Involvement of Chloroplasts in the Programmed Death of Plant Cells
V. D. Samuilov*, E. M. Lagunova, E. V. Dzyubinskaya, D. S. Izyumov, D. B. Kiselevsky, and Ya. V. Makarova
Department of Cell Physiology and Immunology, School of Biology, Lomonosov Moscow State University, Moscow, 119992 Russia; fax: (095) 939-3807; E-mail: vds@8.cellimm.bio.msu.ru* To whom correspondence should be addressed.
Received December 25, 2001; Revision received January 4, 2002
The effect of cyanide, an apoptosis inducer, on pea leaf epidermal peels was investigated. Illumination stimulated the CN--induced destruction of guard cells (containing chloroplasts and mitochondria) but not of epidermal cells (containing mitochondria only). The process was prevented by antioxidants (alpha-tocopherol, 2,5-di-tret-butyl-4-hydroxytoluene, and mannitol), by anaerobiosis, by the protein kinase C inhibitor staurosporine, and by cysteine and serine protease inhibitors. Electron acceptors (menadione, p-benzoquinone, diaminodurene, TMPD, DCPIP, and methyl viologen) suppressed CN--induced apoptosis of guard cells, but not epidermal cells. Methyl viologen had no influence on the removal of CN--induced nucleus destruction in guard cells under anaerobic conditions. The light activation of CN--induced apoptosis of guard cells was suppressed by DCMU (an inhibitor of the electron transfer in Photosystem II) and by DNP-INT (an antagonist of plastoquinol at the Qo site of the chloroplast cytochrome b6f complex). It is concluded that apoptosis initiation in guard cells depends on the simultaneous availability of two factors, ROS and reduced quinones of the electron transfer chain. The conditions for manifestation of programmed cell death in guard and epidermal cells of the pea leaf were significantly different.
KEY WORDS: apoptosis, chloroplasts, reactive oxygen species, plastoquinone, cytochrome b6f complex, staurosporine, protease inhibitors, Pisum sativum L.
Abbreviations: BHT) butylated hydroxytoluene (ionol); BQ) p-benzoquinone; DAD) diaminodurene; DCMU) 3-(3´,4´-dichlorophenyl)-1,1-dimethylurea; DNP-INT) dinitrophenyl ester of iodonitrothymol; DPIP) 2,6-dichlorophenolindophenol; IA) iodoacetamide; LHCII) light-harvesting chlorophyll a/b-protein complex; MV) methyl viologen; NEM) N-ethylmaleimide; PCD) programmed cell death; PMSF) phenylmethylsulfonyl fluoride; PQ) plastoquinone; PS) photosystem; ROS) reactive oxygen species; TMPD) N,N,N´,N´-tetramethyl-p-phenylenediamine.
Programmed cell death (PCD) is the physiological process of cell
destruction that is involved in the accomplishment of the developmental
programs of multicellular organisms, the functioning of the immune
system, the maintenance of tissue homeostasis, and responses to abiotic
stressors. PCD in animal and plants is based on unitary mechanisms [1-3] that presumably evolved in
prokaryotes for protection of their populations against viruses and
were involved in cell differentiation [2, 4-6]. PCD includes apoptosis and
paraptosis [7]. Apoptosis in animals is accompanied
by the transfer of phosphatidylserine from the inner monolayer of the
cytoplasmic membrane to the outer monolayer, a decrease in the cell's
volume, the formation of vesicle-like protrusions of the cytoplasmic
membrane, nucleus condensation, internucleosomal cleavage of the
nuclear DNA, cell fragmentation into membrane vesicles with the
intracellular content (apoptotic bodies) that are engulfed by
macrophages or adjacent cells. Apoptosis in plant cells results in
similar changes. However, plants lack phagocytes. Moreover, cell walls
preclude apoptosis. Therefore, plant cell polymers are degraded by
hydrolytic enzymes to monomers that are utilized by adjacent cells.
The fate of the cell wall is ambiguous. One option is that the cell walls are strengthened by oxidative cross-links between structural proteins and phenolic compounds, the formation of cellulose bulges, and lignification [8-10]. Cell wall strengthening hampers the entry of a pathogen into the cell or bricks up the microbes that have succeeded in penetrating into it. The development of rigid xylem and phloem vessels is accompanied by the formation of bulges that are subsequently strengthened by lignin deposits. Another option is cell wall destruction involving activated hydrolytic enzymes. Aerenchyma formation is accompanied by total cell wall destruction [11]. Hydrolases act locally. They dissolve the cell walls in the segregation layer forming prior to leaf fall and the detachment of ripe fruits [12].
Apoptosis is a multistage process that primarily involves caspases (evolutionarily conserved cysteine proteases) in animals [13, 14]. There are different PCD pathways in animals and humans: a) the cell receptor (e.g., Fas-receptor)-triggered pathway [14, 15]; b) the mitochondrial cytochrome c-dependent pathway [14, 16, 17]; c) the mitochondrial flavoprotein AIF (apoptosis-inducing factor)-dependent, caspase-independent pathway [18]; d) the pathway that involves caspase-12 of the endoplasmic reticulum [19], etc. [2]. Apoptosis in plants can proceed with the participation of vacuolar hydrolases [20]. A mitochondrial cytochrome c-dependent PCD pathway has also been revealed [21-23]. Plant apoptosis is accompanied by the activation of caspase-like proteases [23-26].
Paraptosis, the other PCD form, is distinguished from apoptosis by morphological and biochemical criteria, and by the responses to apoptosis inhibitors [7]. Nucleus fragmentation and vesicle formation that are typical of apoptosis do not occur during paraptosis. This form of cell death is characterized by cytoplasm vacuolization and involves an alternative caspase-9 whose activation does not depend on cytoplasmic protein Apaf-1 (apoptosis protease activating factor-1).
In plants, apoptosis can be caused by cyanide [27-29]. Cyanides are not extraneous compounds for plants [30]. Cyanogenic glycosides are synthesized by plants in order to destroy pathogens, to deter herbivorous animals, and to generate transportable compounds containing reduced nitrogen. Compounds whose hydrolysis results in CN- liberation have been detected in several thousands of plant species including sorghum, almond, cherry, clover, and cassava plants.
Illumination promoted the CN--induced destruction of the nuclei in chloroplast-containing guard cells but not chloroplast-free epidermal cells in the epidermal peels from pea leaves [27]. Guard cells were significantly more CN--resistant than epidermal cells. After only 1-2 h of incubation with CN-, nucleus destruction in epidermal cells in the dark was almost 100%. Nucleus destruction was insignificant in the dark in guard cells, but became 70-80% after 20-24 h of CN- incubation in the light [27].
The goal of this work was to investigate the role of chloroplasts and the involvement of light-dependent processes in plant PCD.
MATERIALS AND METHODS
The studies were conducted with lower epidermal peels from the leaves of 7-15-day-old pea (Pisum sativum L. cv Alpha) seedlings grown under continuous illumination at 20-24°C [27]. Epidermal peels were detached with tweezers and placed into distilled water. To ensure a rapid entry of reagents into cells, an infiltration method was used: epidermal peels were incubated in vacuo for 1-2 min. Epidermal samples were placed into polystyrene plates and incubated in distilled water with the addition of reagents (given in figure captions) at room temperature in the dark or under a luminescent lamp with a light intensity of ~1000 lx. After completing the incubation time, the samples were transferred into distilled water for 5 min and thereupon treated for 5 min with the Battaglia fixer (a mixture of chloroform, 96% ethanol, glacial acetic acid, and 40% formalin at ratio of 5 : 5 : 1 : 1). The samples were subsequently washed with ethanol for 10 min to remove the fixer, incubated for 5 min in water, and stained with Carazzi hematoxylin (a nuclear stain) for 20 min. The stained epidermal peels were washed with tap water and examined under a light microscope. Two (or, in most cases, three) repeats of each of the experiments were done. Three-to-five hundred cells were scanned, and the percentage of cells with destroyed nuclei and nucleus-lacking cells was determined [27].
RESULTS
Rationale of the experimental approaches used in this work. In contrast to many other studies, we used epidermal peels from leaves [27, 31], but not cell cultures, in order to approximate the conditions characteristic of a native system. The advantages of the peels are as follows: a) they represent cell monolayers, and this is convenient for studies using a microscope; b) the peels consist of two types of cells, guard cells (containing chloroplasts and mitochondria) and epidermal cells (containing mitochondria only), enabling us to investigate the role of phototrophic and chemotrophic nutrition in PCD using one preparation only.
Physical and chemical factors seem more advantageous than natural PCD inducers, because they cause synchronous apoptosis with a high yield of dead cells, which facilitates the subsequent assessment of the results. Cyanide, the plant PCD inducer that causes internucleosomal cleavage of the nuclear DNA [28, 29], was used by us [27] as such a chemical factor.
Recently, a number of methods to monitor the PCD process have been developed. The most frequently used method is based on the electrophoretic separation of DNA fragments. A DNA “ladder” is considered a classical sign of apoptosis. However, PCD does not always result in a DNA “ladder”. For instance, mitochondrial flavoprotein AIF-induced apoptosis in animals does not cause the formation of a “ladder” [18]. Another widely used method is based on measuring caspase activity, but apoptosis can also proceed without caspase involvement [18]. There are also other methods, but they do not yield unambiguous results.
Direct microscopic observations supply reliable information on the state of the cell and the cell nucleus, the primary target of apoptosis. Apoptosis in animals results in nucleus destruction and complete cell elimination because of phagocytosis. Apoptosis in plants, similar to that in animals, causes the destruction and disappearance of the nuclei, but the cell walls are either retained or hydrolyzed at the later stages of the process. Hence, the occurrence of nucleus-lacking cells that retain their cell walls is reliable evidence of plant cell death.
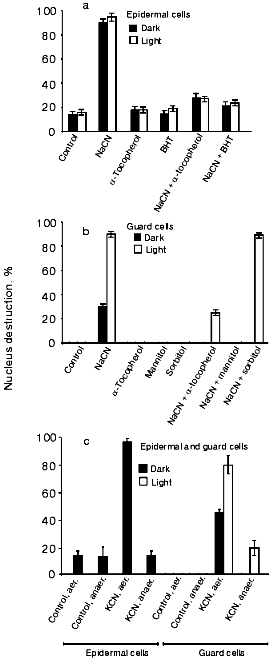
Effects of antioxidants and anaerobiosis on CN--induced cell nucleus destruction. Since the effect of CN-, a nucleus destruction inducer, can be mediated by ROS, we investigated the influence of the antioxidants (free radical traps) alpha-tocopherol, BHT, and mannitol (see, e.g., [32] for a discussion concerning the antioxidant properties of mannitol). All the three compounds suppressed CN--induced cell nucleus destruction in epidermal (Fig. 1a) and guard (Fig. 1b) cells. Sorbitol, a mannitol analog without antioxidant properties, failed to prevent the CN- effect (Fig. 1b). The CN--induced cell nucleus destruction was prevented by anaerobiosis both in the dark and in the light (Fig. 1c).
Effect of electron acceptors on CN--induced destruction of cell nuclei. Light-induced stimulation of CN--induced apoptosis in guard and epidermal cells can be due to the generation of ROS by chloroplasts. In this connection, we tested the effects of the ROS-generating agents MV and menadione (vitamin K3).Fig. 1. Effects of antioxidants (a, b) and anaerobiosis (c) on the CN--induced nucleus destruction in epidermal and guard cells of pea leaves. Additions: 2.5 mM NaCN or KCN, 0.1 mM alpha-tocopherol, 0.1 mM BHT, 125 mM mannitol, and 125 mM sorbitol. Time of incubation with CN-: 1 h and 20 h in the experiments with epidermal and guard cells, respectively. Studies under anaerobic conditions were conducted in Eppendorf test tubes. The incubation medium contained 50 mM glucose, 0.1 mg/ml glucose oxidase, and 0.06 mg/ml catalase. Sunflower oil was layered on top of the solutions in the test tubes, which were stopped so that no air bubble was left over the oil. Epidermal films were preincubated under these conditions for 1 h, and the aqueous phase was then supplemented with KCN.
MV (paraquat), an efficient herbicide, is reduced by PSI (primarily by FeS center FB [33]). By interacting with the respiratory chain, MV also produces multiple effects on mitochondria [34]. Menadione is reduced in chloroplasts by PSII, the cytochrome b6f complex, and PSI [35, 36], and in mitochondria by the NADH:ubiquinone-oxidoreductase [37]. The reduction products, the cation radical of MV and the anion radical of menadione, are spontaneously oxidized by O2, forming O2-. and H2O2. Menadiol is also reduced by O2, forming H2O2 [37, 38]. Besides, menadione elicits the non-photochemical quenching of light-excited chlorophyll [39].
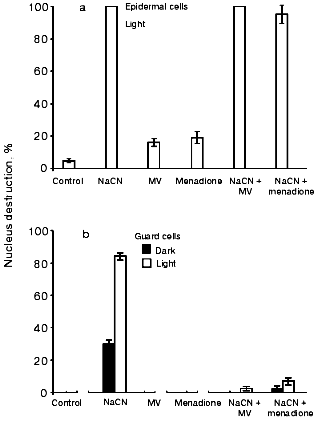
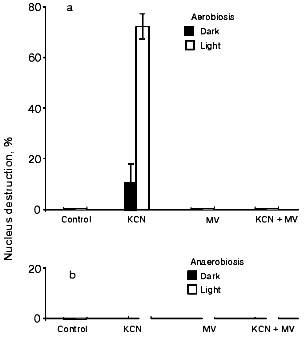
Figure 2a shows that MV and menadione promote nucleus destruction and exert no influence on the CN- effect in epidermal cells. Menadione-induced apoptosis was earlier detected in tobacco protoplasts [21, 40]. MV and menadione per se did not influence the state of the nuclei of guard cells in the light or in the dark, but they prevented the CN--induced nucleus destruction (Fig. 2b). A twofold decrease in the CN- effect on light-incubated guard cells occurred at MV and menadione concentrations of 2.5-3 and ~0.01 mM, respectively (data not shown). CN- effect on guard cells is prevented in anaerobic conditions both in the absence and presence of MV (Fig. 3).
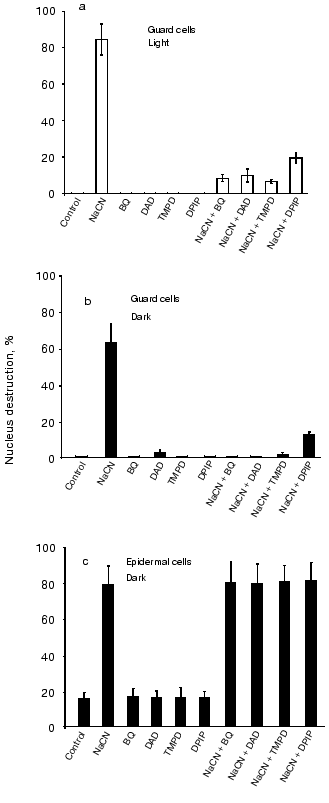
Subsequently, we tested a number of electron acceptors whose reduced forms cannot be spontaneously oxidized by oxygen. BQ, DAD, TMPD, and DPIP prevented CN--induced nucleus destruction in guard cells (in the light, Fig. 4a, and in the dark, Fig. 4b) but not in epidermal cells (Fig. 4c). These compounds interact in chloroplasts with PSII, the cytochrome b6f complex, and PSI [35, 36, 41]. Therefore, they elicit PQ oxidation and suppress NADP+ photoreduction. They also oxidize ubiquinol in mitochondria by interacting with NADH:ubiquinone-oxidoreductase, succinate:ubiquinone-oxidoreductase, and the cytochrome bc1 complex of the respiratory chain.Fig. 2. Effects of MV and menadione on the CN--induced nucleus destruction in epidermal (a) and guard (b) cells. Additions: 2.5 mM NaCN, 5 mM MV, and 0.1 mM menadione. Incubation time: 1 h and 21 h in experiments (a) and (b), respectively.
Fig. 3. Effect of MV on the CN--induced nucleus destruction in epidermal cells under aerobic (a) and anaerobic (b) conditions. Additions: 2.5 mM KCN and 5 mM MV. Incubation time: 20 h. Anaerobic conditions were created by adding 50 mM glucose, 0.1 mg/ml glucose oxidase, and 0.06 mg/ml catalase. Sunflower oil was layered on top of the aqueous solution. Epidermal films were preincubated under these conditions for 1 h. The aqueous solution was then supplemented with KCN.
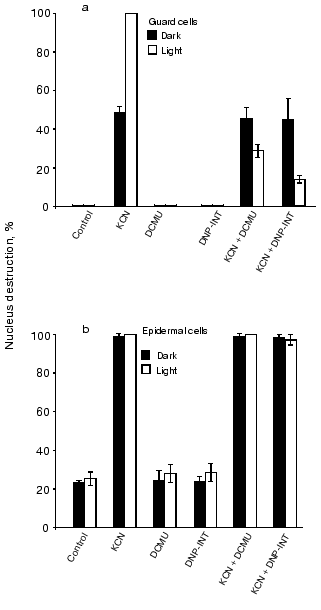
Effects of DCMU and DNP-INT. Figure 5a demonstrates that the CN--induced nucleus reduction in illuminated guard cells is suppressed by DCMU, an inhibitor of the electron transfer between the primary (QA) and the secondary (QB) plastoquinone of chloroplast PSII, and by DNP-INT, an inhibitor of plastoquinol oxidation at site QO of the chloroplast cytochrome b6f complex, but not that of the mitochondrial cytochrome bc1 complex [42]. DCMU and DNP-INT did not influence the CN--induced nucleus destruction in guard cells in the dark (Fig. 5b) and in epidermal cells in the light and in the dark (Fig. 5c).Fig. 4. Effects of BQ, DAD, TMPD, and DPIP on the CN--induced nucleus destruction in guard cells in the light (a) and in the dark (b) and in epidermal cells in the dark (c). Additions: 2.5 mM NaCN, 0.1 mM BQ, DAD, TMPD, or DPIP. Incubation time: 24, 48, and 1 h in experiments (a), (b), and (c), respectively.
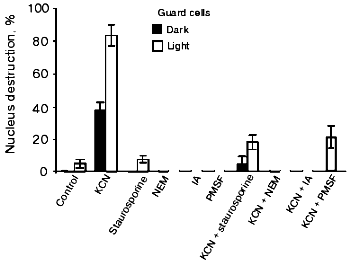
Effects of protein kinase C and protease inhibitors. Staurosporine, an inhibitor of protein kinase C, and the cysteine protease inhibitors NEM and IA suppressed the CN--induced apoptosis of guard cells in the dark and in the light (Fig. 6). PMSF, a serine protease inhibitor, produced the same effect. It was shown earlier that staurosporine, cysteine, and serine protease inhibitors prevent apoptosis induction in tobacco epidermal cells [31] and the cells of soybean (Glycine max) [43] and tomato [26] suspension cultures.Fig. 5. Effects of DCMU and DNP-INT on the CN--induced nucleus destruction in guard (a) and epidermal (b) in the light and in the dark. Additions: 2.5 mM NaCN, 0.01 mM DCMU, and 0.01 mM DNP-INT. Incubation time: 24 and 1 h in experiments (a) and (b), respectively.
Fig. 6. Effects of the inhibitors of protein kinase C and proteases on the CN--induced nucleus destruction in guard cells in the light and in the dark. Additions: 2.5 mM KCN, 2.5 µM staurosporine, 1 mM NEM, 10 mM IA, and 0.5 mM PMSF. Epidermal peels were preincubated with staurosporine, NEM, or PMSF for 1 h before the addition of KCN. The films were incubated with KCN for 23 h.
DISCUSSION
Nucleus destruction occurs in epidermal and guard cells [27] from the epidermal peels of tobacco leaves upon treating them with CN-, an apoptosis inducer in plants [28, 29]. It seems likely that CN- causes H2O2 accumulation in the cells by inhibiting their catalase and peroxidases. Hydrogen peroxide per se causes no effect, but the hydroxyl radical, the product of its one-electron oxidation, is a strong oxidant. The E0´ values of the H2O2/OH* and OH*/H2O systems are 0.32 and 2.31 V, respectively [44]. Due to the high E0´ value of OH* and the free radical properties of OH*, it oxidizes a large number of organic substances, including DNA, proteins, and lipids. It also initiates chain radical reactions.
Illumination accelerated the CN--induced nucleus destruction in guard cells, but not epidermal cells. The stimulatory effect of light can be due to ROS generation by chloroplasts that results in the apoptosis of guard cells. In chloroplasts, H2O2 is formed with the participation of superoxide dismutase from O2-., the product of one-electron reduction of O2 in the electron acceptor branches of PSI and PSII. Hydrogen peroxide also results from incomplete oxidation of water in the electron donor branch of PSII (see review [45]). Apart from heme catalase and peroxidases, CN- inactivates ribuloso-1,5-bisphosphate carboxylase [46] and the mitochondrial cytochrome c oxidase. Inhibition of ribuloso-1,5-bisphosphate carboxylase results in depleting the NADP+ pool and, therefore, should enhance the O2-. generation by the photosynthetic electron transfer chain, due to spontaneous oxidation of ferredoxin by oxygen. Electron transfer to O2 via PSI accounts for up to 10% of the total electron flow through the chloroplast photosynthetic chain even if NADP+ is present in excess [47]. CN--induced nucleus destruction in guard and epidermal cells was suppressed by antioxidants and anaerobiosis (Fig. 1), supportive of the suggestion that this process depends on ROS.
However, the idea that chloroplasts operate as additional ROS generators is at variance with the data obtained concerning the effects of the O2-.-generating reagents MV and menadione (Fig. 2). Illumination and the addition of these reagents produced opposite effects on the CN--induced nucleus destruction in guard cells. The CN- effect was enhanced by illumination and, conversely, relieved or prevented by MV and menadione. In contrast, MV and menadione enhance nucleus destruction in epidermal cells, and these reagents have no effect on the CN- effect (Fig. 2a). The opposite effects of illumination and O2-.-producing reagents suggest that the light-induced stimulation of the CN- induced apoptosis in guard cells is probably due not only to ROS generation, but also to other factors.
Post-translational protein modification is the main mechanism of signal transduction in pro- and eukaryotic organisms. Phosphorylation of proteins and the system of their redox modification are the two main signal systems. They both represent dynamic systems of protein function regulation [48, 49]. Protein phosphorylation is involved in the chromatic adaptation of the photosynthetic apparatus in higher plants, algae, and cyanobacteria [50-53]. PQ reduction via PSII results in protein kinase activation, subsequent phosphorylation of the mobile light-harvesting pigment-protein complex LHCII, dissociation of this complex from PSII, and the redistribution of the electron excitation energy of the light-harvesting antenna from PSII to PSI (“spillover”). With the membrane PQ pool oxidized, the photosynthetic machinery is in state I (LHCII is predominantly bound to PSII). Reduction of the membrane PQ pool results in its transition to state II (LHCII redistribution in favor of PSI).
The molecular mechanism used to signal the PQ redox state to the protein kinase has not yet been elucidated. However, it has been established that the PQ-binding site (QO) that performs the cooperative oxidation of plastoquinol by the Rieske FeS center and the low potential heme bl of cytochrome b6 is involved in activating the protein kinase of LHCII [52-56]. Dynamic structural models of the b6f-cytochrome complex [52, 53, 57] have been suggested that postulate a link between the activation of the LHCII kinase and plastoquinol binding at site QO and the shuttling of the Rieske FeS protein between a b6f complex site adjacent to heme bl (the proximal position of the Rieske protein) and a b6f complex site located near the cytochrome f heme (the distal position of the Rieske protein). Hence, PQ at the quinol oxidation site QO is involved in activating the LHCII kinase. The redox state of PQ also regulates the transcription rate of the genes coding for the apoproteins of the reaction center complexes of PSI and PSII [58].
The results of this work indicate that the redox state of PQ in plants is involved in PCD regulation. By inactivating ribuloso-1,5-bisphosphate carboxylase [48], CN- should cause PQ reduction in the photosynthetic electron transfer chain of chloroplasts. Cyanide can produce a similar effect on the mitochondrial ubiquinone in the dark. We suggest the following sequence of the CN--induced events: quinone reduction --> protein kinase activation --> protein phosphorylation --> PCD initiation. By oxidizing the quinol and a number of other components of the photosynthetic and respiratory electron transfer chains, menadione, BQ, DAD, TMPD, or DPIP disrupt this sequence of events and prevent the CN--induced destruction of cell nuclei (Figs. 2-4). MV that interacts with the electron acceptor complex of PSI and the dehydrogenases of the respiratory chain produces the same effect.
The light-induced activation of the LHCII kinase in chloroplasts is sensitive to DCMU [51]. The CN--induced nucleus destruction in guard cells is also inhibited by DCMU (Fig. 5). The data on the sensitivity of the light-induced activation of the CN--induced nucleus destruction to DNP-INT also support the suggestion that PCD is subject to regulation at the PQ level (Fig. 5). DNP-INT hinders the binding of plastoquinol at site QO of the chloroplast cytochrome b6f complex, but not at that of the mitochondrial bc1 complex [42].
Antioxidants and anaerobiosis prevent the CN--induced nucleus destruction in guard cells (Fig. 1). MV did not influence the CN--induced apoptosis under anaerobic conditions (Fig. 3). MV, an electron acceptor in PSI, should oxidize plastoquinol and the components of the cytochrome b6f complex in the light. The conditions required for the CN--induced nucleus destruction are summarized in the table.
Conditions required for the CN--induced nucleus destruction
in guard cells in epidermal peels from pea leaves
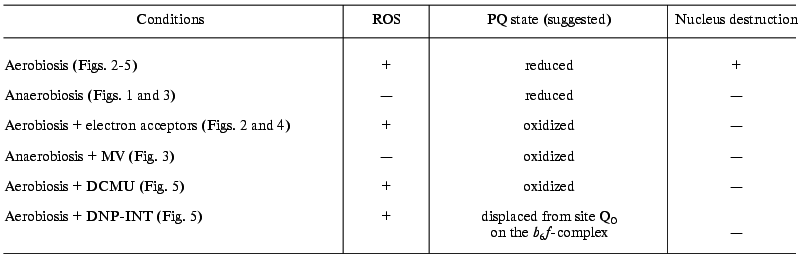
The data obtained suggest that the apoptosis of chloroplast-containing guard cells is induced by the combined action of ROS and reduced quinines of the electron transfer chain. PCD manifests itself in guard and epidermal cells under different conditions. The light-induced activation of the CN--induced nucleus destruction in guard cells is apparently due to the functioning of the protein kinase. This does not imply that the protein kinase involved in PCD is identical to the kinase regulating the transfer of the electron excitation energy between PSI and PSII. Probably, a number of protein kinases are subject to control by the redox state of the quinones. Based on the sensitivity of the process to staurosporine (Fig. 6), protein kinase C is involved in the CN--induced apoptosis of guard cells. The inhibitory effects of NEM and IA (Fig. 6) are also consistent with the involvement of cysteine and serine proteases in PCD.
Of particular importance is the role of the mitochondrial respiratory chain in the apoptosis of guard cells. The data on the CN--induced nucleus destruction in the dark, its prevention by antioxidants, anaerobic conditions (Figs. 1 and 2), and exogenous electron acceptors (Figs. 2-4), and the insensitivity of this process to DCMU and DNP-INT in the dark apparently can be regarded as evidence of the involvement of mitochondria in PCD. However, incubating the system under these conditions (in the dark) causes a change in the redox state of the photosynthetic electron transfer chain also. For instance, the transition of the photosynthetic apparatus of Chlamydomonas reinhardtii to state I can be accomplished in the dark under intense aeration, whereas the transition to state II occurs during the incubation of the cells under anaerobic conditions [57]. Further studies using inhibitors of the respiratory chain are to be conducted to elucidate the mitochondrial role in the apoptosis of guard cells.
This work was supported by the Russian Foundation for Basic Research (grant No. 01-04-48356).
REFERENCES
1.Jabs, T. (1998) Biochem. Pharmacol.,
57, 231-245.
2.Samuilov, V. D., Oleskin, A. V., and Lagunova, E.
M. (2000) Biochemistry (Moscow), 65, 873-887.
3.Lam, E., Kato, N., and Lawton, M. (2001)
Nature, 411, 848-853.
4.Hochman, A. (1997) Crit. Rev. Microbiol.,
23, 207-214.
5.Aravind, L., Dixit, V. M., and Koonin, E. V. (1999)
Trends Biochem. Sci., 24, 47-53.
6.Lewis, K. (2000) Microbiol. Mol. Biol. Rev.,
64, 503-514.
7.Sperandio, S., de Belle, I., and Bredesen, D. E.
(2000) Proc. Natl. Acad. Sci. USA, 97, 14376-14381.
8.Mehdy, M. C. (1994) Plant Physiol.,
105, 467-472.
9.Tenhaken, R., Levine, A., Brisson, L. F., Dixon, R.
A., and Lamb, C. (1995) Proc. Natl. Acad. Sci. USA, 92,
4158-4163.
10.Grant, J. J., and Loake, G. J. (2000) Plant
Physiol., 124, 21-29.
11.Pennel, R. I., and Lamb, C. (1997) Plant
Cell, 9, 1157-1168.
12.Greenberg, J. T. (1996) Proc. Natl. Acad. Sci.
USA, 93, 12094-12097.
13.Thornberry, N. A., and Lazebnik, Y. (1998)
Science, 281, 1312-1316.
14.Chang, H. Y., and Yang, X. (2000) Microbiol.
Mol. Biol. Rev., 64, 821-846.
15.Ashkenazi, A., and Dixit, V. M. (1998)
Science, 281, 1305-1308.
16.Skulachev, V. P. (1998) FEBS Lett.,
423, 275-280.
17.Green, D. R., and Reed, J. C. (1998)
Science, 281, 1309-1312.
18.Daugas, E., Nochy, D., Ravagnan, L., Loeffler,
M., Susin, S. A., Zamzami, N., and Kroemer, G. (2000) FEBS
Lett., 476, 118-123.
19.Nakagawa, T., Zhu, H., Morishima, N., Li., E.,
Xu, J., Yankner, B. A., and Yuan, J. (2000) Nature, 403,
98-103.
20.Jones, A. M. (2001) Plant Physiol.,
125, 94-97.
21.Sun, Y.-L., Zhao, Y., Hong, X., and Zhai, Z.-H.
(1999) FEBS Lett., 462, 317-321.
22.Balk, J., Leaver, C. J., and McCabe, P. F. (1999)
FEBS Lett., 463, 151-154.
23.Korthout, H. A. A. J., Berecki, G., Bruin, W.,
van Duijn, B., and Wang, M. (2000) FEBS Lett., 457,
139-144.
24.Del Pozo, O., and Lam, E. (1998) Curr.
Biol., 8, 1129-1132.
25.Solomon, M., Belenghi, B., Delldonne, M.,
Menachem, E., and Levine, A. (1999) Plant Cell, 11,
131-143.
26.De Jong, A. J., Hoeberichts, F. A., Yakimova, E.
T., Maximova, E., and Woltering, E. J. (2000) Planta,
211, 656-662.
27.Samuilov, V. D., Lagunova, E. M., Beshta, O. E.,
and Kitashov, A. V. (2000) Biochemistry (Moscow), 65,
696-702.
28.Wang, H., Li, J., Bostock, R. M., and Gilchrist,
D. G. (1996) Plant Cell, 8, 375-391.
29.Ryerson, D. E., and Heath, M. C. (1996) Plant
Cell, 8, 393-402.
30.McMahon, J. M., White, W. L. B., and Sayre, R. T.
(1995) J. Exp. Botan., 46, 731-741.
31.Allan, A. C., and Fluhr, R. (1997) Plant
Cell, 9, 1559-1572.
32.Shen, B., Jensen, R. G., and Bohnert, H. J.
(1997) Plant Physiol., 115, 527-532.
33.Fujii, T., Yokoyama, E., Inoue, K., and Sakurai,
H. (1990) Biochim. Biophys. Acta, 1015, 41-48.
34.Palmeira, C. M., Moreno, A. J., and Madeira, V.
M. C. (1995) Biochim. Biophys. Acta, 1229, 187-192.
35.Hauska, G. (1977) in Encyclopedia of Plant
Physiology (Trebst, A., and Avron, M., eds.) Springer-Verlag,
Berlin, Vol. 5, pp. 253-265.
36.Samuilov, V. D., Barskii, E. L., and Kitashov, A.
V. (1997) Biochemistry (Moscow), 62, 909-913.
37.Cadenas, E., Boveris, A., Ragan, C. I., and
Stoppani, A. O. (1977) Arch. Biochem. Biophys., 180,
248-257.
38.Yamashoji, S., Ikeda, T., and Yamashoji, K.
(1991) Biochim. Biophys. Acta, 1059, 99-105.
39.Samuilov, V. D., Borisov, A. Yu., Barsky, E. L.,
Borisova, O. F., and Kitashov, A. V. (1998) Biochem. Mol. Biol.
Int., 46, 333-341.
40.Sun, Y.-L., Zhu, H.-Z., Zhou, J., Dai, Y.-R., and
Zhai, Z.-H. (1999) Cell. Mol. Life Sci., 55, 310-316.
41.Saha, S., Ouitrakul, R., Izawa, S., and Good, N.
E. (1971) J. Biol. Chem., 246, 3204-3209.
42.Rich, P. R. (1984) Biochim. Biophys. Acta,
768, 53-79.
43.Levine, A., Pennel, R. I., Alvarez, M. E.,
Palmer, R., and Lamb, C. (1996) Curr. Biol., 6,
427-437.
44.Koppenol, W. H. (1994) in Free Radical Damage
and Its Control (Rice-Evans, C. A., and Burdon, R. H., eds.)
Elsevier, Amsterdam, pp. 3-24.
45.Samuilov, V. D. (1997) Biochemistry
(Moscow), 62, 451-454.
46.Ishida, H., Shimizu, S., Makino, A., and Mae, T.
(1998) Planta, 204, 305-309.
47.Asada, K., and Takahashi, M. (1987) in
Photoinhibition (Kyle, D. J., Osmond, C. B., and Arntzen, C. J.,
eds.) Elsevier, Amsterdam, pp. 227-287.
48.Stamler, J. S., Lamas, S., and Fang, F. C. (2001)
Cell, 106, 675-683.
49.Matsuzawa, A., and Ichijo, H. (2001) J.
Biochem., 130, 1-8.
50.Bennett, J. (1991) Annu. Rev. Plant Physiol.
Plant Mol. Biol., 42, 281-311.
51.Allen, J. F. (1992) Biochim. Biophys.
Acta, 1098, 275-335.
52.Vener, A. V., Ohad, I., and Andersson, B. (1998)
Curr. Opin. Plant Biol., 1, 217-223.
53.Wollman, F.-A. (2001) EMBO J., 20,
3623-3630.
54.Vener, A. V., van Kan, P. J. M., Gal, A.,
Andersson, B., and Ohad, I. (1995) J. Biol. Chem., 270,
25225-25232.
55.Vener, A. V., van Kan, P. J. M., Rich, P. R.,
Ohad, I., and Andersson, B. (1997) Proc. Natl. Acad. Sci. USA,
94, 1585-1590.
56.Zito, F., Finazzi, G., Delosme, R., Nitschke, W.,
Picot, D., and Wollman, F.-A. (1999) EMBO J., 18,
2961-2969.
57.Finazzi, G., Zito, F., Barbagallo, R. P., and
Wollman, F.-A. (2001) J. Biol. Chem., 276, 9770-9774.
58.Pfannschmidt, T., Nilsson, A., and Allen, J. F.
(1999) Nature, 397, 625-628.