REVIEW: Na+-Translocating NADH:Quinone Oxidoreductase: Progress Achieved and Prospects of Investigations
A. V. Bogachev1* and M. I. Verkhovsky2
1Department of Molecular Energetics of Microorganisms, Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: (7-095) 939-0338; E-mail: sodiumc@genebee.msu.ru2Helsinki Bioenergetics Group, Institute of Biotechnology, P.O. Box 65, (Viikinkaari 1), 00014, University of Helsinki, Helsinki, Finland: fax: +358-9-191-58-001; E-mail: michael.verkhovsky@helsinki.fi
* To whom correspondence should be addressed.
Received September 9, 2004
Structural and catalytic properties of bacterial Na+-translocating NADH:quinone oxidoreductases are briefly described. Special attention is given to studies on kinetics of the enzyme interaction with NADH and the role of sodium ions in this process. Based on the existing data, possible model mechanisms of sodium transfer by Na+-translocating NADH:quinone oxidoreductase are proposed.
KEY WORDS: Na+-translocating NADH:quinone oxidoreductase, sodium potential, Vibrio, respiratory chain
Abbreviations: EPR) electron paramagnetic resonance; Fl) oxidized flavin; FlH2) reduced flavin; FlH·) neutral flavosemiquinone; Fl·-) anionic flavosemiquinone; HQNO) 2-heptyl-4-hydroxyquinoline N-oxide; Na+-NQR) Na+-translocating NADH:quinone oxidoreductase; Na+/e-) number of sodium ions transferred across the membrane by the respiratory chain enzyme normalized to the number of electrons; Q) ubiquinone; QH2) ubiquinol; DeltaµH+) transmembrane difference of H+ electrochemical potentials; Deltapsi) transmembrane electric potential.
Bacteria can grow over a wide range of environmental pH. Under acidic
and neutral conditions, the respiratory chain of prokaryotes can
generate a high proton potential sufficient to provide different
activities. However, if protons were transferred from the cytoplasm
into the external medium by H+-translocating complexes of
the respiratory chain of alkalotolerant or alkalophilic bacteria
against the pH gradient, i.e., when the osmotic component of
DeltaµH+ was opposite in direction
to its electric component, it would result in a large decrease in the
overall proton-motive force. As a result, alkalophilic bacteria would
be unable to perform
DeltaµH+-dependent functions, first
of all oxidative phosphorylation. Skulachev suggested that a
fundamentally new mechanism of energy conservation at high pH values
should exist--the functioning of a sodium cycle. This mechanism implies
that under alkaline conditions the respiratory chain enzymes of
bacteria can translocate sodium ions instead of protons and the sodium
potential generated by primary sodium pumps can be used for energy
support of the main cell activities, such as chemical (synthesis of
ATP), osmotic (accumulation of substrates), and mechanical (motility)
[1-3].
The enzyme responsible for storage of the redox reaction energy as a sodium electrochemical potential was first discovered in the respiratory chain of the marine bacterium Vibrio alginolyticus. In 1977 Unemoto et al. showed that the NADH oxidase reaction catalyzed by subbacterial particles from V. alginolyticus and V. costicola was specifically stimulated by sodium ions [4]. Later, this stimulation was found to occur on the NADH:quinone oxidoreductase domain of the respiratory chain [5]. It was also shown that the Na+-dependence of NADH oxidation in V. alginolyticus was associated not with an allosteric regulation of NADH:quinone oxidoreductase but with coupling of this enzyme activity with the transmembrane transfer of sodium ions [6], i.e., NADH:quinone oxidoreductase of the respiratory chain of V. alginolyticus is a primary sodium pump [7].
The generated sodium potential can be used by V. alginolyticus for mechanical (rotation of the polar flagellum) [8] or osmotic (substrate transfer) [9] activities. Thus, this microorganism exemplifies the possible functioning of the sodium cycle of energy conversion in bacteria [3].
PRIMARY SEQUENCE OF SUBUNITS OF Na+-TRANSLOCATING
NADH:QUINONE OXIDOREDUCTASE
In 1994, the operon encoding Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) in V. alginolyticus was cloned and sequenced independently in two laboratories [10, 11]. This operon was found to contain six open reading frames (nqrA-F). Afterwards, Na+-NQR was shown to consist of six subunits that corresponded to products of six genes of the nqr-operon [12].
Three subunits of Na+-NQR (NqrB, NqrD, and NqrE) are very hydrophobic, whereas the others are relatively hydrophilic (see table). The NqrF subunit is homologous to many proteins responsible for redox reactions. The C-terminal domain of this subunit resembles ferredoxin:NADP+ oxidoreductases of the FNR family. Its primary sequence contains sites responsible for binding NADH and FAD [10, 13]. The N-terminal domain of NqrF is homologous to ferredoxins bearing a 2Fe-2S cluster. Its sequence contains four conservative cysteine residues that seem to be involved in formation of the iron-sulfur cluster [10]. Thus, NqrF is a polypeptide that combines NADH:ferredoxin oxidoreductase and ferredoxin [13]. The other five subunits of Na+-NQR (NqrA-E) do not display a noticeable homology to other proteins with known functions. It should be noted that Na+-NQR is completely different from H+-translocating NADH:quinone oxidoreductases (NDH-1, Complex I), as well as from noncoupled NADH:quinone oxidoreductases (NDH-2) [10, 11, 14]. Thus, proteins of the Na+-NQR type seem to be the third class of NADH:quinone oxidoreductases of respiratory chains that appeared during evolution independently of two other classes of these enzymes.
Physicochemical properties of the Na+-NQR subunits
exemplified by the enzyme from V. harveyi [15] and their possible role in the enzymatic activity
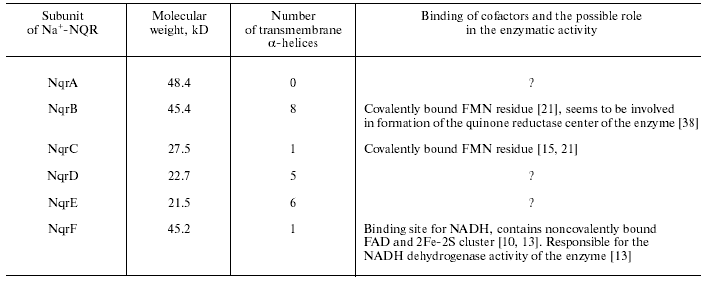
It is interesting that genes homologous to nqrA-F from V. alginolyticus were later found in genomes of various bacteria [15, 16]. Thus, Na+-NQR is widely distributed among prokaryotes, in particular, this protein is present in many pathogenic microorganisms, such as V. cholerae [17], Haemophilus influenzae [18], Klebsiella pneumoniae [14], Neisseria gonorrhoeae, Neisseria meningitidis, Yersinia pestis, Shewanella putrefaciens, Pseudomonas aeruginosa, and Porphyromonas gingivalis.
COFACTOR COMPOSITION OF Na+-NQR
During recent years, the cofactor composition of Na+-NQR has been studied in various laboratories. Results of these studies were very contradictory, and the conclusions seem not final up to now. In 1986, Na+-NQR was isolated from V. alginolyticus and shown to contain only flavins as cofactors: one noncovalently bound FAD and one noncovalently bound FMN [19]. However, only one Na+-NQR subunit (the NqrF subunit) was found to contain a motif for flavin binding [10]. Moreover, a purified preparation of Na+-NQR contained only one noncovalently bound flavin, FAD [20]. This contradiction in the results was resolved later. It was found that Na+-NQR from V. harveyi, in addition to the noncovalently bound FAD, contained another flavin (most likely, FMN), which is covalently bound to the NqrC subunit [15]. The covalent binding of flavin to the protein suggested the existence of another, unlike classical, motif in the primary sequence of the flavin-containing domain and also the impossibility to use the routine extraction procedure for its detection.
Later Nakayama et al. showed that Na+-NQR contains not one but two covalently bound flavins attached to the NqrB and NqrC subunits [21]. These flavins were established to be FMN residues, and in both cases they were bound to threonine residues of the subunits by a phosphoester bond [22, 23]. It should be noted that such covalent binding of flavin is unique and has been found in no other enzymes but Na+-NQR only [23].
Recently, studies on flavin cofactors of Na+-NQR met an unexpected situation. In addition to FAD and FMN, this protein was found to contain also a noncovalently bound riboflavin [24]. Usually riboflavin is used by living organisms only as a precursor for synthesis of FAD and FMN, and Na+-NQR is the only known case of its functioning as a cofactor. However, it is still not established which subunit of Na+-NQR is responsible for binding of riboflavin. Thus, there is a possibility that Na+-NQR contains only three flavins, whereas riboflavin is present in it as a result of hydrolysis of the phosphoester bond of FMN residues.
The analysis of the primary sequences of Na+-NQR subunits predicts that the NqrF contains an iron-sulfur cluster [10]. Indeed, a 2Fe-2S cluster was detected in both the Na+-NQR complex [25, 26] and an isolated fragment of the NqrF subunit [13]. Additionally to flavins and the 2Fe-2S cluster, Na+-NQR also contains a molecule of the tightly bound ubiquinone-8 [15, 26, 27]. Thus, Na+-NQR is now thought to contain the following set of cofactors: one 2Fe-2S cluster, one noncovalently bound FAD, two covalently bound FMN residues, and possibly also one ubiquinone-8 and one noncovalently bound riboflavin.
CATALYTIC ACTIVITIES OF Na+-NQR
During in vivo functioning Na+-NQR oxidizes NADH and transfers two electrons to ubiquinone with production of ubiquinol:
NADH + H+in + Q + 2Na+in --> NAD+ + QH2 + 2Na+out. (1)
This redox reaction is coupled with a vectorial transfer of two sodium ions across the membrane, i.e., the ratio Na+/e- for Na+-NQR is 1 [28]. It is noteworthy that the efficiency of energy conservation by Na+-translocating NADH:quinone oxidoreductases is approximately twofold lower than by H+-translocating NADH:quinone oxidoreductases for which the H+/e- stoichiometry is 2 [29].
Sodium ions are indispensable components of the Na+-NQR-catalyzed reaction; therefore, its rate depends on concentration of Na+, and the reaction virtually does not occur in media depleted in this ion. At physiological values of ionic strength, the dependence of reaction (1) rate on Na+ concentration is hyperbolic [25]. Under these conditions, values of Km(Na+) for Na+-NQR from various sources are in the region of millimolar concentrations [4, 25], and for the enzyme from V. harveyi this value is ~3 mM [25]. Na+-NQR is absolutely specific to Na+. All other ions studied failed to activate the reaction (1). They only nonspecifically stimulated it because increase in the ionic strength was associated with increase in the Na+-NQR affinity for Na+ [4].
It is important that even in the absence of sodium ions Na+-NQR is unable to translocate protons [7, 15]. The sodium transfer by the enzyme is associated with generation of the transmembrane electric potential (Deltapsi) [7, 15]. In the absence of Deltapsi, which can induce electrophoretic current of protons across the membrane, no Na+/H+ exchange catalyzed by Na+-NQR occurs. This suggests that protons released on oxidation of NADH during the catalytic cycle of Na+-NQR enter the cytoplasm, whereas protons required for generation of ubiquinol are also taken up from the cytoplasmic side of the membrane [15]. Thus, Na+-NQR is a primary electrogenic sodium pump.
Na+-NQR is rather specific to NADH. It does not oxidize NADPH and among artificial analogs interacts only with deamino-NADH and thio-NADH [15, 30].
At present, only a few inhibitors of Na+-NQR are known. An antibiotic korormicin has been recently described that specifically inhibits Na+-NQR at the level of its interaction with ubiquinone [23, 31]. Korormicin affects the enzyme without competition with quinone, and its inhibition constant is ~0.1 nM [31]. The effect of HQNO on Na+-NQR is similar [6], but the affinity of this inhibitor to the enzyme is significantly weaker (0.3-0.4 µM) [15, 31]. Na+-NQR is also sensitive to low concentrations of silver ions [32] and some other heavy metals (Cd2+, Pb2+, Zn2+, Cu2+) [30]. These ions influence the initial reactions of the catalytic cycle of Na+-NQR and seem to prevent its interaction with NADH [30].
In addition to reaction (1), Na+-NQR can in vitro catalyze the transdehydrogenase reaction of electron transfer from NADH onto thio-NAD+. This activity does not depend on concentration of sodium ions, is inhibited by heavy metal ions, and is insensitive to HQNO [30].
In addition to the quinone reductase reaction (1), the isolated enzyme can also catalyze so-called NADH dehydrogenase reaction during interaction with soluble quinones [33]. This activity includes a single-electron reduction of soluble quinones (menadione, Q0, Q1, etc.) or some other electron acceptors (hexammineruthenium (III), ferricyanide, etc.). Similarly to the transdehydrogenase activity, the NADH dehydrogenase activity does not depend on concentration of sodium ions, is inhibited by heavy metal ions, and is insensitive to HQNO and korormicin [23, 30, 33]. Only the NqrF subunit seems to be required for this reaction [13].
CATALYTIC CYCLE OF Na+-NQR
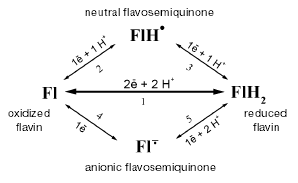
During recent years, the catalytic cycle of Na+-NQR has been studied using various biochemical and biophysical approaches. First of all, the enzyme has been studied by EPR spectrometry. A signal from the iron-sulfur [2Fe-2S]-cluster was detected in a preparation of a reduced Na+-NQR (after the enzyme was preincubated with excess of NADH or dithionite). However, in addition to this signal, the enzyme produced a radical signal with the spin concentration at the ratio 1 : 1 compared to the 2Fe-2S cluster [25]. No signal from the 2Fe-2S cluster was detected in the oxidized preparation of Na+-NQR (oxidized either by air or in the presence of various mediators), because under these conditions the cluster should be in the state [2Fe-2S]2+. Nevertheless, the radical signal was also detected in the oxidized enzyme. Moreover, the spin concentration in the radical signal did not change depending on the degree of reduction of Na+-NQR [25, 34]. Studying the enzyme by EPR and optical spectroscopy this radical signal was shown to be associated with presence in the oxidized Na+-NQR of neutral flavosemiquinone, and the latter became anionic when the enzyme was reduced (see the scheme of possible redox transitions in flavins in Fig. 1) [34]. These conclusions were later supported by data of ENDOR spectrometry [35].
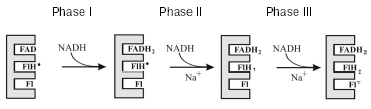
Such an exclusively high stability of flavosemiquinone over a very wide range of redox potentials is extremely unusual, and we took a closer look to the properties of this radical. Using the stop-flow method, we studied fast kinetics of the enzyme reduction in the presence of NADH. This process proceeds in three sequential steps [25, 34] that represent reduction of different flavin cofactors. Figure 2 shows that the first phase is a two-electron reduction of flavin (Fl --> FlH2), the second phase is a one-electron reduction of the neutral flavosemiquinone (FlH* --> FlH2) existing in the oxidized Na+-NQR, and the third phase is a one-electron reduction of the oxidized flavin with production of the anionic flavosemiquinone (Fl --> Fl·-). It should be noted that the reduction of FlH* and production of Fl·- occur during different kinetic phases. Moreover, during the enzyme reduction the neutral flavosemiquinone turns to the completely reduced flavin, whereas the anionic flavosemiquinone is produced from the oxidized flavin. These data suggest that radicals in Na+-NQR are generated from the different flavin cofactors of this protein [34]. This explains the unique redox stability of the radical signal in Na+-NQR. In fact, Na+-NQR includes two different radicals: the neutral flavosemiquinone in the oxidized enzyme and the anionic flavosemiquinone in the reduced enzyme. The neutral flavosemiquinone should have a very high redox potential for the Fl/FlH* pair (transition 2 in Fig. 1), whereas the potential of the anionic flavosemiquinone for the Fl·-/FlH2 pair should be very low (transition 5 in Fig. 1). Therefore, in most cases only one radical signal is detected in Na+-NQR [25, 34].Fig. 1. Scheme of possible redox transitions in flavins. Fl) oxidized flavin; FlH2) completely reduced flavin; FlH·) neutral flavosemiquinone; Fl·-) anionic flavosemiquinone.
Studies on Na+-translocating enzymes present some advantages compared to studies on proton pumps. First of all, studies on Na+ pumps allow us to vary the concentration of the coupling ion without significantly affecting the protein stability, which is impossible in the case of H+-translocating enzymes. In particular one can investigate the catalytic cycle of a sodium-dependent enzyme when its rate is limited by low Na+ concentrations and reveal in it stages which are specifically accelerated by these ions. Such an approach allows one to determine which particular redox transitions in the enzyme are coupled with the translocation of Na+.Fig. 2. Scheme of redox events during the interaction of Na+-NQR with NADH [34].
We used this approach in studies on the above-described kinetics of the Na+-NQR reduction [25, 34]. Sodium ions were found to markedly accelerate some phases of this process. The rate of the phase I (Fig. 2) did not depend on Na+ concentration. Because the rate of this phase and the NADH dehydrogenase activity of Na+-NQR [23, 30, 33] are both very high and independent of sodium, we can identify with certainty this phase as reduction of the noncovalently bound FAD in the NADH dehydrogenase domain of the enzyme (in the NqrF subunit) [25, 34]. However, phases II and III were significantly accelerated with increase in the Na+ concentration. Moreover, the activation constant by Na+ of phase III (Fl --> Fl·-) corresponded to the Km value for Na+ during the reaction (1) [25]. Thus, just this process (Fl --> Fl·-) in the catalytic cycle of the enzyme should be coupled with the translocation of Na+.
Energy-storing enzymes of respiratory chains usually transfer electrons by one-electron pathways via various one-electron cofactors, such as Fe-S clusters (complex I), cytochromes and Fe-S cluster (the bc1 complex), or cytochromes and copper atoms (cytochrome c oxidase) [2]. However, Na+-NQR mainly contains flavins as cofactors (three-four flavins), i.e., potentially two-electron carriers. However, at least two flavins of Na+-NQR transfer only one electron, i.e., under physiological conditions the FlH* --> Fl transition (reaction 2 in Fig. 1) in one of them and the Fl·---> FlH2 transition (reaction 5 in Fig. 1) in the other are fully forbidden because of kinetic or thermodynamic reasons [34]. It is known that sometimes flavin cofactors transfer only a single electron (e.g., in flavodoxins). However, in Na+-NQR two flavins function concurrently in this way, but in different cofactors different redox transitions occur. Thus, despite the composition of cofactors unusual for energy transducing enzymes of respiratory chain, Na+-NQR also performs the one-electron transfer of reducing equivalents.
PROSPECTS IN STUDIES ON THE COUPLING MECHANISM DURING
Na+-NQR FUNCTIONING
Although significant progress has been achieved in studies on Na+-NQR during recent years, the mechanism of the sodium potential generation by this enzyme is still far from being understood. Certainly, some aspects of electron transport are determined and the redox transitions in Na+-NQR that can be coupled with translocation of Na+ are established. However, except for kinetic phase I of the enzyme reduction (which is the FAD reduction to FADH2 in the NADH dehydrogenase domain of the NqrF subunit), other kinetic phases cannot be assigned to particular flavin cofactors. Thus, it is still unclear in what flavin (in FMN in the NqrB subunit, in FMN in NqrC, or, possibly, in riboflavin) the one-electron redox transition is coupled with the Na+ transfer.
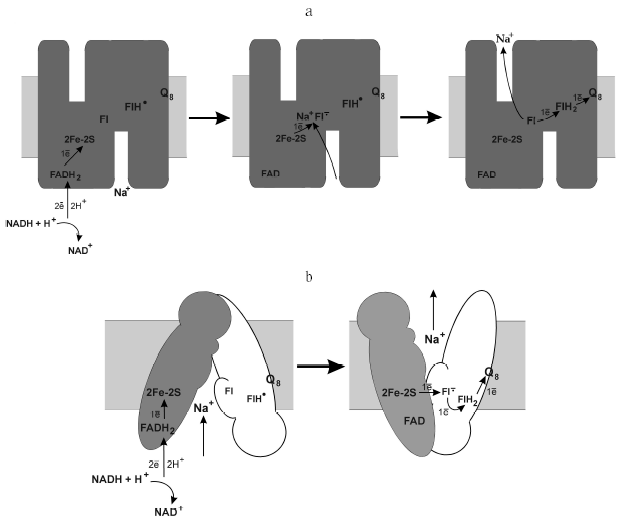
The mechanism of energy conversion of the Na+-NQR-catalyzed redox reactions into sodium potential is also still unknown. The coupling can occur in two fundamentally different ways. The first is the operation by the principle of a “thermodynamic machine”: the charge changes at the one-electron reduction of any cofactor is compensated by the capture of the positive sodium ion from the cytoplasmic side of the membrane [10]. Oxidation of this cofactor has to be accompanied by ejection of sodium, but from the other side of the membrane. 2Fe-2S cluster [10], flavin [36], or ubiquinone [33, 37] have been proposed to be this cofactor. Based on our studies, we conclude that such a “coupling” cofactor can be represented by flavin that stabilizes the anionic flavosemiquinone (Fig. 3a). If Na+-NQR uses just this mechanism of energy conservation (i.e., is a “thermodynamic machine”), the potential of the Fl/Fl·- pair must be dependent on the Na+ concentration ( 60 mV per order of magnitude in [Na+]). Up to now, the effect of Na+ concentration on the midpoint potential has been studied only for one cofactor of Na+-NQR, the 2F-2S cluster. The value of Em for [2F-2S] is about -270 mV (n = 1), and this value does not depend on the Na+ concentration [25].
Na+-NQR might also operate as a “kinetic machine”. In this case, the [Na+]-dependence of the electron transfer to flavin with production of the anionic flavosemiquinone (Fl·-) is not determined thermodynamically but is caused by the necessity of pronounced conformational changes in the protein which brought the cofactors together concurrently with the Na+ transfer across the membrane (Fig. 3b). Unlike the first model, redox potential of the Na+-NQR cofactors should not depend on the Na+ concentration. Thus, redox titration of Na+-NQR at varied concentrations of Na+ seems promising for understanding the operating mechanism of this enzyme.Fig. 3. Possible mechanisms of the sodium potential generation by Na+-translocating NADH:quinone oxidoreductase. a) Scheme of the enzyme operation as a “thermodynamic machine”; b) scheme of the enzyme operation as a “kinetic machine”.
Another important line in further studies has to be the determination of the spatial organization of Na+-NQR using approaches of molecular biology or X-ray crystallography. This stage is also necessary for determination of the operating mechanism of this enzyme.
In conclusion, we would like to once more emphasize the uniqueness of Na+-NQR. This enzyme is extremely unusual in its primary sequence (the absence of homologs in five subunits of Na+-NQR), in the set of cofactors (which seems to include four flavins, one of which is riboflavin), in the mode of their covalent binding to the protein (the phosphoester bond of FMN with threonine residues), and also in their redox characteristics (two flavins function only as one-electron carriers). Nevertheless, the determination of the mechanism of this enzyme will be promising for understanding general mechanisms of energy conversion by enzymes of respiratory chains.
The probable importance of studies on Na+-NQR for medicine has also to be mentioned. This enzyme is widely distributed among pathogenic and conditionally pathogenic microorganisms [15, 16]. Moreover, in the case of V. cholerae Na+-NQR was shown to be significant for the induction of virulence factors [17]. Thus, it is possible that this enzyme could be used as a target in the treatment or prevention of many infectious diseases.
We are grateful to V. P. Skulachev for his constant attention to results of our studies. The work was supported by the Russian Foundation for Basic Research (the project No. 04-04-48101).
REFERENCES
1.Skulachev, V. P. (1984) Trends Biochem.
Sci., 9, 483-485.
2. Skulachev, V. P. (1989) Energetics of Biological Membranes
[in Russian], Moscow, Nauka.
3.Skulachev, V. P. (1989) J. Bioenerg.
Biomembr., 21, 635-647.
4.Unemoto, T., Hayashi, M., and Hayashi, M. (1977)
J. Biochem., 82, 1389-1395.
5.Unemoto, T., and Hayashi, M. (1979) J.
Biochem., 85, 1461-1467.
6.Tokuda, H., and Unemoto, T. (1984) J. Biol.
Chem., 259, 7785-7790.
7.Tokuda, H., Udagawa, T., and Unemoto, T. (1985)
FEBS Lett., 183, 95-98.
8.Dibrov, P. A., Kostyrko, V. A., Lazarova, R. L.,
Skulachev, V. P., and Smirnova, I. A. (1986) Biochim. Biophys.
Acta, 850, 449-457.
9.Tokuda, H., Sugasawa, M., and Unemoto, T. (1982)
J. Biol. Chem., 257, 788-794.
10.Rich, P. R., Meunier, B., and Ward, F. B. (1995)
FEBS Lett., 375, 5-10.
11.Hayashi, M., Hirai, K., and Unemoto, T. (1995)
FEBS Lett., 363, 75-77.
12. Nakayama, Y., Hayashi, M., and Unemoto, T. (1998) FEBS
Lett., 422, 240-242.
13.Turk, K., Puhar, A., Neese, F., Bill, E., Fritz,
G., and Steuber, J. (2004) J. Biol. Chem., 279,
21349-21355.
14.Bertsova, Y. V., and Bogachev, A. V. (2004)
FEBS Lett., 563, 207-212.
15.Zhou, W., Bertsova, Y. V., Feng, B., Tsatsos, P.,
Verkhovskaya, M. L., Gennis, R. B., Bogachev, A. V., and Barquera, B.
(1999) Biochemistry, 38, 16246-16252.
16.Hase, C. C., Fedorova, N. D., Galperin, M. Y.,
and Dibrov, P. A. (2001) Microbiol. Mol. Biol. Rev., 65,
353-370.
17.Hase, C. C., and Mekalanos, J. J. (1999) Proc.
Natl. Acad. Sci. USA, 96, 3183-3187.
18.Hayashi, M., Nakayama, Y., and Unemoto, T. (1996)
FEBS Lett., 381, 174-176.
19.Hayashi, M., and Unemoto, T. (1986) FEBS
Lett., 202, 327-329.
20. Pfenninger-Li, X. D., and Dimroth, P. (1995) FEBS
Lett., 369, 173-176.
21.Nakayama, Y., Yasui, M., Sugahara, K., Hayashi,
M., and Unemoto, T. (2000) FEBS Lett., 474, 165-168.
22.Hayashi, M., Nakayama, Y., Yasui, M., Maeda, M.,
Furuishi, K., and Unemoto, T. (2001) FEBS Lett., 488,
5-8.
23.Hayashi, M., Nakayama, Y., and Unemoto, T. (2001)
Biochim. Biophys. Acta, 1505, 37-44.
24.Barquera, B., Zhou, W., Morgan, J. E., and
Gennis, R. B. (2002) Proc. Natl. Acad. Sci. USA, 99,
10322-10324.
25.Bogachev, A. V., Bertsova, Y. V., Barquera, B.,
and Verkhovsky, M. I. (2001) Biochemistry, 40,
7318-7323.
26.Barquera, B., Hellwig, P., Zhou, W., Morgan, J.
E., Hase, C. C., Gosink, K. K., Nilges, M., Bruesehoff, P. J., Roth,
A., Lancaster, C. R., and Gennis, R. B. (2002) Biochemistry,
41, 3781-3789.
27.Pfenninger-Li, X. D., Albracht, S. P. J., van
Belzen, R., and Dimroth, P. (1996) Biochemistry, 35,
6233-6242.
28.Bogachev, A. V., Murtasina, R. A., and Skulachev,
V. P. (1997) FEBS Lett., 409, 475-477.
29.Galkin, A. S., Grivennikova, V. G., and
Vinogradov, A. D. (2001) Biochemistry (Moscow), 66,
435-443.
30.Bourne, R. M., and Rich, P. R. (1992) Biochem.
Soc. Trans., 20, 577-582.
31.Yoshikawa, K., Nakayama, Y., Hayashi, M.,
Unemoto, T., and Mochida, K. (1999) J. Antibiot., 52,
182-185.
32.Asano, M., Hayashi, M., Unemoto, T., and Tokuda,
H. (1985) Agric. Biol. Chem., 49, 2813-2817.
33.Hayashi, M., and Unemoto, T. (1984) Biochim.
Biophys. Acta, 767, 470-478.
34.Bogachev, A. V., Bertsova, Y. V., Ruuge, E. K.,
Wikstrom, M., and Verkhovsky, M. I. (2002) Biochim. Biophys.
Acta, 1556, 113-120.
35.Barquera, B., Morgan, J. E., Lukoyanov, D.,
Scholes, C. P., Gennis, R. B., and Nilges, M. J. (2003) J. Am. Chem.
Soc., 125, 265-275.
36.Unemoto, T., and Hayashi, M. (1993) J.
Bioenerg. Biomembr., 25, 385-391.
37.Hase, C. C., and Barquera, B. (2001) Biochim.
Biophys. Acta, 1505, 169-178.
38.Hayashi, M., Shibata, N., Nakayama, Y.,
Yoshikawa, K., and Unemoto, T. (2002) Arch. Biochem. Biophys.,
401, 173-177.