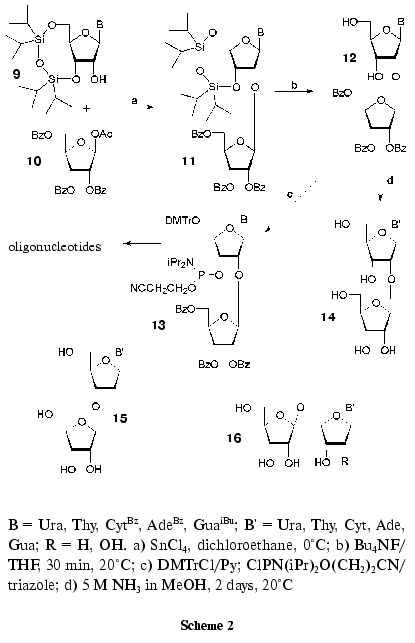REVIEW: Disaccharide Nucleosides and Oligonucleotides on Their Basis. New Tools for the Study of Enzymes of Nucleic Acid Metabolism
E. V. Efimtseva and S. N. Mikhailov*
Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, ul. Vavilova 32, Moscow, 119991 Russia; fax: (095) 135-1405; E-mail: smikh@eimb.ru* To whom correspondence should be addressed.
Received April 17, 2002; Revision received June 7, 2002
The main structural features of an important group of natural compounds, disaccharide nucleosides, are reviewed. The synthesis and properties of modified oligonucleotides on their basis as well as the methods of introduction of reactive aldehyde groups are described. The last part is devoted to the application of these compounds for studies of enzymes of nucleic acid metabolism.
KEY WORDS: disaccharide nucleosides, oligonucleotides, structure, physicochemical and substrate properties, affinity modification, enzymes of nucleic acid metabolism
Disaccharide nucleosides belong to an important group of natural compounds incorporated in tRNA, poly(ADP-ribose), antibiotics, and other physiologically active compounds. To date about a hundred disaccharide nucleoside derivatives have been isolated from various natural sources [1]. These compounds contain an extra carbohydrate residue linked to one of the nucleoside hydroxyl groups via an O-glycoside bond. The presence of a disaccharide residue and a heterocyclic base makes their properties similar to those of carbohydrates and nucleosides. Here we describe structurally specific features of some disaccharide nucleosides and summarize the data on the synthesis and physicochemical and substrate properties of disaccharide nucleosides and oligonucleotides (ONs) on their basis. We also give examples on the preparation of ONs bearing reactive aldehyde groups and their application for studying the enzymes of nucleic acid biosynthesis.
1. DISACCHARIDE NUCLEOSIDES ARE MINOR COMPONENTS OF tRNA
Nucleic acids comprise not only eight standard ribo- and 2´-deoxyribonucleosides, but also a large number of their derivatives, minor nucleosides. McCloskey et al. described the structures of 96 modified nucleosides occurring in various types of RNAs, most of them being found in tRNAs [2-4].
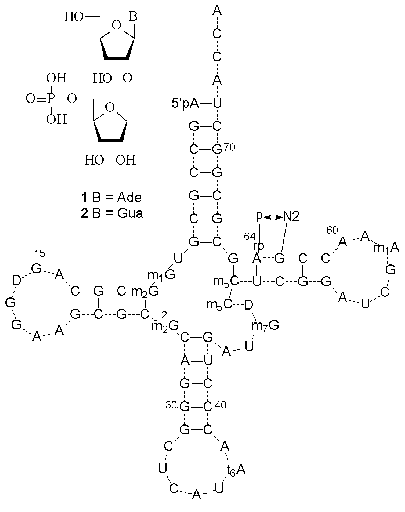
In 1965 Hall isolated a new minor nucleoside from a yeast RNA hydrolyzate and identified it as an adenosine derivative containing an extra ribofuranosyl residue [5]. It was shown later that this minor nucleoside was located in position 64 of the T domain of yeast initiator tRNA (tRNAiMet) [6]. The analysis of primary tRNAiMet structures isolated from some plant and yeast cells showed the presence in position 64 of modified guanosine [7-9]. At the end of the 1980s and beginning of the 1990s the structure of minor nucleosides were identified as O-beta-D-ribofuranosyl-(1´´-2´)-adenosine 5´´-O-phosphate (1) and O-beta-D-ribofuranosyl-(1´´-2´)-guanosine 5´´-O-phosphate (2) (Fig. 1) [8, 10-12]. The presence of a disaccharide minor nucleoside in position 64 is suggested to be a common property of initiator tRNA of lower eukaryotes. It is worth mentioning that only two types of carbohydrate-modified compounds, 2´-O-methyl- and 2´-O-beta-D-ribofuranosylnucleosides, were isolated from RNAs, whereas most minor nucleosides are modified at the heterocyclic base [2-4].
The X-ray analysis showed that a bulky 5´´-phosphoribofuranosyl hydrophilic substituent of nucleosides 1 and 2 (Fig. 1) is strictly fixed on the surface of the tRNAiMet minor groove [13]. The extra phosphate residue is involved in the formation of a hydrogen bond with the 2-amino group of the adjacent guanosine residue. The function of the modified purine nucleoside in position 64 of tRNAiMet was studied by Sprinzl et al. [12]. It was assumed that the modification of nucleoside 64 affected to a certain extent the discrimination of initiation/elongation process of protein biosynthesis. After the selective removal of the extra 5´´-phosphoribofuranosyl residue from tRNAiMet by hydrolysis of the O-glycosidic bond, the elongation function of tRNA was prevalent, although it was less effective than in the case of native tRNAMet [12]. These data suggest that the presence of the modified purine nucleoside in position 64 of tRNAiMet of lower eukaryotes and some plants prevents the formation of the tRNA complex with elongation factor EF-1alpha and GTP, which is indispensable for the elongation process.Fig. 1. The structures of minor nucleosides 1 [11] and 2 [8] and their positions in yeast tRNAiMet [6]. A hydrogen bond between the 5´´-phosphate residue and guanine 2-amino group is shown [13].
The biosynthesis pathway of modified nucleosides 1 and 2 (Fig. 1) is still obscure. However, there is a hypothesis about posttranscriptional ribosylation of adenosine or guanosine residues located in position 64 of the cytoplasmic initiator tRNAMet precursor. It was also presumed in [11] that the 64th purine nucleoside is ribosylated with 5-phosphoribosyl-1-alpha-pyrophosphate to give a beta-glycosidic bond.
2. THE STRUCTURE OF POLY(ADP-RIBOSE)
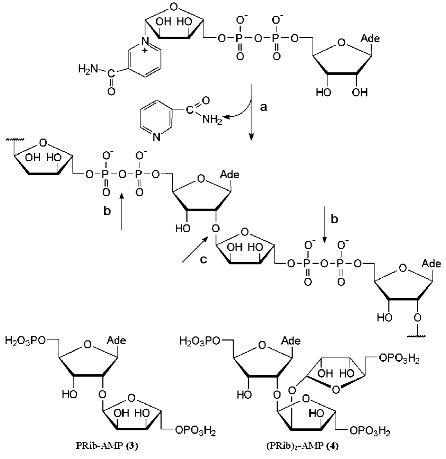
2´-O-alpha-D-Ribofuranosyladenosine is a structural element of a biopolymer poly(ADP-ribose) isolated from eukaryotic cells. In the 1960s it was shown that eukaryotic nuclei contain an enzyme poly(ADP-ribose) polymerase, which transforms NAD+ into a homopolymer composed of repeating ADP-ribose residues while releasing nicotinamide (Fig. 2) [14-17]. Linear poly(ADP-ribose) molecules are from several to 30 monomeric residues in length and are covalently bound to histones and other nuclear proteins [18, 19]. The biological role of poly(ADP-ribose) is still unclear, although it is assumed that it is involved in DNA replication, recombination, and repair and cellular differentiation [17, 19].
Enzymes of two classes are involved in the degradation of poly(ADP-ribose) (Fig. 2) [19-21]. Glycohydrolase of poly(ADP-ribose) cleaves the bond between adenosine and a ribose residue to give adenosine diphosphate ribose (ADP-ribose). The digestion of the pyrophosphate bond by phosphodiesterases yields 2´-(5´´-phosphoribosyl)-5´-adenosine monophosphate (PRib-AMP) (3). Using NMR spectroscopy, Oppenheimer and Ferro determined the structure of the monomeric unit 3 of poly(ADP-ribose) [22]. Compound 3 can be regarded as an isomer of minor nucleoside 1. The crucial structural difference between them lies in the configuration of the O-glycosidic bond. It is worth mentioning that PRib-AMP (3) functions as a prosthetic group of Klebsiella aerogenes citrate lyase [23].Fig. 2. Structure and biosynthesis of poly(ADP-ribose) and the enzymes involved in its cleavage: a) poly(ADP-ribose) polymerase; b) phosphodiesterase; c) poly(ADP-ribose) glycohydrolase.
In addition to a linear polymer poly(ADP-ribose), branched biopolymers related to branched polysaccharides were also isolated [19, 24]. The structure of the fragment (PRib)2-AMP (4) located in the branching site was confirmed by physicochemical methods [25]. Three ribofuranose residues of compound 4 are linked via alpha(1-->2) O-glycosidic bonds. It is worth noting that the disaccharide fragments of compounds 3 and 4 (Fig. 2) have not been synthesized yet.
3. DISACCHARIDE NUCLEOSIDES AS COMPONENTS OF ANTIBIOTICS AND
OTHER NATURAL COMPOUNDS
The structure, biological activity, and biosynthesis of nucleoside antibiotics have been comprehensively discussed in Isono's reviews [26, 27]. Garner's [28], Lerner's [29], and Knapp's [30] reviews summarize data on the synthesis of disaccharide nucleosides and complex nucleoside antibiotics. To date a large number of disaccharide nucleoside antibiotics of various structures have been isolated and characterized. The structures of purine antibiotics containing phosphate groups can serve as examples.
Adenosine and alpha-D-glucopyranose linked to phosphorylated allaric acid are structural elements of thuringiensin (5) [31-33]. This antibiotic isolated from Bacillus thuringiensis by two research groups displays insecticide and antibacterial properties [34, 35]. Thuringiensin (5) is an inhibitor of DNA-dependent RNA polymerase of both prokaryotes and eukaryotes [36].
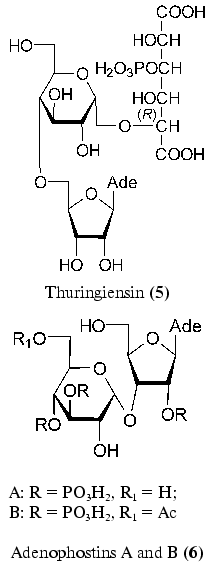
Of the recently isolated disaccharide nucleoside, adenophostins A and B (6) produced by Penicillum brevicompactum can be mentioned [37, 38]. These compounds are most powerful agonists of inositol 1,4,5-triphosphate receptors [39, 40]. This compound plays an important role in Ca2+ release. A structurally specific feature of adenophostins (6) is the presence of an alpha-glycosidic bond between ribofuranose and glucopyranose residues.
Recently found guanofosfocins (7) are related to phosphorylated antibiotics [41, 42]. Guanofosfocin A was isolated from Streptomyces sp. cultural medium, whereas guanofosfocins B and C areproduced by Trichoderma sp. This antibiotic contains additional O-glycosidic bond between mannopyranose and 8-hydroxyguanine to form a unique cyclic structure. Guanofosfocins (7) inhibit chitin synthetase and are effective against the fungus Candida albicans. However, due to a low stability, their study as potential antimycotic agents is hampered [42].
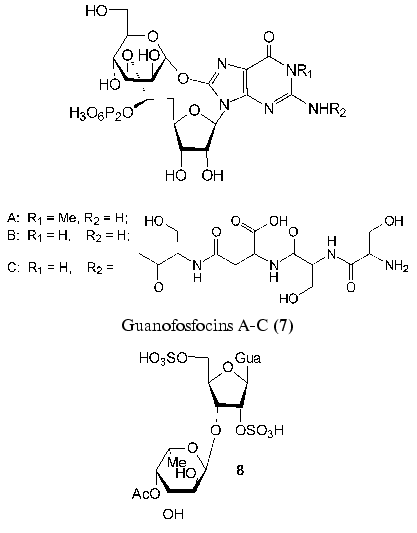
A new neuroactive compound HF-7 (8) was isolated from the venom of the funnel-web spider, Hololena curta. The chemical synthesis of disulfo-derivative of 3´-O-alpha-L-fucopyranosylguanosine (8) and the comparison of its physicochemical properties with those of the natural compound confirmed the structure of HF-7 [43]. HF-7 is an anionic nucleoside and, like adenofostins (6), affects the intracellular concentration of Ca2+ in nerve cells.
4. METHODS OF SYNTHESIS OF DISACCHARIDE NUCLEOSIDES AND THEIR
PROPERTIES
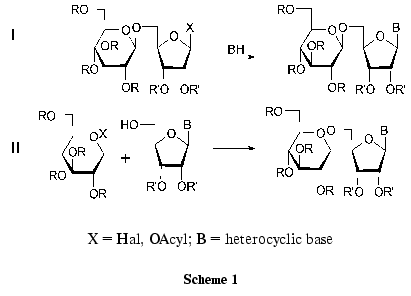
Disaccharide nucleosides can be synthesized using the two methods presented in Scheme 1 [29]. The first method is the coupling of protected disaccharide and the corresponding heterocyclic base derivative. Most nucleoside antibiotics were synthesized in this way. This scheme is multistage, since it includes the preparation of the properly protected disaccharide component. If the target compound bears a residue of a natural nucleoside, the synthesis is considerably shorter.
The other method of preparation of disaccharide nucleosides involves the formation of a new O-glycosidic bond between the properly protected nucleoside and the activated monosaccharide using classical procedures of glycoside synthesis. The use of nucleosides as starting compounds can substantially simplify the preparation of disaccharide nucleosides. Some examples of the formation of O-glycoside bonds between the partially protected nucleoside and the monosaccharide have been published (Scheme 1, route II). However, the product yields in these reactions did not exceed 20-30% because of by-product formation [29].
A general method of synthesis of 2´-O-beta-D-ribofuranosylnucleosides (14) comprising tRNA (Fig. 1) was suggested recently [44, 45]. The method consists in the coupling of N-substituted 3´,5´-O-tetraisopropyldisiloxane-1,3-diyl-ribonucleosides (9) with a small excess of 1-O-acetyl-2,3,5-tri-O-benzoyl-beta-D-ribofuranose (10) activated with tin tetrachloride in 1,2-dichloroethane (Scheme 2) [46]. The reaction conditions were similar to those of the nucleoside synthesis according to Vorbruggen [47] and that of alkyl beta-D-ribofuranosides [48]. O-Glycosylation was stereospecific to form beta-glycosides 11 in yields of 70-80% [44, 45]. After deprotection of silyl groups, compounds 12 were converted to the corresponding phosphoramidite derivatives 13, which were used in the standard automatic ON synthesis. Deacylation of compounds 12 resulted in disaccharide nucleosides 14 in high yields [44, 45].
The developed method was used for the preparation of pyrimidine 3´-O-beta-D-ribofuranosyl-2´-deoxyribonucleosides (15) [49], 5´-O-beta-D-ribofuranosyl-2´-deoxyribonucleosides (16, R = H) [50, 51], and 5´-O-beta-D-ribofuranosylnucleosides (16, R = OH) [51, 52].
To study the scope of the method, some other acylated sugars were used in the O-glycosylation reaction. In this way, 2´-O-beta-D-ribofuranosylnucleoside analogs containing in 2´-O position alpha-D- (and beta-L)-arabinofuranosyl, beta-D-erythrofuranosyl, beta-D-ribopyranosyl [53, 54], and 5-amino-5-deoxy-beta-D-ribofuranosyl residues [54] were synthesized. Recently on the basis of the general method for synthesis of 2´-O-beta-D-ribofuranosylnucleosides [44, 45], minor nucleoside 1 was first obtained (Fig. 1) [55].
The study of disaccharide nucleosides 14 (Scheme 2) in crystal and solution showed that the introduction of an extra ribofuranosyl residue only insignificantly affected the nucleoside fragment conformation [45]. It is worth mentioning that according to the X-ray analysis and NMR spectroscopy data, an exocyclic 5´-CH2OH group of the extra ribofuranosyl residue of compound 14 (B = Ura) is located close to the heterocyclic base [45]. A similar conformation was observed for the synthesized minor nucleoside 1 [55].
It is known that N- and O-glycosidic bonds are unstable under acidic conditions. The hydrolytic instability can be regarded as a factor effecting the use of compounds of this type. It was shown for purine disaccharide nucleosides that the hydrolysis of the O-glycosidic bond is accompanied by apurinization, whereas pyrimidine nucleosides are only cleaved at the O-glycosidic bond to give a nucleoside and ribose [50]. The O-glycosidic bond in compound 14 (B = Ade) is twice as stable as in the corresponding uridine derivative 14 (B = Urd). It is important that the N-glycosidic bond in adenine nucleoside 14 (B = Ade) is three times more stable than in the natural nucleoside. It is known that apurinization proceeds via protonation of the heterocyclic base and formation of a cyclic oxocarbenium ion [56].
We developed convenient preparative methods for synthesis of 2´-O-beta-D-ribofuranosylnucleosides starting from natural ribonucleosides. Using this scheme, a large set of disaccharide nucleosides was prepared, which proved the all-purpose character and efficacy of the method.
5. OLIGONUCLEOTIDES CONTAINING DISACCHARIDE NUCLEOSIDES
When starting the preparation of ON containing disaccharide nucleosides, we based it on the following grounds.
1. Modified ONs should retain all functional groups and distances between them to ensure potential interactions with nucleic acids and proteins. It should be expected that parameters of the complex formation with nucleic acids and binding to enzymes will be similar for natural and modified ON.
2. The presence of an extra cis-diol group of the ribofuranose residue enables smooth introduction of dialdehyde group using a well known reaction of periodate oxidation in aqueous medium at room temperature.
3. Reactive dialdehyde groups are located close to the sugar-phosphate backbone, which ensures the interaction with lysine residues of DNA-binding proteins. One or more modified residues can be thereby located in a predetermined position of the ON chain.
4. It should be mentioned that the resulting ONs can be used for the preparation of various conjugates and the attachment to polymeric supports.
The aforementioned structural features of modified ONs are required for their use as affinity reagents and development on their basis of selective inhibitors of the enzymes of nucleic acid biosynthesis. As will be seen from the later discussion, most of these general statements were realized.
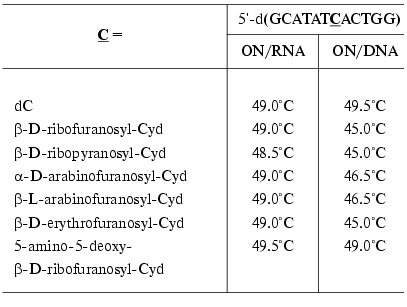
Synthons 13 bearing routine protective groups were used in the standard variant of automatic synthesis for the preparation of oligodeoxyribonucleotides [54, 57-61] and oligoribonucleotides [62]. The parameters of their complex formation with complementary DNA and RNA were studied. The ON containing 2´-O-beta-D-ribofuranosylcytidine formed stable complexes with RNA (DeltaTm = 0°C) (Table 1) [54], whereas melting temperatures of the DNA complexes were somewhat lower than in the case of natural compounds [54]. To increase the affinity towards complementary DNA and RNA, a large number of ONs modified at the 2´-O-position of the carbohydrate residue, such as hydroxyalkyl and aminoalkyl derivatives, were obtained during the last decade [63]. In order to ensure the stability of duplexes, the analogs containing other monosaccharide residues in the 2´-O-position were also used (Table 1) [54]. As seen in Table 1, all modified ONs formed stable complexes with RNA. In the case of DNA/DNA duplexes, the highest stability was observed for the ON containing 5-amino-5-deoxy-beta-D-ribofuranosyl residue.
Table 1. Melting temperatures of the
modified ON complexes with complementary DNA and RNA [54]
The effect of 2´-O-beta-D-ribofuranosyladenosine on the stability of the duplex formation was studied by high resolution NMR spectroscopy and methods of molecular dynamics on the example of a double-stranded RNA fragment (self-complementary oligoribonucleotide 5´-r(GCGA-RibAUUCGC)) [62]. It was shown that an extra bulky ribofuranose residue was located in the minor groove of the double-stranded RNA and did not significantly affect the RNA structure in solution (DeltaTm = 0°C). It is worth noting that these results coincide well with the X-ray analysis data about the position of the minor nucleoside 1 in tRNAiMet [13]. This implies that ONs containing various monosaccharide residues in the 2´-O-position are promising for studying the enzymes of nucleic acid biosynthesis.
Substrate properties of the ON containing 2´-O-beta-D-ribofuranosylnucleoside 14 (Scheme 2) were studied in the reaction of DNA synthesis. Modified ONs I-VI (X is a residue of 2´-O-beta-D-ribofuranosyladenosine) were used as primers in the reaction of DNA synthesis catalyzed by HIV-1 reverse transcriptase (RT HIV-1) on an RNA template [64]. The synthesis was performed in the presence of all four dNTP. It should be mentioned that primers I-VI were effectively bound to the enzyme. The position of a modified unit in the ON chain substantially affected the primer elongation. Primer II was elongated with the same efficacy as the natural ON I. Primers III and IV were elongated considerably worse, and ON V was not elongated (Table 2). It is interesting to note that ON VI was effectively elongated only on one unit, that is, the presence of 2´-O-beta-D-ribofuranosyladenosine in positions -3 or -4 of from the 3´-end prevented the primer elongation.
Table 2. RT HIV-1-catalyzed elongation of
modified primers in the reaction of DNA synthesis on an RNA template
[64]
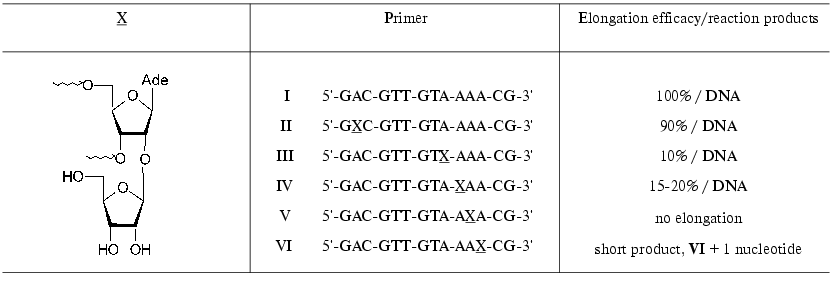
Computer modeling of the template-primer-enzyme complex was performed. The coordinates of RT HIV-1 tertiary structure [65] and decameric RNA duplex containing 2´-O-beta-D-ribofuranosyladenosine [62] were taken as a basis. For protein complexes of primer V considerable steric contacts with protein amino acid residues were observed, which did not escape when the extra ribose residue rotated around the O-glycosidic bond [64]. At the same time, in the complex formed by primer VI these contacts were insignificant. After the elongation of this primer by one nucleotide unit the modification was shifted into position -4, which topologically corresponded to primer V and resulted in the termination of the enzymatic reaction. The inhibition was a consequence of steric contacts of the extra bulky 2´-O-beta-D-ribofuranosyl residue with amino acid residues of the enzyme.
The preparation of modified ONs containing disaccharide nucleosides is promising for the development of selective inhibitors of polymerizing enzymes.
6. INTRODUCTION OF REACTIVE ALDEHYDE GROUPS INTO OLIGONUCLEOTIDES
AND APPLICATION OF THESE COMPOUNDS FOR AFFINITY MODIFICATION OF
ENZYMES
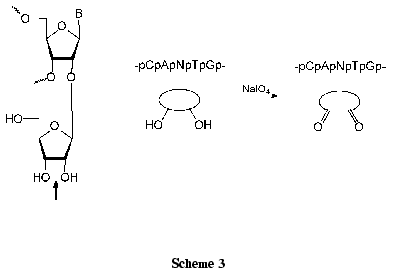
Regioselective introduction of additional functional groups into ONs open new possibilities for functionalization of nucleic acids. A cis-diol group of the extra ribofuranosyl residue can be smoothly transformed into a dialdehyde one (Scheme 3) [66]. The reaction procedure is very simple: nucleosides (at the concentration of about 5*10-2 M) are quantitatively oxidized with 1.2-1.3 equivalents of NaIO4 for 10-30 min at 20°C in an aqueous medium [66, 67]. Dinucleoside monophosphates and ONs are commonly oxidized with 10-100-fold excess of NaIO4 using HPLC control [60, 66, 68].
Aldehyde derivatives of nucleosides and nucleotides are widely used for affinity modifications of lysine residues of proteins [69-71]. Three major types of interaction of dialdehyde derivatives with proteins are known, but chemical mechanisms of these processes have not been conclusively elucidated [66]
These interactions have been reviewed [66].
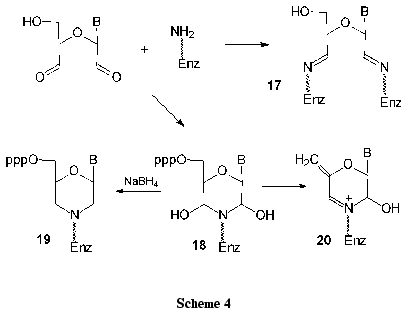
1. In the reaction of nucleoside dialdehyde derivatives with lysine amino groups of serum albumins, protein chains are joined to form conjugates 17 of high molecular mass. It is noteworthy that borohydride reduction is not compulsory for the preparation of stable complexes (Scheme 4).
2. Competitive inhibition can be referred to the second type. When unstable dihydroxymorpholine derivative 18 is formed, it can be reduced with borohydrides to give a stable morpholine derivative 19 (Scheme 4). After specific enzymatic or chemical cleavage of the modified protein, amino acids or protein regions of the active or nucleotide-binding sites can be localized.
3. In some cases the irreversible inhibition of enzymes accompanied by elimination of phosphate groups was observed. It is assumed that rather stable derivatives 20 are thereby formed.
Till recently oxidized nucleoside and nucleotide derivatives are used as affinity ligands. Due to the lack of regioselective methods of introduction of dialdehyde groups into various positions of the ON chain, the use of modified ONs was hampered and before our studies was limited to RNA derivatives modified at the 3´-end [66].
Recently we elaborated some general methods for introduction of dialdehyde groups into selected ON position. To this end, we used the incorporation of both disaccharide nucleosides [59-61] and galactopyranosylnucleosides [72, 73] into the ON chain. DNA duplexes bearing extra dialdehyde groups were used for affinity modification of T7 RNA polymerase [59] and restriction-modification enzymes EcoRII and MvaI [60, 61, 72-75]. It is worth mentioning that the authors used borohydride reduction for stabilization of enzyme complexes with modified DNA duplexes. The data obtained revealed the localization of DNA-binding sites of these enzymes.

Duplexes VII and VIII were obtained, in which one of the dC residues was replaced by an oxidized derivative of 2´-O-beta-D-ribofuranosylcytidine in flanking nucleotide sequences or in the recognition site. It was shown that modified ONs formed stable complexes with complementary DNA (complexes VII and VIII) (DeltaTm = 0-3°C) [61, 74]. All the duplexes were EcoRII and MvaI substrates independently of the modification position. The yields of affinity modification of these enzymes with duplexes VII and VIII were 2-6%. The subsequent chemical cleavage of the conjugates showed that the covalent addition took place at MvaI Val381-Met422 region [61] and EcoRII Gly268-Met391 region [75] irrespective of the position of the modification in the ON duplex.
To study T7 RNA polymerase and its mutants, modified ON duplexes IX and X, containing the T7 RNA polymerase promoter and five transcript bases in the message region were used as templates [59].
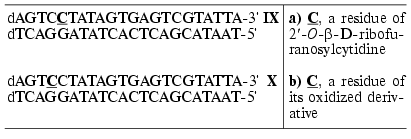
It was shown that duplexes IXa,b-Xa,b bearing 2´-O-beta-D-ribofuranosylcytidine or its oxidized derivative were competitive inhibitors of T7 RNA polymerase, the values of binding constants being 2-3.5 times lower than in the case of the reference unmodified duplex.
Duplexes IXb and Xb did not form covalent complexes with the enzyme of wild type. Affinity modification of T7 RNA polymerase mutant Tyr639Lys was only observed for duplex Xb. On this basis, it was concluded that Tyr639 was located in the protein region directly involved in transcription initiation.
The developed methods of introduction of extra functional groups into ONs, such as cis-diol and aldehyde groups, open new possibilities for functionalization of nucleic acids. In the modified ONs all the natural nucleic bases and the corresponding distances are retained, which is required for providing potential interactions with nucleic acids and proteins. The possibility of various labeling of the ON can also be mentioned. It can be assumed with confidence that the synthesized compounds will be widely used for studying protein-nucleic interactions.
The work was supported by the Russian Foundation for Basic Research and the program “Studies and Engineering on Foreground Branches of Science and Technology”.
REFERENCES
1.Dictionary of Natural Products on CD-ROM Version
9.2, February 2001. Chapman&Hall/CRC.
2.Limbach, P. A., Crain, P. F., and McCloskey, J. A.
(1994) Nucleic Acids Res., 22, 2183-2196.
3.Rozenski, J., Crain, P. F., and McCloskey, J. A.
(1999) Nucleic Acids Res., 27, 196-197.
4.The RNA Modification Database, University of Utah.
http://medlib.med.utah.edu/RNAmods/.
5.Hall, R. H. (1965) Biochemistry, 4,
661-670.
6.Simsek, M., and RajBhandary, U. L. (1972)
Biochem. Biophys. Res. Commun., 49, 508-514.
7.Ghosh, H. P., Ghosh, K., Simsek, M., and
Rajbhandry, U. L. (1982) Nucleic Acids Res., 10,
3241-3247.
8.Glasser, A.-L., Desgres, J., Heitzler, J., Gehrke,
C. W., and Keith, G. (1991) Nucleic Acids Res., 19,
5199-5203.
9.Keith, G., Heitzler, J., Adlouni, C. E., Glasser,
A.-L., Fix, C., Desgres, J., and Dirheimer, G. (1993) Nucleic Acids
Res., 21, 2949.
10.Desgres, J., Keith, G., Kuo, K. C., and Gehrke,
C. W. (1989) Nucleic Acids Res., 17, 865-882.
11.Keith, G., Glasser, A.-L., Desgres, J., Kuo, K.
C., and Gehrke, C. W. (1990) Nucleic Acids Res., 18,
5989-5993.
12.Kiesewetter, S., Ott, G., and Sprinzl, M. (1990)
Nucleic Acids Res., 18, 4677-4682.
13.Basavappa, R., and Sigler, P. B. (1991) J.
EMBO, 10, 3105-3111.
14.Chambon, P., Weil, J. D., Doly, J., Strosser, M.
T., and Mandel, P. (1966) Biochem. Biophys. Res. Commun.,
25, 638-643.
15.Fujimura, S., Hasegawa, S., and Sugimura, T.
(1967) Biochim. Biophys. Acta, 134, 496-499.
16.Nishizuka, Y., Ueda, K., Nakazawa, K., and
Hayaishi, O. (1967) J. Biol. Chem., 242, 3164-3171.
17.Suhadolnik, R. J., Baur, R., Lichtenwalner, D.
M., Uematsu, T., Roberts, J. H., Sudhakar, S., and Smulson, M. (1977)
J. Biol. Chem., 252, 4134-4144.
18.Sugimura, T. (1973) Progr. Nucleic Acids Res.
Mol. Biol., 13, 127-151.
19.Sugimura, T., and Miwa, M. (1994) Mol. Cell.
Biochem., 138, 5-12.
20.Miwa, M., and Sugimura, T. (1971) J. Biol.
Chem., 246, 6362-6364.
21.Ueda, K., Oka, J., Narumiya, S., Miyakawa, N.,
and Hayaishi, O. (1972) Biochem. Biophys. Res. Commun.,
46, 516-523.
22.Ferro, A. M., and Oppenheimer, N. J. (1978)
Proc. Natl. Acad. Sci. USA, 75, 809-813.
23.Oppenheimer, N. J., Sing, M., Sweeley, C. C.,
Sung, S. J., and Srere, P. A. (1979) J. Biol. Chem., 254,
1000-1002.
24.Tanaka, M., Hayashi, K., Sakura, H., Miwa, M.,
Matsushima, T., and Sugimura, T. (1978) Nucleic Acids Res.,
5, 3183-3194.
25.Miwa, M., Ishihara, M., Takishima, S., Takasuka,
N., Maeda, M., Yamaizumi, Z., Sugimura, T., Yokoyama, T., and Miyazawa,
T. (1981) J. Biol. Chem., 256, 2916-2921.
26.Isono, K. (1988) J. Antibiot., 41,
1711-1739.
27.Isono, K. (1991) Pharmacol. Ther.,
52, 269-286.
28.Garner, P. (1988) in Studies in Natural
Products Chemistry (Atta-ur-Rahman, ed.) Vol. 1, Elsevier,
Amsterdam, pp. 397-434.
29.Lerner, L. M. (1991) in Chemistry of
Nucleosides and Nucleotides (Townsend, L. B., ed.) Vol. 2, Plenum
Press, New York, pp. 27-79.
30.Knapp, S. (1995) Chem. Rev., 95,
1859-1876.
31.Farkas, J., Sebesta, K., Horska, K., Samek, Z.,
Dolejs, L., and Sorm, F. (1969) Coll. Czech. Chem. Commun.,
34, 1118-1120.
32.Kalvoda, L., Prystas, M., and Sorm, F. (1973)
Tetrahedron Lett., 1873-1876.
33.Farkas, J., Sebesta, K., Horska, K., Samek, Z.,
Dolejs, L., and Sorm, F. (1977) Coll. Czech. Chem. Commun.,
42, 909-929.
34.Sebesta, K., Horska, K., and Vancova, J. (1969)
Coll. Czech. Chem. Commun., 34, 891-900.
35.Bond, R. P. M., Boyce, C. B. C., and French, S.
J. (1969) Biochem. J., 114, 477-488.
36.Sebesta, K., and Sternbah, H. (1970) FEBS
Lett., 8, 233-235.
37.Takahashi, M., Kagasaki, T., Hosoya, T., and
Takahashi, S. (1993) J. Antibiot., 46, 1643-1647.
38.Takahashi, S., Kinoshita, T., and Takahashi, M.
(1994) J. Antibiot., 47, 95-100.
39.Takahashi, M., Tanzawa, K., and Takahashi, S.
(1994) J. Biol. Chem., 269, 369-372.
40.Marchant, J. S., Beecroft, M., Riley, A.,
Jenkins, D., Marwood, R., Taylor, C., and Potter, B. (1997)
Biochemistry,36, 12780-12790.
41.Katoh, H., Yamada, H., Iida, K., Aoki, M.,
Itezono, Y., Nakayama, N., Suzuki, Y., Watanabe, M., and Shimada, H.
(1996) Chem. Abst., 125, 322575y.
42.Sugimura, H., and Stansfield, K. (1998)
Synlett, 985-986.
43.McCormick, J., Li, Y., McCormick, K., Duynstee,
H. I., van Engen, A. K., van der Marel, G. A., Ganem, B., van Boom, J.
H., and Meinwald, J. (1999) J. Am. Chem. Soc., 121,
5661-5665.
44.Mikhailov, S. N., de Bruyn, A., and Herdewijn, P.
(1995) Nucleosides, Nucleotides, 14, 481-484.
45.Mikhailov, S. N., Efimtseva, E. V., Gurskaya, G.
V., Zavodnik, V. E., de Bruyn, A., Janssen, G., Rozenski, J., and
Herdewijn, P. (1997) J. Carbohydr. Chem., 16, 75-92.
46.Markiewicz, W. T., and Wiewiorowski, M. (1986) in
Nucleic Acid Chemistry, Improved and New Synthetic
Procedures, and Techniques. Part 3 (Townsend, L. B., and
Tipson, R. S., eds.) Wiley, New York, p. 229.
47.Vorbruggen, H., and Ruh-Pohlenz, C. (2000)
Organic Reactions. Synthesis of Nucleosides (Paquette, L.
A., et al., eds.) Vol. 55, Wiley, New York.
48.Hanessian, S., and Banoub, J. (1980) in
Methods in Carbohydrate Chemistry, Academic Press, New York, p.
243.
49.Mikhailov, S. N., de Clercq, E., and Herdewijn,
P. (1996) Nucleosides, Nucleotides, 15,
1323-1334.
50.Mikhailov, S. N., Rodionov, A. A., Efimtseva, E.
V., Fomitcheva, M. V., Padyukova, N. Sh., Herdewijn, P., and Oivanen,
M. (1997) Carbohydr. Lett., 2, 321-328.
51.Rodionov, A. A., Efimtseva, E. V., Fomitcheva, M.
V., Padyukova, N. Sh., and Mikhailov, S. N. (1999) Bioorg.
Khim., 25, 204-211.
52.Mikhailov, S. N., Rodionov, A. A., Efimtseva, E.
V., Ermolinsky, B. S., Fomitcheva, M. V., Padyukova, N. Sh.,
Rothenbacher, K., Lescrinier, E., and Herdewijn, P. (1998) Eur. J.
Org. Chem., 2193-2199.
53.Efimtseva, E. V., Rodionov, A. A., Bobkov, G. V.,
Ermolinsky, B. S., Fomitcheva, M. V., Mikhailov, S. N., and Herdewijn,
P. (1999) Collection, Symp. Ser., 2, 22-26.
54.Efimtseva, E. V., Bobkov, G. V., Mikhailov, S.
N., van Aerschot, A., Schepers, G., Busson, R., Rozenski, J., and
Herdewijn, P. (2001) Helv. Chim. Acta, 84, 2387-2397.
55.Rodionov, A. A., Efimtseva, E. V., Mikhailov, S.
N., Rozenski, J., Luyten, I., and Herdewijn, P. (2000)
Nucleosides, Nucleotides Nucleic Acids, 19,
1847-1859.
56.Oivanen, M., Hovinen, J., Lehikoinen, P., and
Lonnberg, H. (1993) Trends Org. Chem., 4, 397-412.
57.Efimtseva, E. V., Ermolinsky, B. S., Fomitcheva,
M. V., Meshkov, S. V., Padyukova, N. Sh., Mikhailov, S. N., van
Aerschot, A., Rozenski, J., and Herdewijn, P. (1996) Coll. Czech.
Chem. Comm. Spec. Issue, 61, 206-209.
58.Efimtseva, E. V., Victorova, L. S., Rodionov, A.
A., Ermolinsky, B. S., Fomitcheva, M. V., Mikhailov, S. N., Oivanen,
M., van Aerschot, A., and Herdewijn, P. (1998) Nucleosides,
Nucleotides, 17, 1681-1684.
59.Tunitskaya, V. L., Rusakova, E. E., Memelova, L.
V., Kochetkov, S. N., van Aerschot, A., Herdewijn, P., Efimtseva, E.
V., Ermolinsky, B. S., and Mikhailov, S. N. (1999) FEBS Lett.,
442, 20-24.
60.Mikhailov, S. N., Ermolinsky, B. S., Efimtseva,
E. V., Moiseyev, G. P., Tunitskaya, V. L., Rusakova, E. E., Kochetkov,
S. N., Gritsenko, O. M., Gromova, E. S., van Aerschot, A., and
Herdewijn, P. (1999) Collection, Symp. Ser.,2,
141-144.
61.Gritsenko, O. M., Mikhailov, S. N., Efimtseva, E.
V., van Aerschot, A., Herdewijn, P., and Gromova, E. S. (2000)
Nucleosides, Nucleotides Nucleic Acids, 19,
1805-1820.
62.Luyten, I., Esnouf, R. M., Mikhailov, S. N.,
Efimtseva, E. V., Michiels, P., Heus, H. A., Hilbers, C. W., and
Herdewijn, P. (2000) Helv. Chim. Acta, 83, 1278-1289.
63.Manoharan, M. (1999) Biochim. Biophys.
Acta, 1489, 117-130.
64.Andreeva, O. I., Golubeva, A. S., Kochetkov, S.
N., van Aerschot, A., Herdewijn, P., Efimtseva, E. V., Ermolinsky, B.
S., and Mikhailov, S. N. (2002) Bioorg. Med. Chem. Lett.,
12, 681-684.
65.Jacobo-Molina, A., Ding, J., Nanni, R. G., Clark,
A. D., Jr., Lu, X., Tantillo, C., Williams, R. L., Kamer, G., Ferris,
A. L., Clark, P., Hizi, A., Hughes, S. H., and Arnold, E. (1993)
Proc. Natl. Acad. Sci. USA, 90, 6320-6324.
66.Ermolinsky, B. S., and Mikhailov, S. N. (2000)
Bioorg. Khim., 26, 483-504.
67.Mikhailov, S. N., and Yakovlev, G. I. (1987)
Khim. Prirod. Soedin., 40-43.
68.Ermolinsky, B. S. (2000) Candidate's dissertation
[in Russian], IMB, Moscow.
69.Gritsenko, O. M., and Gromova, E. S. (1999)
Uspekhi Khim., 68, 267-278.
70.Vlasov, V. V., and Kazakov, S. A. (1983) in
Affinity Modification of Polymers (Knorre, D. G., ed.) Nauka,
Novosibirsk, pp. 7-56.
71.Colman, R. F. (1983) Ann. Rev. Biochem.,
52, 67-91.
72.Mikhailov, S. N., Efimtseva, E. V., Ermolinsky,
B. S., Fomitcheva, M. V., Gritsenko, O. M., Brevnov, M. G., Gromova, E.
S., Schepers, G., van Aerschot, A., and Herdewijn, P. (1996)
Collection. Spec. Issue, 61, 210-212.
73.Brevnov, M. G., Gritsenko, O. M., Mikhailov, S.
N., Efimtseva, E. V., Ermolinsky, B. S., van Aerschot, A., Herdewijn,
P., Repyk, A. V., and Gromova, E. S. (1997) Nucleic Acids Res.,
25, 3302-3309.
74.Gritsenko, O. M. (2000) Candidate's dissertation
[in Russian], Moscow State University, Moscow.
75.Gritsenko, O. M., Koudan, E. V., Mikhailov, S.
N., van Aerschot, A., Herdewijn, P., and Gromova, E. S. (2002)
Nucleosides, Nucleotides Nucleic Acids, 21,
753-764 (2002).