Molecular Cloning of Kunitz-Type Proteinase Inhibitor Group B Genes from Potato
A. S. Speranskaya1*, A. A. Krinitsina1, P. Poltronieri2, P. Fasano2, A. Santino2, A. B. Shevelev1, and T. A. Valueva1
1Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, 119071 Moscow, Russia; fax: (7-095) 954-2732; E-mail: annatar@aha.ru2National Research Council of Italy, ISPA-CNR, Via Monteroni 14, Lecce, 73100, Italy; fax: +39(0)832420000
* To whom correspondence should be addressed.
Received August 17, 2004; Revision received October 19, 2004
Eighteen clones representing copies of four Kunitz-type proteinase inhibitor group B genes (PKPI-B) obtained by polymerase chain reaction cloning of potato (Solanum tuberosum L. cv. Istrinskii) genomic DNA were sequenced and analyzed. Three new genes were found and named PKPI-B1, PKPI-B2, and PKPI-B10: these were represented by five, two, and seven clones, respectively. The remaining four clones corresponded to the formerly characterized PKPI-B9 gene. These data show that at least four PKPI-B encoding genes are harbored in the genome of potato cv. Istrinskii. Their analysis suggests that variability of PKPI-B encoding genes in potato is limited and could be explained by cross-hybridization events in the ancestor forms rather than by random mutagenesis.
KEY WORDS: Kunitz-type proteinase inhibitors, gene sequences, genome, potato
Potato (Solanum tuberosum L.) tubers contain a vast group of water-extractable proteins comprising several families of proteinase inhibitors [1]. Some of these proteins of 20-24 kD molecular mass belong to soybean trypsin inhibitor structural family (SKTI) in which they compose the subfamily of potato Kunitz-type proteinase inhibitors (PKPIs) [2].
The physiological role of PKPIs still remains unclear, and their in vivo expression regulation mechanisms have not yet been determined. However, the protective action of these proteins against pathogens is important [3] since plant tissue degradation caused by pathogen-derived secretory proteinases is considered one of the principal mechanisms of the spread of infection in the host-pathogen interaction [4]. At least some PKPIs are reported to suppress Phytophthora infestans (Mont.) de Bary (phytophthorosis causative agent) hyphae growth and development [5, 6]. Therefore, the expression of a particular set of PKPI genes, participating in plant defense and immunity, may be considered a relevant tool for the production of new highly resistant potato varieties either by conventional breeding methods or by innovative biotechnologies.
More than 70 PKPI gene sequences from different potato cultivars are known. As derived from primary structure comparison, PKPIs are divided into three groups denoted A, B, and C [2]. PKPIs from different structural groups exhibit different specificity towards proteinases. For instance, PKPI-A group includes inhibitors of aspartic proteinases such as cathepsin D [2]. Group PKPI-B contains inhibitors of chymotrypsin clan serine proteinases [2] exemplified by trypsin, chymotrypsin, and elastase [14]. PKPI group C includes proteins that inhibit bacillary subtilisins and plant cysteine proteinases as well as some non-proteinase enzymes such as invertase and alpha-amylase [2, 8].
It has been proposed that at least 12 allelic variants of PKPI encoding genes can be harbored in the tetraploid potato genome. As estimated by Heibges et al. [7], the total number of individual PKPI sequences from the A-B-C groups can range between 12 and 43 in the potato cv. Provita genome. At present 26 PKPI-B genes are known to be distributed in six potato cultivars such as Danshaku, Saturna, Provita, Keszthelyi 855 (White Lady), Elkana, and Pentland squire [2, 7-11]. Previously we reported the structure of the PKPI-B9 gene from S. tuberosum cv. Istrinskii [12] encoding for the PSPI-21-6.3 protein isolated from tubers and characterized earlier [5, 13].
Since newly discovered nucleotide sequences of known PKPI genes rarely coincided with sequences already reported, a unique PKPI gene set in each potato cv. was presumed [7]. This hypothesis as well as functional variability of PKPIs from different structural groups is in a good agreement with putative defense role of PKPIs since PKPI multiplicity extends the spectrum of pathogens potentially inhibited. Therefore, it can be hypothesized that different potato varieties (particularly of divergent origin) harbor PKPI isoforms with unique anti-proteinase activity that could be different from the PKPIs characterized previously. According to this hypothesis, PKPIs with new features can thus be discovered. It is noteworthy that Kunitz-family proteinase inhibitors have been found in seeds and storage organs in other plant species of different taxonomic groups such as cereals (Gramineae) [15, 16]. For instance, bifunctional subtilisin/alpha-amylase inhibitors from barley, wheat, rye, and triticale caryopses belong to the structural family of soybean Kunitz trypsin inhibitor [15, 16]. So far bifunctional inhibitors have not been found in species from the Solanaceae family, namely in potato tubers. From this point of view, PKPI genes from different potato cultivars may be a promising source of proteins with new and unique activity.
The goal of the present work was the cloning and analysis of new PKPI-B genes from potato cv. Istrinskii.
MATERIALS AND METHODS
Total genomic DNA was isolated from two-three-week-old shoots of Solanum tuberosum L. cv. Istrinskii as described earlier [17]. The tubers were germinated in a humid chamber under natural illumination at room temperature.
Specific primers 5´(GGGAGCTCAACCTACCTAGTGATGCTACT)3´ and 5´(GGAAGCTTACTGGACTTGC(T/A)TGAAGGAGA)3´ designated as PKPI-B1 and PKPI-B2 were designed mainly on the basis of PKPI-B sequences previously published [2, 7].
Polymerase chain reaction (PCR) was carried out in a Tercyc MC-16 thermocycler (DNA-Technology, Russia). Taq DNA polymerase was purchased from Bionem (Russia). Total genomic DNA template was added to the reaction mixture in the final concentration 0.01 µg/µl. PCR conditions were: 1) 1 cycle: 94°C for 1 min; 2) 5 cycles: 94°C for 30 sec, 56°C for 10 sec, 72°C for 10 sec; and 3) 25 cycles: 94°C for 5 sec, 60°C for 5 sec, 72°C for 30 sec. The amplified DNA fragments were analyzed by electrophoresis in 1% agarose gel and purified using a DNA extraction kit (MBI Fermentas, Lithuania).
The eluted fragments were cloned to pGem-T-Easy or pGEM vectors supplied by Promega (USA). E. coli BMH 71-18 (mutS) strain (Promega) was used for genetic engineering manipulations. DNA purifications and restrictase mapping were carried out as described in [18]. Standard primers T7-promotor, sp6 (Promega) or specific primers PKPI-B1 and PKPI-B2 were used for sequencing.
Sequencing data accumulation, processing, and sequence alignment were performed using the DNASTAR package (http://www.dnastar.com). Cluster analysis of the sequences was performed using the DNASTAR package and MEGA 2.1 program (www.megasoftware.net).
RESULTS AND DISCUSSION
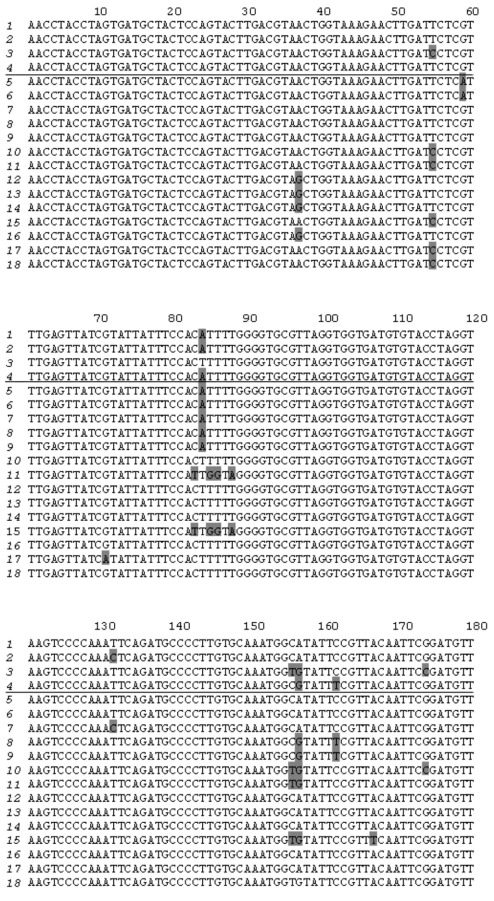
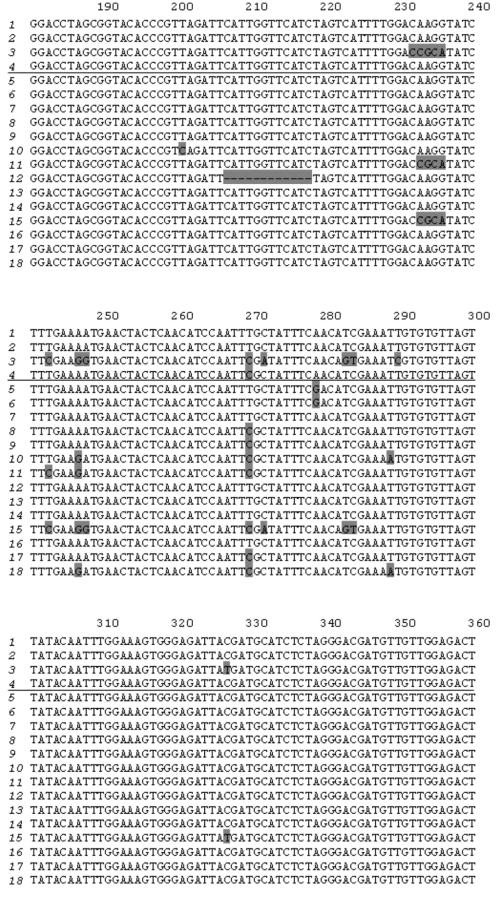
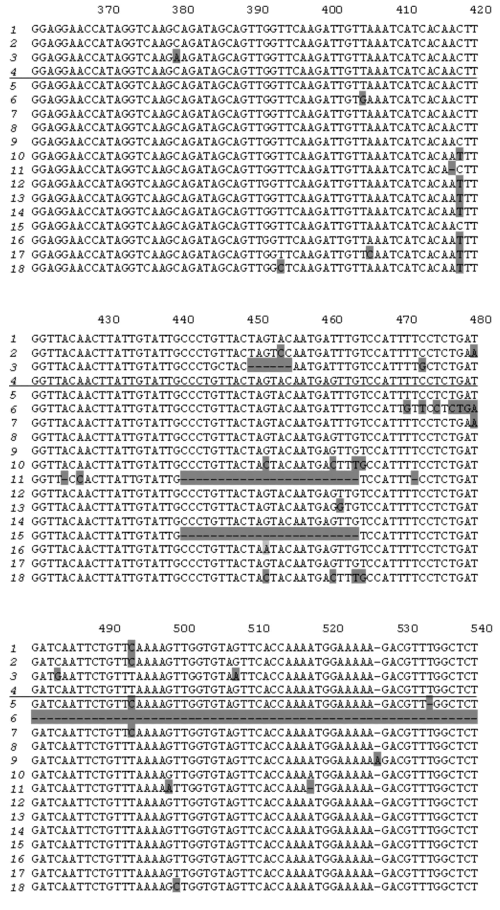
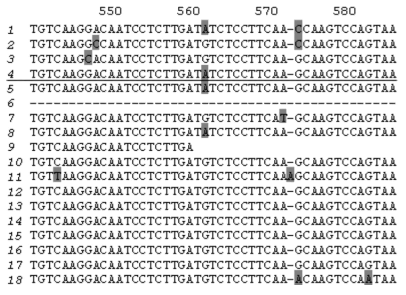
Cloning PCR products obtained using potato genomic DNA as template and primers PKPI-B1 and PKPI-B2 yielded 19 plasmid clones bearing inserts of about 600 bp each. Both strands of these clones were sequenced. PKPI genes and nucleotide sequences were processed using the DNASTAR software package to obtain alignment and clustering of individual sequences. Copies of divergent genes were distinguished from each other by the position of variable amino acids representative of each cluster. Cluster analysis demonstrated that the 18 clones represented multiple copies of four genes, one of which corresponded to PKPI-B9 (NCBI Acc. No. [AY693424]) described in a previous work [12]. Three other genes were found to be unique and were named PKPI-B1 [AY692479], PKPI-B2 [AY693423], and PKPI-B10 [AF536175]. It is noteworthy that the number of copies used for reconstruction of each gene sequence varied from two to seven. Namely, PKPI-B9 was represented by four copies, and PKPI-B1, PKPI-B2, and PKPI-B10 occurred in five, two, and seven copies, respectively. Copies of the same gene differed from each other by a few stochastically located changes presumably caused by Taq DNA-polymerase inaccuracy. There was a single clone bearing a hybrid sequence derived from PKPI-B10 and PKPI-B1 putatively considered as an artifact.
To compare new sequences with the published PKPI genes, NCBI Gene Bank Blast analysis was carried out. Results of this are shown in Fig. 1. PKPI-B2 and PKPI-B9 coincided completely with two cDNAs cloned from potato cv. Provita (P4D11 [AF495584] and P7D8 [AF495585], respectively [7]). PKPI-B1 exhibited 99% identity with three cDNA clones from potato cv. Provita, P1G7 [AF495581], P4D11 [AF495584], and P2G2 [AF495583] [7]. PKPI-B10 gene sequence showed the highest degree of identity (96%) toward P1D4 cDNA [AF492358] from potato cv. Provita [7]. PKPI-B1, PKPI-B2, and PKPI-B10 nucleotide sequences exhibited identity (ranging between 92 to 98%) toward the PKPI-B genes pKEN14-28 [D17330], pF4 [D17329], gCDT-B1 [D17332], S1C1 [AF492769], pAM66 [U30814], PI-2 [AY166690], and x64370 [X64370] encoded in potato cv. Danshaku, Saturna, Keszthelyi 855, Elkana, and Pentland squire, respectively [2, 7-10].
A high degree of identity of PKPI-B1, PKPI-B2, and PKPI-B9 genes from cv. Istrinskii (except PKPI-B10) with known sequences from cv. Provita and Danshaku (belonging to European and Japanese selections) makes doubtful the hypothesis of high intra-cultivar and inter-cultivar PKPI hypervariability proposed by Hiebges et al. [7].Fig. 1. Nucleotide sequence comparison of PKPI-B1 genes (1), PKPI-B2 (2), PKPI-B10 (3), and PKPI-B9 (4) from potato cv. Istrinskii (separated by horizontal line) and PKPI-B genes from other potato cultivars: P1G7 (5), P2G2 (6), P4D11 (7), P7D8 (8), P2F10 (9), P1H5 (10), P1D4 (11) (cv. Provita) [7]; x64370 (12) (cv. Pentland squire) [11]; pKEN14-28 (13), pF4 (14), gCDT-B1 (15) (cv. Danshaku) [2]; S1C1 (16) (cv. Saturna) [7]; pAM66 (17) (cv. Keszthelyi 855 (White Lady)) [9]; PI-2 (18) (cv. Elkana) [10]. The different amino acid residues are shaded.
The occurrence of four variants of PKPI-B genes in potato cv. Istrinskii allows two possible hypotheses. Since cultivated potato varieties bear a tetraploid genome, these genes may represent allelic copies of the same gene locus in the case of complete heterozygosity: conversely, in the case of complete homozygosity, four loci per haploid genome (one sequence per allele) could be present. It is noteworthy that the authors of work [7] estimated nine possible PKPI-B genes in the potato cv. Provita genome with the hypothesis of complete allele homozygosity. Therefore, the exact number of potato genomic loci bearing PKPI-B genes remains undetermined.
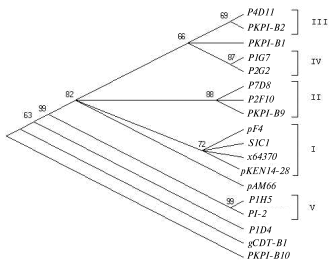
Analysis of single nucleotide positions in PKPI-B sequences demonstrates that polymorphism usually appears in the same sites in any gene (Fig. 1). Figure 2 shows a dendrogram constructed using the MEGA 2.1 program for all published PKPI-B genes using the maximum parsimony method [19]. Analysis of the phylogenetic tree divides the genes into at least five divergent groups (Fig. 2). The genes found in the same potato cultivar are never clustered together, demonstrating the absence of tendency to horizontal dissemination of the same mutations.
Figure 3 illustrates amino acid sequences of PKPI-B proteins. From this analysis, the strict conservancy of biochemically characterized pivotal amino acids is deduced. First of all, the Cys49-Cys98 and Cys147-Cys164 pairs forming the first and the second peptide loops are conserved as well as potential key amino acids of the enzyme-binding reactive centers. Namely, putative trypsin binding site Arg68-Phe69, chymotrypsin-leukocyte elastase binding site Met116-Leu117 [13], and chymotrypsin binding site Met153-Xxx154 are conserved. Previously we presumed involvement of the second disulfide bond Cys147-Cys164 into a peptide loop critical for chymotrypsin-leukocyte elastase binding site Met116-Leu117 formation [13]. This hypothesis was essentially confirmed by data of work [8]. Namely, protein P1D4 lacking Cys149 and unable to form the second loop (Fig. 3) inhibited chymotrypsin 5-6-fold more weakly than P1H5 where this loop was conserved [8]. On the other hand, protein P4B1 showed a 5-6-fold lower chymotrypsin inhibition compared to P1H5, although the Cys147-Cys164 pair was present [8]. It is noteworthy that P4B1 protein was deprived of Met153 in the enzyme-binding center and potentially implicated in the binding of proteinases. This amino acid change might be the principal reason for the loss of anti-chymotrypsin activity.Fig. 2. Phylogenetic tree of PKPI-B gene nucleotide sequences. The tree is constructed by the maximal parsimony algorithm [19]. Numbers near the nodes show bootstrap index of support calculated at 100-fold replicate. Gene groups are shown in Roman numbers. Gene descriptions are given in the legend to Fig. 1.
PKPI-B10 protein bears a deletion in the strand between Cys147 and Cys164 (Fig. 3). A similar deletion was found previously in genome-derived clone gCDT-B1 [2] and cDNA clones P1D4 and P4B1 [7, 8]. This suggests that absence or modification of certain residues may contribute to the inhibition specificity.Fig. 3. Comparison of amino acid sequences encoded by PKPI-B genes: 1) PKPI-B1; 2) PKPI-B2; 3) PKPI-B10; 4) PKPI-B9 [12]; 5) P1G7 [7]; 6) P2G2 [7]; 7) P4D11 [7]; 8) P7D8 [7]; 9) P2F10 [7]; 10) X64370 [11]; 11) pKEN14-28 [2]; 12) pF4 [2]; 13) S1C1 [7]; 14) pAM66 [9]; 15) P1H5 [7]; 16) PI-2 [10]; 17) P1D4 [7]; 18) gCDT-B1 [2]. The homologous regions are shaded with gray color of various intensity. The different amino acid residues are highlighted in gray. The reactive center amino acid residues (Arg68, Met116, Met153) are colored in black; the area removed as result of posttranslational modification of protein encoded by genes PKPI-B9 is marked by box; disulfide bonds (Cys49-Cys98 and Cys147-Cys164) are marked with one and two asterisks, conformably; v, the site of posttranslational modification of the precursor after vacuolar translocation.
Post-translational modification of PKPIs might be another way to generate gaps and variability in amino acid chains. For instance, PSPI-21-6.3 protein, encoded by PKPI-B9 gene, is found to be composed by two chains joint by a disulfide bond [13]. In the course of maturation, the precursor of this protein undergoes proteolysis at sites Ser153-Thr154 and Phe159-Ser160, which leads to excision of a strand within the peptide loop confined by the second disulfide bond [13]. PSPI-21-6.3 protein exhibited a high inhibitory activity towards chymotrypsin and elastase [13].
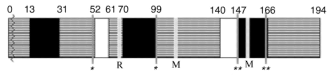
Figure 4 delineates a scheme of conserved and moderately and highly variable regions in proteins encoded by PKPI-B genes. Sequences between Ala32-Cys49 and Cys98-Ser138 are contained in highly conserved domains. The stretches between Cys49-Ser58 and Ser138-Cys147 exhibit a moderate conservancy. Variable strands are located in the Arg68-Cys98 and Cys147-Cys164 regions.
High PKPI-B variability in Arg68-Cys98 and Cys147-Cys164 regions is in a good agreement with their functional importance [13]. It is general opinion that the appearance of changes in these regions could be due to independent mutations. On the other hand, the polymorphism of moderately variable regions could have originated from recombination of ancestor PKPI-B genes. This kind of polymorphism may be attributed to Ser19-->Pro19 codon (TCT-->CCT) first base substitution, as observed in x64370, pKEN14-28, pF4, S1C1, and pAM66 genes and to Ile53-->Val53 (ATA-->GTA) base substitution in PKPI-9, P1G7, pKEN14-28, pF4, S1C1, pAM66, PI-2, and gCDT-B1 genes. Potential recombination points in PKPI-B should be located in the conserved regions or at borders of regions with lower conservancy.Fig. 4. Scheme of distribution of conserved (gray), moderately variable (white), and highly variable (black) regions in PKPI-B protein molecules. Numbers delineate addresses in amino acid sequences. R and M residues are involved in the reactive centers (Arg68, Met116, Met153); disulfide bonds Cys49-Cys98 and Cys147-Cys164 are denoted as one and two asterisks, respectively; vertical black bar delineates site of posttranslational modification of the precursor after vacuolar translocation.
In conclusion, four PKPI-B encoding genes (B1, B2, B9, and B10) are harbored in the genome of potato cv. Istrinskii. Their analysis and clustering into a phylogenetic tree suggest interesting considerations on the domain structure and recombination events causing evolution of PKPI class-B genes in Solanum tuberosum.
This work was partially supported by the Russian Foundation for Basic Research (projects 04-04-48644, 04-04-48694, and 04-04-48693) and NATO-Russia Joint Research Support program (grant JSTC.RCLG 98102).
REFERENCES
1.Mosolov, V. V., and Valueva, T. A. (1993) Plant
Protein Inhibitors of Proteolytic Enzymes [in Russian], VINITI,
Moscow.
2.Ishikawa, A., Ohta, S., Matsuoka, K., Hattori, T.,
and Nakamura, K. (1994) Plant Cell Physiol., 35,
303-312.
3.Revina, T. A., Speranskaya, A. S., Kladnitskaya, G.
V., Shevelev, A. B., and Valueva, T. A. (2004) Biochemistry
(Moscow), 69, 1092-1098.
4.Poussereau, N., Creton, S., Billon-Grand, G.,
Rascl, C., and Fevre, M. (2001) Microbiology, 147,
717-726.
5.Valueva, T. A., Revina, T. A., Kladnitskaya, G. V.,
and Mosolov, V. V. (1998) FEBS Lett., 426, 131-134.
6.Valueva, T. A., Revina, T. A., Gvozdeva, E. L.,
Gerasimova, N. G., and Ozeretskovskaya, O. L. (2003) Bioorg
Khim., 29, 499-504.
7.Heibges, A., Glaczinski, H., Ballvora, A.,
Salamini, F., and Gebhardt, C. (2003) Mol. Gen. Genom.,
269, 526-534.
8.Heibges, A., Glaczinski, H., Ballvora, A.,
Salamini, F., and Gebhardt, C. (2003) Mol. Gen. Genom.,
269, 535-541.
9.Banfalvi, Z., Molnar, A., Molnar, G., Lakatos, L.,
and Szabo, L. (1996) FEBS Lett., 383, 159-164.
10.Pouvreau, L., Gruppen, H., van Koningsveld, G.
A., van den Broek, L. A. M., and Voragen, A. G. J. (2003) J. Agric.
Food Chem., 51, 5001-5003.
11.Maganja, D. B., Strukelj, B., Pungercar, J.,
Gubensek, F., Turk, V., and Kregar, I. (1992) Plant Mol. Biol.,
20, 311-313.
12.Speranskaya, A. S., Dorokhin, A.V., Novikova, S.
I., Shevelev, A. B., and Valueva, T. A. (2003) Genetika,
39, 1490-1497.
13.Valueva, T. A., Revina, T. A., Mosolov, V. V.,
and Mentele, R. (2000) Biol. Chem., 381, 1215-1221.
14.Barrett, A. J., Rawlings, N. D., and O'Brien, E.
A. (2001) J. Struct. Biol., 134, 95-102.
15.Mosolov, V. V., and Shul'gin, M. N. (1986)
Planta, 167, 595-600.
16.Garcia-Olmedo, F., Salcedo, G., Sanchez-Monge,
R., Gomez, L., Royo, J., and Carbonero, P. (1987) Oxford Surv. Plant
Mol. Cell Biol., 4, 275-334.
17.Ezhova, T. A., Soldatova, O. P., Penin, A. A.,
and Shestakov, S. V. (2002) The Plant Genome Molecular-Genetic
Mapping [in Russian], MAKS Press, Moscow.
18.Sambrook, J., Fritch, E., and Maniatis, T. (1989)
Molecular Cloning: A Laboratory Manual, 2nd Edn., Vol. 3, Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
19.Felsenstein, J. (1988) Ann. Rev. Genet.,
22, 521-565.