Peptide Derivatives of Antibiotics Tylosin and Desmycosin, Protein Synthesis Inhibitors
N. V. Sumbatyan1, G. A. Korshunova2, and A. A. Bogdanov1,2*
1Faculty of Chemistry, Lomonosov Moscow State University, Moscow 119992, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow 119992, Russia; fax: (7-095) 939-3181; E-mail: bogdanov@belozersky.msu.ru
* To whom correspondence should be addressed.
Received July 1, 2003
Biologically active peptide derivatives of 16-member macrolide antibiotics were synthesized as potential probes for the investigation of nascent peptide chain topography in the ribosomal exit tunnel. The tylosin and desmycosin aldehyde groups at the C6 position of the lactone ring were modified by the aminooxyacetyl-L-alanyl-L-alanine methyl ester.
KEY WORDS: antibiotics, macrolides, tylosin, desmycosin, peptide derivatives of macrolides, ribosomal tunnel
During the protein biosynthesis process, the nascent polypeptide chain moves step by step through the ribosomal tunnel (RT), which is located inside of the large ribosomal subunit [1]. The RT is constructed predominantly of nucleotide residues of ribosomal RNA, and its upper region is formed by the nucleotides included in peptidyl transferase center (PTC) of the ribosome. The mechanism of transfer and conformation of the growing polypeptide in the RT are unknown. The only fact revealed is that its C-terminal dipeptide portion has beta-strand structure stabilized by the specific contact with the walls of the RT [2].
Recently, X-ray analysis was used to determine that inside the RT, approximately 20 Å from the PTC, there is a binding center of macrolides, which are widely known translation inhibitors [3, 4]. Macrolides are the group of natural or semi-synthetic antibiotics whose structure is based on 14-16-member lactone rings with carbohydrate substitutes attached. Macrolides are bound to RT in the way that their lactone ring is located orthogonally to the long axis of the RT, covering most of its cleft. Carbohydrate residues of the macrolides are located along the walls of the RT. Hence, the mechanism of protein synthesis inhibition by macrolides relies on the mechanical obstruction they provide to the passage of nascent polypeptide chain through the RT [3-5].
The goal of this study was to obtain peptide derivatives of macrolides where the peptide part would model the growing chain, while the antibiotic would serve as an “anchor” for positioning the peptide at the specific site of the RT. Tylosin and desmycosin were used as macrolides. Tylosin (Scheme, structural formula 1) contains two carbohydrate substitutes: disaccharide formed of mycaminose and mycarose residues at position 5 of the 16-member lactone ring, and mycinose residue at position 14; desmycosin (2) differs from tylosin by the absence of the mycarose residue. These macrolides contain a reactive acetaldehyde group at position 6 of the ring. To date, there are a large number of tylosin and desmycosin derivatives modified by the aldehyde group; it was also shown that these compounds retain antibacterial activity [6-9]. However, peptide derivatives of tylosin and desmycosin were yet not obtained.
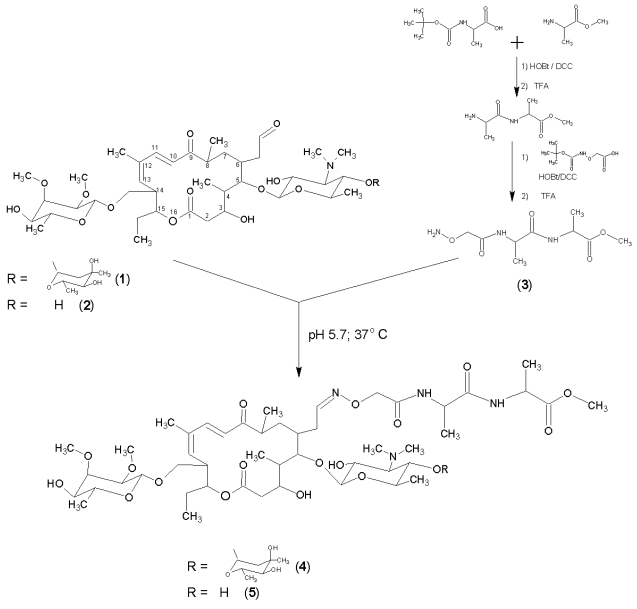
An aldehyde group can be modified via the formation of a substituted oxime. For this purpose a peptide containing a hydroxylamine function was synthesized: based on Boc-alanine and alanine methyl ester a methyl ester of the protected dipeptide was obtained, which (after the removal of the Boc-group) was condensed with aminooxyacetic acid. Aminooxyacetyl-L-alanyl-L-alanine methyl ester (3) was obtained after the unblocking of the peptide. The synthesis of this derivative was performed in the solution of dimethylformamide using N,N´-dicyclohexylcarbodiimide as a condensing agent in the presence of 1-hydroxybenzotriazole. Boc-protecting groups were removed by trifluoroacetic acid.
It was previously revealed that tylosin is stable in water solution within the pH range 4 to 9, and at pH < 4 it transforms into desmycosin [10]. This property was utilized by us to obtain the latter: tylosin water solution, pH 3, was kept for a day at room temperature, then the solution was neutralized and the product extracted with chloroform, crystallized from ether, and the compound structure was verified by TLC and HPLC; MALDI MS (found/calc.) 772.2/771.83. The chemical yield was 87%.
Condensation of antibiotics with aminooxyacetyl-L-alanyl-L-alanine methyl ester via aldehyde group was performed under conditions analogous to those developed for ligation with the formation of oxime for other types of compounds [11-13]. The reaction was performed in water-organic medium at pH 5.7 (0.4 M Na-acetate-dimethylformamide 1 : 1); initial concentrations of tylosin and desmycosin were 0.02 M, the mixture was incubated at 37°C for 12 h. The resulting tylosin (4) and desmycosin (5) derivatives were extracted with chloroform and crystallized from ether; the yields were 55 and 47%, accordingly. Products 4 and 5 were further purified for the biological studies employing preparative TLC on silica gel plates in chloroform-methanol system (7 : 3 v/v). The purity of the obtained compounds was verified by TLC and HPLC. The structure was confirmed by mass-spectrometry (MALDI MS (found/calc.): 4) 1145.3/1145.17; 5) 1001.2/1001.05) and quantitative amino acid analysis techniques, which indicated the presence of alanine residues in products 1 and 2 in required proportions.
Preliminary analysis revealed that the peptide derivatives of tylosin and desmycosin inhibit protein synthesis in non-cellular transcription-translation system with efficiency comparable to the unmodified antibiotics. Hence, the obtained compounds are promising for use as probes for topography studies of nascent polypeptide chain in the ribosomal tunnel.
The authors are grateful to Dr. Paola Fucini (Max Planck Institute for Molecular Genetics, Berlin, Germany) for the determination of biological activity of the obtained compounds. A. A. Bogdanov thanks the Alexander von Humboldt Foundation for support.
REFERENCES
1.Nissen, P., Hansen, J., Ban, N., Moore, P. B., and
Steitz, T. A. (2000) Science, 289, 920-930.
2.Bogdanov, A. A. (2003) Mol. Biol. (Moscow),
37, 1-4.
3.Schlunzen, F., Zarivach, R., Harms J., Bashan, A.,
Tocilj, A., Albrecht, R., Yonath, A., and Franceschi, F. (2001)
Nature, 413, 814-821.
4.Hansen, J., Ippolito, J. A., Ban, N., Nissen, P.,
Moore, P. B., and Steitz, T. A. (2002) Moll. Cell, 10,
117-128.
5.Gaynor, M., and Mankin, A. S. (2003) Curr. Top.
Med. Chem., 3, 949-961.
6.Matsubara, H., Inokoshi, J., Nakagawa, A., Tanaka,
H., and Omura, S. (1983) J. Antibiot. (Tokyo), 36,
1713-1721.
7.Debono, M., Willard, K. E., Kirst, H. A., Wind, J.
A., Crouse, G. D., Tao, E. V., Vicenzi, J. T., Counter, F. T., Ott, J.
L., and Ose, E. E. (1989) J. Antibiot. (Tokyo), 42,
1253-1267.
8.Kirst, H. A., Toth, J. E., Debono, M., Willard, K.
E., Truedell, B. A., Ott, J. L., Counter, F. T., Felty-Duckworth, A.
M., and Pekarek, R. S. (1988) J. Med. Chem., 31,
1631-1641.
9.Hranjec, M., Starcevic, K., Zamola, B., Mutak, S.,
Derek, M., and Karminski-Zamola, G. (2002) J. Antibiot. (Tokyo),
55, 308-314.
10.The Merck Index (9 ed.) (1976) Merck and
Co., Inc., Rahway, New York.
11.Vandersse, R., Thevenet, L., Marraud, M.,
Boggetto, N., Reboud, M., and Corbier, C. (2003) J. Peptide
Sci., 9, 282-299.
12.Ingallinella, P., Di Marco, A., Taliani, M.,
Fattori, D., and Pessi, A. (2001) Bioorg. Med. Chem. Lett.,
11, 1343-1346.
13.Zatsepin, T. S., Stetsenko, D. A., Arzumanov, A.
A., Romanova, E. A., Gait, M. J., and Oretskaya, T. S. (2002)
Bioconjug. Chem., 13, 822-830.