REVIEW: Biochemical Problems of Regulation by Oligopeptides
A. A. Zamyatnin
Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, Moscow 119071, Russia; fax: (7-095) 954-2732; E-mail: aaz@inbi.ras.ruCurrent address: Universidad Tecnica Federico Santa Maria, Avenida Espana 1680, Valparaiso, Chile; E-mail: aaz@inf.utfsm.cl
Received July 9, 2004
Some problems are considered which arise in biochemical studies on structure and function of natural oligopeptides consisting of 2-50 amino acid residues. The problems under consideration include the generation of oligopeptides from precursors, chemical structure, the role of functionally important radicals and spatial configuration, and structure-function relationships. Different types of regulation are shown mainly for oligopeptides involved in muscle contraction.
KEY WORDS: oligopeptide, precursor, content, receptor, chemical structure, homology, conformation, function, activity, regulation, interaction, enzyme, muscle contraction, EROP-Moscow knowledge database, alignment
Natural oligopeptides consisting of 2-50 amino acid residues [1, 2] are found in virtually all living organisms. Their functions are so diverse that it is difficult to point out a vitally important process that occurs without their involvement. These oligopeptides are generally called regulatory, because the majority of them are involved in regulation of physiological processes in all regulatory systems: nervous, endocrine, and immune [3]. Moreover, their regulatory functions extend beyond the limits of a separate organism or biological species. Thus, oligopeptide toxins of eukaryotes obviously play an important role in the regulation of interspecies relationships, and antimicrobial oligopeptides of prokaryotes regulate competition in occupation of ecological niches and act as signal molecules in intercellular communications [4].
About 6000 natural oligopeptides are known that are produced by natural ribosomal template synthesis [5]. This number has to be increased by addition of many natural structures produced in bacteria and fungi as a result of non-ribosomal synthesis and which, as a rule, contain nonstandard amino acid residues often indescribable by the standard one-letter amino acid code. In particular, more than 300 bacterial oligopeptides with a high content of aminoisobutyric acid residue (peptaiboles) are already known [6]. Such studies are reported in more than 20,000 publications per year, with descriptions of about 500 new structures of this chemical class.
Most oligopeptides can be isolated and purified, and their chemical structure can be directly determined. But many amino acid sequences are also revealed using the translation of the corresponding nucleotide sequences.
The standard strategy of studies on natural oligopeptides usually includes the determination of a functionally important component, its purification, isolation, determination of chemical structure, synthesis, and verification of functional properties of the substance. The spatial structure of some oligopeptides can also be determined. Thus, the variety of these natural substances is studied with participation of biochemists, physiologists, physicists, and many other specialists who have an interest in problems of the structure and functions of oligopeptides. Physicians and pharmacologists also pay increasing attention to oligopeptides.
Study of the relationships between the structure and function of natural compounds is one of central topics of biochemistry and biophysics. We have discussed earlier the biophysical aspects of regulation by oligopeptides [7], in particular, the functionally significant size of oligopeptides has been considered, as well as difficulties arising in attempts to classify them. The time of delivery of an oligopeptide molecule to its receptor structure was also calculated theoretically. But problems of regulation by oligopeptides are not limited by this. Therefore, the present work considers the unique chemical and structural features of oligopeptide molecules that provide their involvement in primary mechanisms of functional reactions of a living organism.
In the following description of biochemical problems of regulation by oligopeptides, we have used data of the EROP-Moscow (Endogenous Regulatory OligoPeptides) [5] computer databank, which contains functional and other characteristics of about 6000 natural oligopeptides mainly prepared by template synthesis. Such a great number of known natural oligopeptides and the problems associated with their investigation cannot be described in a single article; therefore, we shall consider the most urgent problems and the most typical examples. In memory of B. F. Poglazov, who largely dedicated his life to the problem of muscle contraction, we shall most often focus our attention on myotropic oligopeptides.
BIOGENESIS
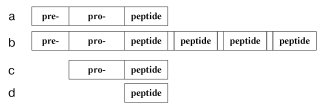
Oligopeptide regulation is started by production of oligopeptides as they are. In general, the production of oligopeptide molecules is characterized by some stages. First, translation results in a pre-pro-peptide (Fig. 1) that can contain several hundreds of amino acid residues. Then a fragment pre- (signaling peptide) is detached which is located on the N-terminal and usually consists of 15-30 amino acid residues. The remaining pro-peptide, which is a precursor of oligopeptides, can include one (Fig. 1a) and more (Fig. 1b) identical or different regions often separated by dipeptide pairs (the flanking) containing lysine or arginine residues. The fragments resulting by detachment of these pairs are oligopeptides. For example, a precursor of the mollusk's tetrapeptide FMRFamide contains 28 identical copies of this oligopeptide [8], whereas from a precursor of the mammalian oligopeptide four molecules of met-enkephalin and one molecule of leu-enkephalin are released [9]. However, the pre- and/or pro-regions can be absent in prokaryotes. Then either a pro-peptide (Fig. 1c) is produced, or the peptide chain is a finished oligopeptide (Fig. 1d). Biogenesis of a pro-peptide free of the pre-region is exemplified by generation of bacteriocins piscicolin JG126 (Cronobacterium piscicola) [10] and sakacin P (Lactobacillus sakei) [11]. The complete coincidence of the peptide and precursor structures is shown by generation of the ATP-binding peptide lceA in the bacterium Lactococcus lactis [12] and entericidin B isolated from Salmonella typhimurium [13].
However, this is not the end of oligopeptide production. Many oligopeptides display a concurrent existence in multiple forms produced as a result of enzymatic reactions. The coexistence of a finished oligopeptide and various proteolytic enzymes leads to further degradation resulting in still shorter fragments of the initial molecule.Fig. 1. Scheme of the structure of a natural oligopeptide precursor: a) the general simplified image; b) precursor of several oligopeptides flanked by arginine/lysine residues (empty regions); c) pro-peptide of prokaryotes; d) an oligopeptide precursor of prokaryotes.
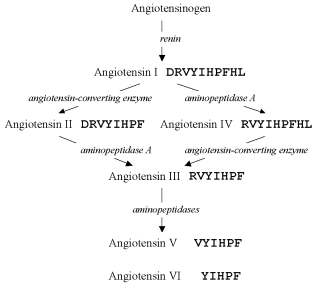
The amino acid sequence of one of the first described oligopeptides, angiotensin (earlier called hypertensin), was deciphered, and in 1956 two structures were found containing ten and eight amino acid residues which were designated angiotensin I and II, respectively [14]. But at least four forms of this oligopeptide, angiotensins III-VI, were found later [15]. Comprehensive studies have revealed the pathways of enzymatic degradation of preceding structures, which result in these forms (Fig. 2), and their concurrent existence has been confirmed.
Production of multiple oligopeptide forms was later shown to be rather a common phenomenon, for example, for somatostatin [16-18], atrial natriuretic peptide [19, 20], etc. Therefore, thousands of the known oligopeptides probably also exist in multiple forms, but these forms are not yet identified.Fig. 2. Pathways of angiotensin conversions. Enzymes involved in the process are shown in italics.
Note, that other types of multiple oligopeptide forms are also widely distributed. The same organism can present coexisting homologous oligopeptide structures with identical functional properties. Their amino acid sequences contain unit substitutions, however, and they are encoded by different genes with different nucleotide sequences and are produced independently, and not as a result of posttranslational modification. Such are, for example, three homologs of the peptide YY of sturgeon [21]. Moreover, alternative splicing resulting in multiple forms of mRNA can produce oligopeptides with similar structure and functions, such as calcitonins [22] or glucagons [23].
CHEMICAL STRUCTURE
Comprehensive knowledge of the chemical structure of an oligopeptide is based on determination of its amino acid sequence, detection of modifications of its N- and C-terminals, other posttranslational modifications, and also of S-S and other internal chemical bonds between distant amino acid residues, including the additional peptide bond produced upon cyclization.
The direct determination of exact chemical structure of oligopeptides is in many cases associated with great difficulties because their natural content often is only 10-15-10-12 M [24]. The location of oligopeptides in the cell is also associated with problems. Therefore, highly sensitive biochemical and immunohistochemical techniques are used for analysis.
However, determination of the amino acid sequence produced by translation from the corresponding nucleotide sequence needs no highly precise analytical biochemical methods. But many probable posttranslational modifications, which occur in about half of the known oligopeptides, remain unfound by this approach (Table 1).
Table 1. Chemical modifications of natural
oligopeptides (% of the total number of known oligopeptides)
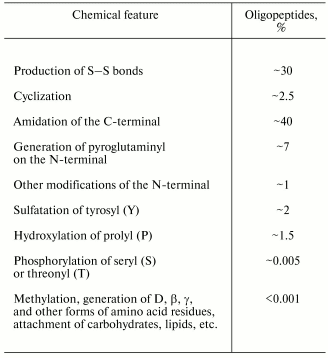
The simplest modification (but not the commonest one) is the binding of the N- and C-terminal amino acid residues of an oligopeptide with a new peptide bond. This results in the maximally possible macrocycle for the molecule. In this case, concepts of the N- and C-terminals became meaningless, and if the primary structure of the precursor is unknown, the problem arises of the starting-point for reading the amino acid sequence.
About one third of known oligopeptides contains cysteinyl pairs, i.e., disulfide bonds, and there can be six such bonds in one oligopeptide. In the case of many S-S bonds the structure presents several macrocycles and appears to form a complicated knot, which freezes the majority of degrees of freedom. Some cyclic oligopeptides also have S-S bonds.
Amidation of the C-terminal is the commonest post-translational modification in oligopeptides. This modification is realized with involvement of different enzymes [25] or non-enzymatically [26], and may be detected without analytical biochemical techniques because usually amidation covers the amino acid residue X which is followed by glycine (G), arginine (R), or lysine (K) residues that results in the reaction:
A little less often the N-terminal is modified: the lateral radical in the glutamine residue (Q) on this terminal is non-enzymatically locked on the N-terminal group, with production of pyroglutaminyl [27, 28]:
This modification occurs in the majority of N-terminal glutamine residues; therefore, as a rule, no additional biochemical analysis is needed to predict it after translation of the nucleotide sequence.
Altogether, ~100 natural chemical modifications of amino acid residues are known in oligopeptides.
SPATIAL STRUCTURE
Oligopeptides, especially those without covalent bonds between distant amino acid residues, have great conformational mobility. The same molecule in a free state can change its configuration with time. And in the totality of similar molecules, each one can have varied conformation at the same moment. This, in particular, explains difficulties in attempts to crystallize oligopeptides [29] for X-ray diffraction analysis.
The possible conformational variety of the same oligopeptide molecule is also shown by computerized modeling with many programs providing minimization of energy and optimization of the molecular configuration [7, 30].
X-Ray diffraction analysis of enkephalins is an example of experimental study on the spatial structure of oligopeptides. Thus, four spatial shapes, from elongated to packed, were found in crystals of the pentapeptide leu-enkephalin [31]. Based on this finding, it was suggested that this molecule, depending on its conformation, could selectively bind to one or another subtype of opiate receptor.
Varied shapes of the same molecule were revealed by NMR-spectroscopy for another opioid pentapeptide, met-enkephalin [32]. Moreover, by the same method this compound in solution was shown to exist as a dimer. But it should be noted that during both X-ray diffraction analysis and NMR-spectroscopy molecules of oligopeptides under study were under conditions very unlike those that are specific for a living organism. Therefore, it is still unlikely that we know all possible conformational states of these and other oligopeptides which are important for their regulatory functions.
Structures obtained either experimentally or by theoretical modeling fail to represent all specific features of configuration of oligopeptide molecules under real conditions. X-Ray diffraction analysis reveals only the conformations prepared by crystallization, and data of computerized modeling strongly depend on the quality of programs, which use approximate estimates in calculations for various types of intramolecular interactions in oligopeptides. Therefore, most of these data cannot give a reliable concept about molecular configurations, especially those taken by oligopeptides on interactions with receptor structures.
FUNCTIONALLY IMPORTANT CHEMICAL GROUPS
Considering the specificity of functional activity of oligopeptides, it was supposed that amino acid compositions of oligopeptide and protein molecules should be different [33, 34]. The average amino acid composition of regulatory oligopeptides was compared with the total composition of proteins [35-37], and for some functional classes a rather noticeable difference was found. Some of these classes were characterized by a significant number of amino acid residues containing positively charged (K, R) and cyclic (F, H, P, W, Y) radicals. The prevailing of some of these residues, especially of positive ones, along with a decreased content of negatively charged residues, was the most pronounced in various antimicrobial oligopeptides [38] and oligopeptide toxins [39]. Similar specific features, although less pronounced, were also found in neuropeptides [40] and the oligopeptide hormones liberins and statins [41].
The existence of certain receptor structures (receptors, ion channels, etc.) suggests a specific interaction between an oligopeptide (e.g., a hormone or neuropeptide) and the corresponding target. The interaction can be two-step [40]. At first, electrostatic interaction of positive charges of the oligopeptide ligand with negative charges of the receptor structure results in their nonspecific approximation. Then an adequate conformational rearrangement of the ligand occurs (the recognition) which includes the adjustment of the elements involved in the recognition of functionally important radicals using various weak interactions, e.g., by production of intermolecular hydrogen bonds, by stacking, and hydrophobic interactions.
However, no such feature was found in oligopeptide immunoregulators [42]. This is possibly because the immune response involves many organs and specialized cells, with a great variety of still insufficiently known functional relationships between them. These relationships are realized by both specific oligopeptide immunoregulators and oligopeptides of other functional classes, in particular, neuropeptides. This circumstance prevents the elucidation of specific features of the totality of oligopeptide regulators of the immune system. Such specific features can probably exist, but they are not recognizable on the level of immune regulation as a whole, although they can be revealed on the level of individual binding of oligopeptides with the definite structures involved in this regulation. This problem will be solvable when a clear concept is proposed about structural organization of the whole immune system and of its separate components including oligopeptides.
RELATIONSHIP OF STRUCTURE AND FUNCTION
The concepts of “structure” and “function” are widely used in studies of biologically active substances. However, considering structural and functional properties of oligopeptides, we conclude that these concepts are not always accurately defined and are rather ambiguous. Thus, the concept of structure more often concerns only the chemical structure (the primary one), not paying attention to the spatial structure. As to function, the definition of this term meets significant difficulties, and a generalized definition of function is absent in the relevant dictionaries [43, 44]. The possible reason of this is that specific and multiform activities, which underlie biological functions, occur at different levels of organization, from molecular to interspecies, and cannot be correctly generalized.
Nevertheless, elucidation of the dependence of function on the structure of oligopeptide, which includes various testing of natural structures and also their synthetic fragments and analogs, is the subject of the majority of both in vivo and in vitro studies. However, in both cases the functional reaction seems to start from a certain elementary interaction of an oligopeptide with a certain available receptor structure. The biological function can consist either of separate acts exemplified by in vitro tests on contraction of muscle strips or many elementary events for in vivo experiments. In the latter case, it is difficult to interpret in detail the mechanism of oligopeptide effect.
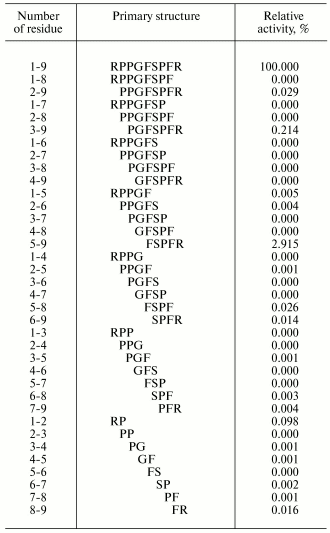
Attempts have been made in many works to find the region of an oligopeptide responsible for the functional reaction induced. For example, effects of various regions of the bradykinin molecule on contraction of smooth muscles of guinea pig ileum have been thoroughly studied. For this study, all 35 bradykinin fragments were synthesized (Table 2) [45]. And these fragments were either completely inactive, or their activities were insignificant as compared to the initial bradykinin. Similar findings were obtained also upon various replacements of individual amino acid residues [46]. Consequently, the whole bradykinin molecule consisting of nine amino acid residues determines the functional response.
Table 2. Primary structures and activities
of bradykinin (1-9) and all its possible fragments in the test of
muscle contraction of the guinea pig ileum [45]
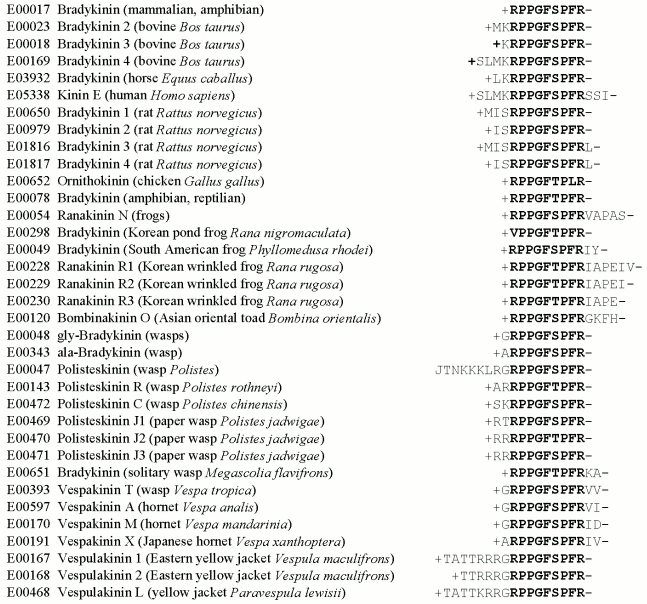
Computer-aided studies result in the same conclusion. To obtain it, one can isolate from the EROP-Moscow oligopeptide databank structures of all known bradykinins prepared from various living organisms and perform the alignment procedure. The result (Fig. 3) suggests that this bradykinin family should have a quasi-conservative region of nine amino acid residues, and this region is essentially unchanged in different bradykinins, in spite of the evolutionary remoteness of the organisms from which these bradykinins have been isolated. The natural elongation of the bradykinin chain virtually has no effect on its activity. Note also, that the quasi-conservative region of bradykinins includes amino acid residues containing functionally important groups (positively charged and cyclic radicals) that are virtually absent outside this region.
It seems that families of more homologous oligopeptides should be able to induce the same type of activity. In most cases this really occurs. However, there are also structurally homologous families of oligopeptides with strongly different functional activities. In particular, such are the endothelins (vasoconstrictor substances of mammals) and the strongest toxins (sarafotoxins) of snake venoms [47], which as a result of the alignment form the same family. It may be that the structural homology of the family resulted by the alignment concerns only the amino acid residue sequence and not the spatial configuration taken by the molecule on interaction with the receptor structure.Fig. 3. The structurally homologous family of bradykinins. The oligopeptide number in the EROP-Moscow databank, name, the source of isolation, and the primary structure are presented. Names of oligopeptides and animals are given correspondingly to international notations. Amino acid residues are shown by one-letter abbreviations; J is the pyroglutaminyl residue, “plus” is the N-terminal and “minus” is the C-terminal radicals. The quasi-conservative region is shown in bold.
And likeness of spatial structure elements seems to underlie manifestations of the same activity by oligopeptides from different families. Thus, contraction of muscles of the guinea pig ileum can be caused not only by bradykinin, but also by neuropeptides, such as neurotensin, angiotensin, or cholecystokinin 8 [7].
The continuous appearance of many new data on the structure of oligopeptides and receptors allows us to expect that mechanisms of their functioning will be soon be found in detail, and this will permit detecting the most effective structures of physiologically active substances promising for medicine.
Obviously, the problems described do not exhaust all challenges for biochemistry in the field of regulation by oligopeptides. Studies on isolated systems seem to be the most successful. However, in real living systems this regulation is much more complicated, because they have concurrently co-located not one ligand type and not one receptor type [48]. And not only the oligopeptide under consideration can exist in multiple conformations, moreover, it is encircled by water molecules, inorganic ions, and many other substances, including other oligopeptides, proteins, nucleotides, lipids, sugars. Besides, many oligopeptides are polyfunctional (pluripotent), and they may be considered as regulators of nervous, endocrine, and immune systems and are responsible for sophisticated connections between these systems. Complicated regulations are also displayed by a number of known cascade processes [49] when a single oligopeptide molecule provides for release of many other molecules, and this process is repeated.
Thus, the solution of some problems of regulation by oligopeptides is associated with a scrupulous taking into account of mechanisms involved in functioning of the pool of peptide and non-peptide endogenous substances, i.e., these problems are of higher order and are waiting to be solved.
The author is deeply grateful to A. S. Borchikov and M. G. Vladimirov for participation in development of specialized computer programs for the alignment procedure based on given similarity parameters and for obtaining the results of computer-aided analysis.
This work was supported by the Russian Foundation for Basic Research, project No. 02-07-90175, and the Russian Academy of Sciences Program RAN-10P “Molecular and Cellular Biology”.
REFERENCES
1.Zamyatnin, A. A. (1990) Neirokhimiya,
9, 71-82.
2.Zamyatnin, A. A. (1991) Prot. Seq. Data
Anal., 4, 49-52.
3.Sewald, N., and Jakubke, H.-D. (2002) Peptides:
Chemistry and Biology, Wiley-VCH Verlag GmbH, Weinheim.
4.Woo, P. C., To, A. P., Lau, S. K., and Yuen, K. Y.
(2003) Med. Hypotheses, 61, 503-508.
5.Zamyatnin, A. A. (2004) Endogenous Regulatory
OligoPeptide Knowledgebase, http://erop.inbi.ras.ru/.
6.Whitmore, L., and Wallace, B. A. (2004) Nucleic
Acids Res., 32, D593-D594.
7.Zamyatnin, A. A. (2003) Biofizika,
48, 1030-1039.
8.Price, D. A., Davies, N. W., Doble, K. E., and
Greenberg, M. J. (1987) Zool. Sci., 4, 395-410.
9.Noda, M., Teranishi, Y., Takahashi, H., Toyosato,
M., Notake, M., Nakanishi, S., and Numa, S. (1982) Nature,
297, 431-434.
10.Jack, R. W., Wan, J., Gordon, J., Harmark, K.,
Davidson, B. E., Hiller, A. J., Wettenhall, R. E., Hickey, M. W., and
Coventry, M. J. (1996) Appl. Environ. Microbiol., 62,
2897-2903.
11.Holk, A. L., Axelsson, L., Huehne, K., and
Krokel, L. (1994) FEMS Microbiol. Lett., 115,
143-149.
12.Stoddard, G. W., Petzel, J. P., van Belkum, M.
J., Kok, J., and McKay, L. L. (1992) Appl. Environ. Microbiol.,
58, 1952-1961.
13.McClelland, M., Sanderson, K. E., Spieth, J.,
Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J.,
Dante, M., Du, F., Hou, S., Layman, D., Leonard, S., Nguyen, C., Scott,
K., Holmes, A., Grewal, N., Mulvaney, E., Ryan, E., Sun, H., Florea,
L., Miller, W., Stoneking, T., Nhan, M., Waterson, R., and Wilson, R.
K. (2001) Nature, 413, 852-856.
14.Skeggs, L. T., Lentz, K. L., Kahn, J. R.,
Shumway, N. P., and Woods, K. R. (1956) J. Exp. Med.,
104, 193-197.
15.Tsai, B.-S., Peach, M. J., Khoshla, M. C., and
Bumpus, F. M. (1975) J. Med. Chem., 18, 1180-1183.
16.Brazeau, P., Vale, W., Burgus, R., Ling, N.,
Butcher, M., Rivier, J., and Guillemin, R. (1973) Science,
179, 77-79.
17.Arakawa, Y., and Tachibana, S. (1984) Life
Sci., 35, 2529-2536.
18.Pradayrol, L., Jornvall, H., Mutt, V., and Ribet,
A. (1980) FEBS Lett., 109, 55-58.
19.Geller, D. M., Currie, M. G., Wakitani, K., Cole,
B. R., Adams, S. P., Fok, F. K., Siegel, N. R., Eubanks, S. R.,
Galluppi, G. R., and Needleman, P. (1984) Biochem. Biophys. Res.
Commun., 120, 333-338.
20.Iimura, O., Shimamoto, K., Ando, T., Ura, N.,
Ishida, H., Nakagawa, M., Yokoyama, T., Fukuyama, S., Yamaguchi, Y.,
and Yamaji, I. (1987) Can. J. Physiol. Pharmacol., 65,
1701-1705.
21.Kim, J. B., Gadboll, V., Whittaker, J., Barton,
B. A., and Conlon, J. M. (2000) Gen. Comp. Endocrinol.,
120, 353-363.
22.Rosenfeld, M. G., Lin, C. R., Amara, S. G.,
Stolarsky, L., Roos, B. A., Ong, E. S., and Evans, R. M. (1982)
Proc. Natl. Acad. Sci. USA, 79, 1717-1721.
23.Irwin, D. M., and Wong, J. (1995) Mol.
Endocrinol., 9, 267-277.
24.Ashmarin, I. P., and Obukhova, M. F. (1985)
Zh. Vyssh. Nervn. Deyat., 35, 211-221.
25.Kizer, J. S., Busby, W. H., Cottle, C., and
Youngblood, W. W. (1984) Proc. Natl. Acad. Sci. USA, 81,
3228-3232.
26.Bateman, R. C., Yougblood, W. W., Busby, W. H.,
and Kizer, J. S. (1985) J. Biol. Chem., 260,
9088-9091.
27.Dimarchi, R. D., Tam, J. P., Kent, S. B. H., and
Merrifield, R. B. (1982) Int. J. Pept. Prot. Res., 19,
88-93.
28.Abracham, G. A., and Podell, D. N. (1984) Mol.
Cell. Biochem., 38, 181-190.
29.Karle, I. L. (1981) in The Peptides:
Analysis, Synthesis, Biology (Gross, E., and
Meienhofer, J., eds.) Academic Press, New York, pp. 1-54.
30.Schulz, C. P. (2000) Nature Struct. Biol.,
7, 7-10.
31.Camerman, A., Mastropaolo, D., Karle, I., Karle,
J., and Camerman, N. (1983) Nature, 306, 447-450.
32.Higashijima, T., Kobayashi, J., Nagai, U., and
Miyazawa, T. (1979) Eur. J. Biochem., 97, 43-57.
33.Sabesan, M. N. (1980) Fed. Proc.,
39, 1946.
34.Sabesan, M. N., and Harper, E. T. (1980) J.
Theor. Biol., 83, 457-467.
35.Zamyatnin, A. A. (1984) Ann. Rev. Biophys.
Bioeng., 13, 145-165.
36.Zamyatnin, A. A. (1990) Biofizika,
35, 555-559.
37.Zamyatnin, A. A. (1991) Prot. Seq. Data
Anal., 4, 57-60.
38.Zamyatnin, A. A., and Voronina, O. L. (1998)
Usp. Biol. Khim., 38, 165-197.
39.Zamyatnin, A. A. (1996) Neirokhimiya,
13, 243-259.
40.Zamyatnin, A. A., and Voronina, O. L. (1997)
Neirokhimiya, 14, 263-272.
41.Zamyatnin, A. A., and Voronina, O. L. (1997)
Biofizika, 43, 438-446.
42.Voronina, O. L., and Zamyatnin, A. A. (2001)
Neirokhimiya, 18, 163-181.
43.Gazenko, O. G. (ed.) (1987) Dictionary of
Physiological Terms [in Russian], Nauka, Moscow.
44.Gilyarov, M. S. (ed.) (1986) Biological
Encyclopedic Dictionary [in Russian], Sovetskaya Entsiklopediya,
Moscow.
45.Kameyama, T., Sasaki, K.-I., and Nabeshima, J.
(1970) Yakugaku Zasshi, 90, 1006-1011.
46.Erdos, E. G., and Wilde, A. F. (eds.) (1970)
Handbook for Experimental Pharmacology, Vol. 25,
Springer-Verlag, Berlin-Heidelberg-New York.
47.Lee, C. Y., Chiappinelli, V. A., Takasaki, C.,
Kimura, S., Goto, K., and Masaki, T. (1988) Nature, 335,
303.
48.Sundler, F., Ekbald, E., Bottcher, G., Alumets,
J., and Hakanson, R. (1985) Biogenetics of Neurohormonal
Peptides (Hakanson, R., and Thorell, J., eds.) Academic Press,
London, pp. 213-243.
49.Ashmarin, I. P., Kulaichev, A. P., and Chepurnov,
S. A. (1989) Fiziol. Zh. SSSR, 75, 627-632.