|
DNA Methylation and EpigenotypesR. Holliday12 Roma Court, West Pennant Hills, Sydney, NSW 2125, Australia; E-mail: RandL.Holliday@bigpond.com
|
Received October 9, 2004
The science of epigenetics is the study of all those mechanisms that control the unfolding of the genetic program for development and determine the phenotypes of differentiated cells. The pattern of gene expression in each of these cells is called the epigenotype. The best known and most thoroughly studied epigenetic mechanism is DNA methylation, which provides a basis both for the switching of gene activities, and the maintenance of stable phenotypes. The human epigenome project is the determination of the pattern of DNA methylation in multiple cell types. Some methylation sites, such as those in repeated genetic elements, are likely to be the same in all cell types, but genes with specialized functions will have distinct patterns of DNA methylation. Another project for the future is the study of the reprogramming of the genome in gametogenesis and early development. Much is already known about the de novo methylation of tumor suppressor genes in cancer cells, but the significance of epigenetic defects during ageing and in some familial diseases remains to be determined.
KEY WORDS: epigenetics, cancer, DNA methylation
WHAT IS AN EPIGENOTYPE?
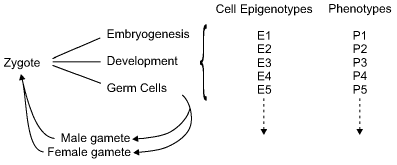
In classical genetics, the words genotype and phenotype are commonly used in single factor inheritance, but sometimes in two or more factor inheritance. For a simple recessive, the three possible genotypes are AA, Aa, and aa. The genotypes AA and Aa produce the same dominant phenotype, and aa produces the recessive phenotype. In a complex organism with many differentiated cells, there are many cell phenotypes, but in all these the genotype derived from the fertilized egg is the same (with rare exceptions). What then determines the phenotypes of specialized cells? It is due to all those mechanisms that ensure that a given set of genes will be active in any particular cell type, and that another set will be inactive. These mechanisms are epigenetic, and they determine the epigenotype of that particular cell. Thus, a complex organism has many cell epigenotypes.
Waddington [1] introduced the term epigenotype, but he had a very broad definition, namely: “The total developmental system consisting of interrelated developmental pathways through which the adult form of an organism is realized”. This is too general and all-embracing to be useful, hence the more specific definition introduced here.
Each complex organism is derived from the fusion of two haploid cells (gametes) to form the zygote. The mammalian zygote divides to form a number of seemingly identical totipotent cells, but soon these diverge into different cell types. They first form the three primordial layers of the embryo: the ectoderm, the mesoderm, and the endoderm, together with the extra-embryonic tissues. Although the inherited genes are the same in all the cells, the products of those genes are different in distinct cell types. We can say that the phenotypes of the cells diverge as the embryo develops: there are muscle cells, nerve cells, connective tissue cells, and so on. These can only arise from differential gene expression.
All cells (apart from anucleate erythrocytes) have the standard set of active genes required for normal metabolism. The products of such genes are commonly referred to as housekeeping proteins or enzymes. When specialized differentiated cells are formed, another set of proteins is synthesized, which are commonly known as luxury proteins. Thus, cell type A has a set of luxury proteins A´, and cell type B has a set of B´ proteins, and both have the same or similar housekeeping proteins. Cell A does not synthesize B´ proteins, and cell B does not synthesize A´ proteins. In some cases, both cells may synthesize C´ luxury proteins, or there may be none of these common to both types of cell. Thus, we can say that the distinct phenotypes of the two cell types A and B are due to their different epigenotypes.
Epigenetics includes all those mechanisms that give rise to differential gene expression in specialized cells, and these may include DNA methylation, or chromatin configurations, or combinations of the two. The upshot is a spectrum of both genes that are active and genes that are silent in any given cell type. Epigenetics also encompasses all those mechanisms that are responsible for the unfolding of the genetic program for development, and this depends on events such as cell signaling and many other cellular interactions. It includes the behavior of stem line cells, which divide to produce one identical daughter, and another cell which will produce one or more differentiated cell types. Thus, a bone marrow stem cell produces all the cell types in blood, but a skin stem cell may produce only keratinocytes. Epigenetic mechanisms are also responsible for genomic imprinting, whereby some genes derived from the paternal gamete and maternal gametes are differentially expressed.
When the adult organism is formed, it contains the totality of epigenotypes in its various cells, tissues, and organs. So far as we know all these epigenotypes have the same genotype, that is, the complete set of inherited maternal and paternal genes, with one important exception. The cells that will synthesize antibodies re-organize germ line DNA to assemble many different immunoglobulin genes, such that any one cell lineage has one particular set of such genes (reviewed in [2]). In addition, there are changes in base sequence through the activity of enzymes that have the effect of altering DNA sequences in the hypervariable regions of immunoglobulin genes [3, 4]. We should be cautious in asserting that such mechanisms do not occur in other specialized tissues, such as brain tissue. Nevertheless, one can formulate the generalization that the many distinct epigenotypes in a complex organism have a common genotype.
GENETIC VARIABILITY
So far, I have discussed the development of the fertilized egg to the adult, but a biological species comprises many male and female individuals, each of which has a unique genotype. There are multiple genetic polymorphisms throughout the genome, and there are also minisatellite regions of variable length that produce the DNA fingerprints characteristic of every individual. Remarkably, all these distinct genotypes produce normal individuals with the same set of epigenotypes (apart from those in the reproductive organs). The exceptions, which will be discussed later, are inherited defects that produce abnormalities in metabolism, or pre-disposition to certain diseases. Leaving these aside, we can say that in terms of anatomy and physiology, all males of the species, and all females, are much the same. Nevertheless, every individual is unique in his or her appearance. Thus, we can conclude that the genotype of every individual produces the outward phenotype, which is instantly recognizable. We know this from the fact that identical twins, which have the same genotypes, are also phenotypically identical, or at least so similar that they cannot be distinguished by casual observers. Exceptions to this may be very informative, and will be considered later.
Natural selection acts on inherited variation, and results over many generations in the appearance of new adaptations. Thus, in primate evolution there has been selection for brain size, and also for the anatomical differences between different species. We know that the genomes of man and chimpanzees have innumerable genes in common, and the epigenotypes of muscle cells, lymphocytes, fibroblasts, and so on, could only be distinguished by careful molecular scrutiny. Nevertheless some of the genetic differences produce distinct epigenetic outcomes, affecting such features as limb length, brain size, bodily hair, and so on. Thus the genome of any given species determines the phenotype of that species, and it is this that has allowed taxonomists to determine the degree of similarity or difference between species. Nowadays, however, DNA sequences provide a much better means of establishing relatedness, and evolutionary trees. Ultimately, the comparison of the human and chimpanzee genomes should provide us with the means to determine which DNA sequences are responsible for those uniquely human characteristics, including higher brain functions.
INHERITANCE AND STABILITY OF EPIGENOTYPES
Some specialized cells are post-mitotic, such as brain neurons or heart muscle cells. Their epigenotype and phenotype are remarkably stable, and in fact must last a lifetime. Some genes retain activity permanently, and others are permanently shut down. Only during ageing do the cells become dysfunctional, probably through the failure to maintain the normal integrity of macromolecules. Therefore to understand the ageing of these cells, we almost certainly need to understand the nature of the controls of gene activities. Some specialized cells divide, such as fibroblasts or lymphocytes. Again, their epigenotypes are stably maintained through innumerable mitotic divisions, although they also eventually become senescent and cease division [5].
The stability of dividing specialized cells demands a special mechanism, and this was one of the reasons for proposing that a heritable pattern of DNA methylation may be required [6, 7]. Maintenance methylases recognize hemi-methylated DNA at the replication fork, and methylate the new DNA strand. Such enzymes do not normally act on non-methylated DNA. This type of inheritance can also explain the stability of active and inactive X chromosomes in female mammals. Genes on the inactive X have methylated CpG islands, whereas the same CpG islands on the active X are non-methylated. This provides a model system, which may well explain the stability of the epigenotypes of dividing cells. Since the inactivation of one X chromosome is random, clones of cells are formed which may have different phenotypes, such as the black and ginger patches in a tortoiseshell cat. There is no reason to believe that clones are important during normal development. Indeed, all the evidence suggests that important epigenetic changes occur in groups of cells, for example a group of mesoderm cells may respond to a given diffusible signal and differentiate into muscle tissue. However, a bone marrow stem cell gives rise to a daughter cell which through clonal expansion gives rise to all the cell types in blood. This type of clonal inheritance is in some ways similar to the clone produced from the fertilized egg, with its diversity of cell types. Every human being consists of a clone of cells, together with all the extracellular material they produce.
THE HUMAN EPIGENOME
The determination of the molecular details of epigenotypes is a huge undertaking. At present, it would be true to say that the specificity of the processes at work is so poorly understood that the whole problem is a mysterious black box. We have some clues, which come from the study of DNA methylation. It has already been demonstrated that the pattern of methylation in certain genes in somatic cells is invariant between individuals [8, 9]. This suggests that the pattern is part of an important specificity defining the epigenotype of those cells. If the methylation was variable between individuals, it would probably not be important. In these same studies and many others it was found that the cells in which a gene is active, it is unmethylated, and methylated when it is inactive (reviewed in [10, 11]).
The epigenome project is enormously ambitious. The aim is to use modern methods of detecting cytosine methylation in the whole genome in a variety of differentiated cell types [11]. The results should show whether gene silencing can be attributed primarily to DNA methylation or whether other mechanisms are more important. Provided the gene product is known there are several methods to determine, whether or not a cell is or is not synthesizing it. These include Northern blots, Western blots, immunofluorescence, microchip arrays, or proteome analysis. A first step would be to sequence genes or gene regions of a cell type that is known to have a luxury protein, and the same region on another cell type in which the gene is silent. Many such comparisons should soon reveal the role of methylation in defining epigenotypes. If it does not have central role, then chromatin configurations are presumably involved, and new methods will be necessary to probe these. When the patterns of methylation are uncovered in the DNA of different tissues, it will generate the additional problem of understanding the instructions used for producing those patterns. Recently RNA has been implicated in methylating promoter sequences and silencing genes, and therefore another huge area of research has been opened up [12]. In the epigenome project it is to be expected that there will be many sites in the genome which are methylated in all types of cell. These will be the multiple genetic elements and ancient transposable elements which were probably silenced by methylation as a defense mechanism against intruding foreign DNA.
As well as the epigenome sequencing in somatic cells, it will also be essential to examine events in gametogenesis and early embryogenesis, where it is known that many methylation changes occur. At present very little is known about the reprogramming of the genome, which occurs during these stages of the life cycle. This is another major problem for the future, and it will provide the key to an understanding of the totipotency of embryonic stem cells (ES cells), and the whole process of early development. It will also provide the means to change ES cells into various differentiated ones, which many believe is necessary for the treatment or cure of diseases, which at present are incurable.
MENDELIAN AND EPIGENETIC INHERITANCE IN MAN
McKusick's Mendelian Inheritance in Man [13] documents thousands of inherited traits. Many of these are in genes coding for housekeeping proteins, and they commonly result in a standard metabolic defect. Others are in genes for specialized function, and one of the earliest known examples was sickle cell anemia, and the many other known mutations in hemoglobins. A third category is defects in genes of unknown function; the phenotype may be well characterized but the protein or enzyme involved may be unknown. This information will eventually come from the sequencing of the human genome. Indeed, one of the main driving forces, which launched that project, was the necessity of obtaining new insights into human disease. This includes an ever increasing list of genes that predispose individuals to various cancers, dementias, or other very significant human defects. At present some are well understood, such as the mutation causing retinoblastoma, but others are in genes of unknown function. Finally, there is a category of inherited traits, which have complex features. They are sometimes labeled “multifactorial with variable penetrance”. It is very likely that many of these actually have an epigenetic basis, rather than being due to standard gene mutations.
When a mutation in a gene coding for a luxury protein occurs, then the appropriate epigenotype, or epigenotypes, will be affected. Examples are inherited blood diseases, those that occur in the immune system, the digestive tract, and so on. We can also confidently predict that some alterations in epigenotypes will come from defects that are epigenetic.
This is one area in which the study of identical twins is so useful. It is known that their genotypes are identical, so any differences seen may have an epigenetic basis. The list of discordant features of identical twins is continually growing, and one of the most interesting is lifespan [14-16]. Although identical twins have a more similar lifespan that non-identical twins or siblings of the same sex, there is a surprising extent of difference. Sometimes this is assumed to be due environmental influences, which probably provides a partial explanation. However, it is more likely that stochastic changes, including those in epigenotypes, are of greater significance. One can be confident about this, because inbred mice kept under identical environmental conditions, have quite variable lifespans [17]. Nowadays it is commonly stated that disease is either genetic or environmental, when in reality stochastic events are equally important. In the absence of strong predisposing factors, such as asbestos-related mesothelioma, the cumulative changes that characterize many carcinomas are the result of chance. It is simply bad luck if a mutation occurs in an oncogene or tumor suppressor gene, and the same can be said of epigenetic defects in these or other genes.
GENOMIC IMPRINTING
The mammalian genome is subject to imprinting. This a process whereby information is added to DNA sequences, such that imprinted genes may be active in the male gametes, and inactive in female, or vice versa. In this regard, male and female gametes have different epigenotypes. The imprinted genes complement each other in the zygote, and normal development proceeds. Two male derived genomes, or two female derived genomes produce an abnormal embryo or fetus. Differential DNA methylation of specific cytosine residues is known to be one essential feature of imprinting, and this methylation is erased and re-established during gametogenesis (reviewed in [18]).
Imprinted genes usually function early in development, and a plausible argument can be made that there is a necessity for these genes to be present in single copy. If we assume that switches in a gene's activity are important in unfolding the program for development, then it is easy to see that the switching of a singe copy to + and - is very much simpler than the switching of two copies, which would produce a +/- outcome in both daughter cells half the time. This interpretation provides a specific function for imprinting, which is otherwise elusive [19]. If imprinting signals are important in early development, they may not be long lasting. The loss of the signal would be an epigenetic change normally without significance. This may well be the reason why the cloning of mammalian species is only rarely successful, and the cloned animals have abnormalities. Much more information will become available about the fate of imprinting signals from the epigenome project, which can compare gamete genomes, those of the early and late embryo, and those of somatic cells.
EPIGENOTYPES, CANCER, AND AGEING
Cancer is a disease, which changes the phenotype of normal cells. Usually this is a multistep process, so that some features of the normal cell are lost and others are preserved. Finally, the malignant cell is the most de-differentiated. Thus, during tumorigenesis the initial epigenotype is progressively altered. It is also evident that genetic stability is lost, because the karyotype becomes very variable. The same can be said of the pattern of DNA methylation. Thus, tumor progression is due to the cellular selection of subclones from the initial clonal event. It is obvious that the cells that are selected that are most resistant to the normal cellular signals that preserve cell and tissue homeostasis. Five base genomic sequencing of tumor cells demonstrates that methylation is highly heterogeneous. In some DNA sequences, there is decreased methylation, whereas some CpG islands, which are normally unmethylated, become subject to de novo methylation. A very large number of tumor suppressor genes have been shown to be methylated and silenced in a wide array of tumors [20].
It is also evident that epigenotypes are much more stable in large long-lived mammals than in small short-lived ones [17, 21, 22]. This makes biological sense because epigenotypes must last a lifetime. One of the features of long life is the stability of various macromolecules and cellular components. There must be an efficient buffer against changes in epigenotypes, which is lacking in short lived animals such as rodents. This is also seen in such epigenetic controls as X chromosome inactivation, which is far more stable in man than in mouse. Ageing is the eventual failure of all those maintenance, repair, and control processes which preserve the integrity of the soma, with all its epigenotypes [17]. To fully understand ageing much more information is needed about the way all the maintenance mechanisms operate, and how they eventually fail. Much has been written about ageing, somatic mutations, and oxygen free radical damage. It is very likely that epigenetic defects are important in ageing. One way of examining this is to use powerful new methods to look for ectopic expression of proteins. Thus cells of a given epigenotype may occasionally synthesize illegitimate proteins, that is, a protein normally found in some other epigenotype. This could arise by abnormal changes in DNA methylation. So far, such ectopic gene expression has not been detected in ageing cells.
CONCLUSION
In the last decade or so, the use of the word epigenetics has rapidly spread. It has been realized that there is a set of mechanisms that are responsible for the unfolding of the genetic program for development, and also for maintaining the stability of differentiated cells, whether dividing or post-mitotic. The totality of these mechanisms constitutes the new science of epigenetics, and one of the best known and now best documented is DNA methylation. This provides the means to switch gene activities and also to maintain the stability of differentiated cells.
The term epigenotype is used to describe the pattern of gene and gene inactivation in any one cell type (figure). In a given species, normal individuals have the same set of epigenotypes (apart from those determining sexual differences). The epigenome project is a huge undertaking which will document the pattern of DNA methylation in the whole human genome in multiple differentiated cells. One can expect that the methylation of repetitive or ancient transposable elements will be constant between cells, but genes with specialized functions will be very different. The complexity will be such that some new form of notation will be required to describe each epigenotype.
I thank Julian Sale and Lily Huschtcha for providing up-to-date information.Another major undertaking in the future will be to determine the reprogramming of the mammalian genome during gametogenesis and early development. It is already known that the patterns of DNA methylation in tumor cells are abnormal and also heterogeneous, and that tumor suppressor genes often become methylated and silenced. A challenge is to determine the extent of epigenetic defects during ageing. This will be difficult because such defects will be heterogeneous in the cells of any one tissue. Another area for future study is the assessment of the importance of heritable epigenetic defects in diseases known to have a familial component, but not one that is due to normal Mendelian inheritance.
REFERENCES
1.Waddington, C. H. (1939) An Introduction to
Modern Genetics, Allen and Unwin, London.
2.Hozumi, N., and Tonegawa, S. (2004) J.
Immunol., 173, 4260-4264.
3.Neuberger, M. S., Harris, R. S., Di Noia, J., and
Petersen-Mahrt, S. K. (2003) Trend Biochem. Sci., 28,
305-312.
4.Pham, P., Bransteitter, R., Petruska, J., and
Goodman, M. F. (2003) Nature, 424, 103-107.
5.Holliday, R. (2001) in Stem Cell Biology
(Marshak, D. R., Gardner, R. L., and Gottleib, D., eds.) Cold
Spring Harbor Laboratory Press, New York, pp. 95-109.
6.Holliday, R., and Pugh, J. E. (1975)
Science, 187, 226-232.
7.Riggs, A. D. (1975) Cytogenet. Cell Genet.,
14, 9-25.
8.Kochanek, S., Toth, M., Dehmal, A., Renz, D., and
Doerfler, W. (1990) Proc. Natl. Acad. Sci. USA, 87,
8830-8834.
9.Behn-Krappa, A., Holker, I., Sandaradura de Silva,
U., and Doerfler, W. (1991) Genomics, 11, 1-7.
10.Doerfler, W. (2003) in The Epigenome:
Molecular Hide and Seek (Beck, S., and Olek, A., eds.) Wiley-VCH,
Weinheim, Germany, pp. 23-38.
11.Novick, K. L., Nimmrich, L., Gene, B., Mair, S.,
Piepenbrock, C., Olek, A., and Beck, S. (2002) Curr. Issues Mol.
Biol., 4, 111-128.
12.Kawasaki, H., and Taira, K. (2004) Nature,
431, 211-217.
13.McKusick, V. A. (1998) Mendelian Inheritance
in Man, 12th Edn., Johns Hopkins University Press, Baltimore.
14.Singh, S. M., Murphy, B., and O'Reilly, R. (2002)
Clin. Genet., 62, 97-107.
15.Ljungquist, B., Berg, S., Lanke, J., McLearn, G.
E., and Pedersen, N. L. (1998) J. Geront. A Biol. Sci. Med.
Sci., 53, M441-446.
16.Carmelli, D. (1982) Hum. Biol., 54,
525-537.
17.Holliday, R. (1995) Understanding Ageing,
Cambridge University Press, Cambridge.
18.Ferguson-Smith, A. (2003) in The Epigenome:
Molecular Hide and Seek (Beck, S., and Olek, A., eds.) Wiley-VCH
Weinheim, Germany, pp. 81-99.
19.Holliday, R. (1990) in Genomic Imprinting
(Monk, M., and Surani, A., eds.) The Company of Biologists Ltd.,
Cambridge, pp. 125-129.
20.Millar, D. S., Holliday, R., and Grigg, G. W.
(2003) in The Epigenome: Molecular Hide and Seek (Beck, S., and
Olek, A., eds.) Wiley-VCH, Weinheim, Germany, pp. 3-20.
21.Holliday, R. (1987) Science, 238,
163-170.
22.Holliday, R. (1996) in Genetic Instability in
Cancer (Lindahl, T., ed.) Cancer Surveys, 28, Cold
Spring Harbor Laboratory Press, New York, pp. 103-115.