ribosome needs to be brought to this same atomic level of resolution, and the proposed reaction mechanism deserves critical testing. Finally, the molecular basis of the mRNA translocation step that must occur after each peptidyltransferase event remains obscure.
Thus, although the current crystal structure provides one beautiful frame, we still look forward to seeing the entire movie”. [Footnote: Separate elements of the ribosome dynamics have been already discussed at the International Conference dedicated to A. S. Spirin (August 27-September 1, 2001, Pushchino-on-Oka, Moscow Region). After the work on this manuscript had been finished, a number of important publications appeared on the same theme [38-43].]
The author is grateful to A. A. Bogdanov, R. Brimacombe, K. Nierhaus, H. Noller, V. Ramakrishnan, and the members of the RNP group for useful discussions; to A. A. Bogdanov for his reading the manuscript and valuable remarks; to T. Rassokhin for his computerized drawings. The author is grateful to various foundations for supporting his works in the field of structure and functions of nucleoproteins: the Russian Foundation for Basic Research (RFBR), grant No. 01-04-48643, the RFBR-the National Science Foundation of China, grant No. 99-04-39072, the RFBR-OEAD, grant No. 00-04-02007, the University of Russia, grant Nos. 99-2185 and 99-1700, the Ministry of Science, grant No. 415/99.
REFERENCES
1.Clemons, W. M., May, J. L., Wimberly, B. T.,
McCutcheon, J. P., Capel, M. S., and Ramakrishnan, V. (1999)
Nature, 400, 833-840.
2.Ban, N., Nissen, P., Hansen, J., Capel, M., Moore,
P. B., and Steitz, T. A. (1999) Nature, 400, 841-847.
3.Cate, J. H., Yusupov, M. M., Yusupova, G. Zh.,
Earnest, T. N., and Noller, H. F. (1999) Science, 285,
2095-2104.
4.Garrett, R. (1999) Nature, 400,
811-812.
5.Pennisi, E. (1999) Science, 285,
2048-2051.
6.Liljas, A. (1999) Science, 285,
2077-2078.
7.Ramakrishnan, V., Capel, M. S., Clemons, W. M.,
Jr., May, J. L. C., and Wimberly, B. T. (2000) in The Ribosome.
Structure, Function, Antibiotics, and Cellular Interactions
(Garrett, R. A., Douthwaite, S. R., Liljas, A., Matheson, A. T.,
Moore, P. B., and Noller, H. F., eds.) ASM Press, Washington DC, pp.
3-9.
8.Ban, N., Nissen, P., Hansen, J., Moore, P. B., and
Steitz, T. A. (2000) in The Ribosome. Structure, Function,
Antibiotics, and Cellular Interactions (Garrett, R. A., Douthwaite,
S. R., Liljas, A., Matheson, A. T., Moore, P. B., and Noller, H. F.,
eds.) ASM Press, Washington DC, pp. 11-20.
9.Bashan, A., Pioletti, M., Bartels, H., Janell, D.,
Schluenzen, F., Gluehmann, M., Levin, I., Harms, J., Hansen, H. A. S.,
Tocilji, A., Auerbach, T., Avila, H., Simitsopoulou, M., Peretz, M.,
Bennett, W. S., Agmon, I., Kessler, M., Weinstein, S., Franceschi, F.,
and Yonath, A. (2000) in The Ribosome. Structure, Function,
Antibiotics, and Cellular Interactions (Garrett, R. A., Douthwaite,
S. R., Liljas, A., Matheson, A. T., Moore, P. B., and Noller, H. F.,
eds.) ASM Press, Washington DC, pp. 21-33.
10.Yonath, A., and Franceschi, F. (2000) in Abst.
RNA 2000, Madison, USA, p. 2.
11.Nissen, P., Ban, N., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) in Abst. RNA 2000, Madison, USA, p.
1.
12.Brodersen, D., Carter, A., Clemons, W., Jr.,
Morgan-Warren, R., Wimberly, B., and Ramakrishnan, V. (2000) in
Abst. 18th Int. Congr. of Biochemistry and Molecular Biology,
Birmingham, UK, p. 91.
13.Nissen, P., Ban, N., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) in Abst. 18th Int. Congr. of Biochemistry
and Molecular Biology, Birmingham, UK, p. 91.
14.Yonath, A. (2000) in Abst. 18th Int. Congr. of
Biochemistry and Molecular Biology, Birmingham, UK, p. 97.
15.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) Science,289, 905-920.
16.Nissen, P., Hansen, J., Ban, N., Moore, P. B.,
and Steitz, T. A. (2000) Science, 289, 920-930.
17.Schluenzen, F., Tocilj, A., Zarivach, R., Harms,
J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I.,
Franceschi, F., and Yonath, A. (2000) Cell, 102,
615-623.
18.Wimberly, B. T., Brodersen, D. E., Clemons, W.
M., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T., and
Ramakrishnan, V. (2000) Nature, 407, 327-339.
19.Carter, A. P., Clemons, W. M., Brodersen, D. E.,
Morgan-Warren, R. J., Wimberly, B. T., and Ramakrishnan, V. (2000)
Nature, 407, 340-348.
20.Moore, P. B. (1985) Proc. The Robert A. Welch
Foundation Conf. on Chemical Research. XXIX Genetic Chemistry: The
Molecular Basis of Heredity, Houston, Texas, USA, pp. 185-215.
21.Spirin, A. S. (1986) Molecular Biology:
Ribosome Structure and Protein Biosynthesis [in Russian], Vysshaya
Shkola, Moscow.
22.Spirin, A. S. (1998) Soros Educat. J.,
11, 65-70.
23.Spirin, A. S. (1999) Soros Educat. J.,
4, 2-9.
24.Fersht, A. (1999) Structure and Mechanism in
Protein Science: a Guide to Enzyme Catalysis and Protein Folding,
W. H. Freeman and Company, USA.
25.Spirin, A. S., and Gavrilova, L. P. (1971) The
Ribosome [in Russian], Nauka, Moscow.
26.Agrawal, R. K., and Frank, J. (1999) Curr.
Opin. Struct. Biol., 9, 215-221.
27.Karpova, E. A., Serdyuk, I. N., Tarakhovskii, Yu.
S., and Orlova, E. V. (1986) Dokl. Akad. Nauk SSSR, 289,
1263-1266.
28.Glotz, C., Mussig, J., Gewitz, I. I. S.,
Makowski, I., Arad, T., Yonath, A., and Wittmann, H. G. (1987)
Biochem. Int., 15, 953-960.
29.Nomura, M. (1990) in The Ribosome, Structure,
Function, and Evolution (Hill, W. E., Dahlberg, A., Garrett, R. A.,
Moore, P. B., Schlessiger, D., and Warner, J. R., eds.) ASM Press,
Washington DC, pp. 3-55.
30.Brimacombe, R. (2000) Structure, 8,
R195-R200.
31.Culver, G. M., Cate, J. H., Yusupova, G. Zh.,
Yusupov, M. M., and Noller, H. F. (1999) Science, 285,
2133-2135.
32.Noller, H. F., Kop, J., Wheaton, V., Brosius, J.,
Gutell, R. R., Kopylov, A. M., Dohme, F., Herr, W., Stahl, D. A.,
Gupta, R., and Woese, C. R. (1981) Nucleic Acids Res.,
9, 6167-6189.
33.Cech, T. R. (2000) Science, 289,
878-879.
34.Gesteland, R. F., Cech, T. R., and Atkins, J. F.
(eds.) (1999) The RNA World, CSHL Press, N. Y.
35.Yusupov, M. M., Yusupova, G. Zh., Baucom, A.,
Lieberman, K., Earnest, T. N., Cate, J. H. D., and Noller, H. F. (2001)
Science, 292, 883-896.
36.Lancaster, L., Culver, G. M., Yusupova, G. Z.,
Cate, J. H., Yusupov, M. M., and Noller, H. F. (2000) RNA,
6, 717-729.
37.Sergiev, P. V., Dontsova, O. A., and Bogdanov, A.
A. (2001) Mol. Biol. (Moscow), 35, 559-583.
38.Pioletti, M., Schlunzen, F., Harms, J., Zarivach,
R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T.,
Jacobi, C., Hartsch, T., Yonath, A., and Franceschi, F. (2001) EMBO
J., 20,1829-1839.
39.Ogle, J. M., Brodersen, D. E., Clemons, W. M.,
Jr., Tarry, M. J., Carter, A. P., and Ramakrishnan, V. (2001)
Science, 292,897-902.
40.Ramakrishnan, V., and Moore, P. B. (2001)
Curr. Opin. Struct. Biol., 11,144-154.
41.Carter, A. P., Clemons, W. M., Jr., Brodersen, D.
E., Morgan-Warren, R. J., Hartsch, T., Wimberly, B. T., and
Ramakrishnan, V. (2001) Science, 291,498-501.
42.Brodersen, D. E., Clemons, W. M., Jr., Carter, A.
P., Morgan-Warren, R. J., Wimberly, B. T., and Ramakrishnan, V. (2000)
Cell, 103, 1143-1154.
43.Nissen, P., Ippolito, J. A., Ban, N., Moore, P.
B., and Steitz, T. A. (2001) Proc. Natl. Acad. Sci.
USA,98,4899-4903.
44.Wimberly, B. T., White, S. W., and Ramakrishnan,
V. (1997) Structure, 5, 1187-1198.
45.Dragon, F., and Brakier-Gingras, L. (1993)
Nucleic Acids Res., 21, 1199-1203.
46.Rassokhin, T. I., Golovin, A. V., Petrova, E. V.,
Spiridonova, V. A., Karginova, O. A., Rozhdestvenskii, T. S., Brozius,
Yu., and Kopylov, A. M. (2001) Mol. Biol. (Moscow), 35,
617-627.