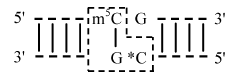REVIEW: DNA Methyltransferases and Structural-Functional Specificity of Eukaryotic DNA Modification#
Ya. I. Buryanov* and T. V. Shevchuk
Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Pushchino Branch, Russian Academy of Sciences, pr. Nauki 6, 142290 Pushchino, Moscow Region, Russia; fax: (8-27) 33-0527; E-mail: buryanov@fibkh.serpukhov.su* To whom correspondence should be addressed.
# This article was not published in the journal special issue devoted to the 70th anniversary of B. F. Vanyushin (Biochemistry (Moscow) (2005) 70, No. 5) because of limiting volume of the journal.
Received October 27, 2004
Properties of the main families of mammalian, plant, and fungal DNA methyltransferases are considered. Structural-functional specificity of eukaryotic genome sequences methylated by DNA methyltransferases is characterized. The total methylation of cytosine in DNA sequences is described, as well as its relation with RNA interference. Mechanisms of regulation of expression and modulation of DNA methyltransferase activity in the eukaryotic cell are discussed.
KEY WORDS: DNA, methylation, DNA methyltransferases, CpG, CpNpG, and CpNpN sequences, structure, functions, eukaryotes
Abbreviations: DNA methylase) DNA methyltransferase; Dnmt1, Dnmt2, Dnmt3) DNA methylases of different mammalian families; TRD) the site-recognizing domain of DNA methylase; Masc) DNA methylases of different families of the fungus Ascobolus; MET, CMT, DRM) DNA methylases of different plant families; MIP) methylation induced premeiotically; m5C) 5-methylcytosine; N) any nucleoside; PTGS) post-transcriptional gene silencing; TGS) transcriptional gene silencing; RIP) repeat-induced point mutation.
Although enzymatic methylation of eukaryotic DNA has been under
investigation for more than 50 years, we are still far from a
comprehensive understanding of the functional role of this modification
of the genome. There are two main stages in studies on the enzymatic
methylation of eukaryotic DNA.
In the first stage the species-, cell-, tissue-associated [1-4], and intragenomic specificities [5-11] of animal and plant DNA methylation were shown by classical biochemical methods. There were also found changes in the 5-methylcytosine content in the eukaryotic genome during ontogeny [6, 7] and changes in DNA methylation during carcinogenesis and under the influence of various physiological factors [8-11]. Russian scientists of A. N. Belozersky's school have a significant priority in the contribution to the structural-functional studies on eukaryotic genome methylation and establishing the involvement of DNA methylation in the regulation of genetic expression. During this stage the CpG-type methylation of animal and plant DNA [12-16] was shown, as well as the CpNpG-type (N is any nucleoside) methylation of higher plant DNA [17, 18].
The second and still current stage of studies on the enzymatic methylation of eukaryotic DNA is associated with revolutionary achievements in molecular genetics, sequencing of methylated DNAs, and also with isolation and investigation of DNA methyltransferases (DNA methylases) as they are. Voluminous information has been obtained about the distribution of methylated CpG sequences and unmethylated CpG islands in the genome of vertebrates [19-21] and plants [22], and the first data have also been obtained about the CpNpG-type DNA methylation in mammals [23] and the CpNpN-asymmetric methylation in cells of fungi [24], animals [25], and plants [26].
At present, enzymatic DNA methylation in eukaryotes is known to be involved in the regulation of gene transcription, cell differentiation, and embryogenesis [27], epigenetic control of genome imprinting [28], and inactivation of mobile genetic elements [27, 29]. This methylation is found in both mammals and higher plants. The normal pattern of DNA methylation is disturbed in carcinogenesis and human hereditary diseases. Significant progress in studies on the role of DNA methylation was achieved after the discovery of proteins binding to methylated CpG sequences of DNA and mobilizing into these regions histone deacetylases, which produce the transcriptionally inactive chromatin structure. In this context, “methylation meets acetylation” [30], i.e., DNA methylation and histone deacetylation seem to be strongly coupled with generation of an untranscribable chromatin structure.
But there is a pronounced gap in structural-functional studies on the enzymatic methylation of eukaryotic DNAs, because only the CpG-type of this modification has been investigated. At present, the association of other site-specific types of eukaryotic DNA methylation with various genetic functions becomes clearer. But analysis of the methylation pattern of the whole eukaryotic genome, which is the background of normal and disordered cell life activities, seems promising for understanding the functional role of these types of DNA methylation and elucidating the role of individual DNA methylases in specific genetic processes.
The present review considers features of the main families of eukaryotic DNA methylases, the structural-functional specificity of the genome sequences modified by them, and also possible mechanisms of regulation of DNA methylase activities in the cell.
EUKARYOTIC CYTOSINE(C5)-DNA METHYLTRANSFERASES
Cytosine(C5)-DNA methyltransferases catalyze the transfer of a methyl group from S-adenosyl-methionine onto cytosine residues in specific sequences of duplex DNA, with production of 5-methylcytosine and S-adenosyl-homocysteine. This reaction is irreversible. A comparison of the primary structures of prokaryotic and eukaryotic DNA methylases allows us to assign them to the same class of enzymes with identical catalytic structure. All these enzymes are monomeric proteins with some conservative homologous regions (motifs) in the structure, which determine the specific enzymatic functions. For most proteins, cytosine(C5)-DNA methyltransferases have up to 10 conservative regions arranged in a strictly defined sequence. Comparison of the primary structures of cytosine(C5)-DNA methyltransferases reveals the association of their major functions with their conservative motifs, whereas the site-specific recognition belongs to a variable region of the target-recognizing domain (TRD) [31]. Among ten conservative blocks of amino acids in cytosine(C5)-DNA methyltransferases, four moderately homologous motifs (II, III, V, and VII) are found which can be absent in some of the enzymes and also six highly homologous motifs (I, IV, VI, VIII, IX, and X) [32]. The variable TRD region located between motifs VIII and IX significantly varies in amino acid sequence and its length in site-specific methyltransferases. The conservative motifs are responsible for the common function of all methyltransferases, i.e., the catalytic transfer of a methyl group from S-adenosyl-methionine onto DNA, whereas the variable TRD region determines recognition of the specific DNA sequence and methylation in it of the heterocyclic base [33].
Eukaryotic DNA methylases methylate cytosine in the half-methylated replicating DNA in the symmetric sequences CpG and CpNpG, and this presents a semiconservative inheritance of the methylation pattern of the parental DNA (the so-called maintenance methylation). They also methylate fully unmethylated sequences, i.e., perform de novo DNA methylation.
DNA METHYLTRANSFERASES OF MAMMALS
The Dnmt1 family. Structural-functional studies on genes of eukaryotic DNA methylases were started markedly later than similar studies in prokaryotes. The cDNA of the DNMT1 gene encoding the full-size gene of mouse DNA methylase was first cloned in 1988 [34], and then it was expressed in mammalian cells [35] and in E. coli [36]. The expressed cDNA encodes a 190-kD protein of 1620 amino acid residues, which manifests optimal methyltransferase activity on half-methylated DNA. This enzyme is significantly larger than the prokaryotic enzymes due to presence on the N-terminal part of the molecule of a rather elongated additional region of about two thirds of the whole molecule [34, 37]. The DNA methylase Dnmt1 performs in the cell maintenance methylation, and this function is controlled by its N-terminal domain. The N-terminal domain of the DNA methylase Dnmt1 provides for the discrimination by the enzyme of unmethylated and half-methylated CpG sequences in DNA and in vivo and in vitro methylates preferentially these half-methylated sites. Deprived of its N-terminal domain, the enzyme loses this ability and changes to a typical prokaryotic DNA methylase [38]. However, recently this principle was revised, and the activity of the Dnmt1 methylase was shown to need also a significant part of the N-terminal domain without the first 300 amino acids [39]. The N-terminal domain is suggested to be necessary for correct formation of the tertiary structure of the Dnmt1 methylase [40]. The DNMT1 gene seems to be produced by fusion of the prokaryotic DNA methylase gene with one [27] or two [39] genes that encode proteins binding to DNA.
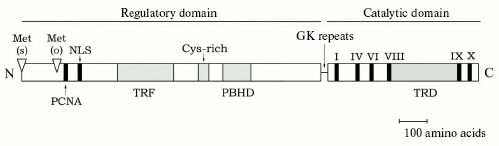
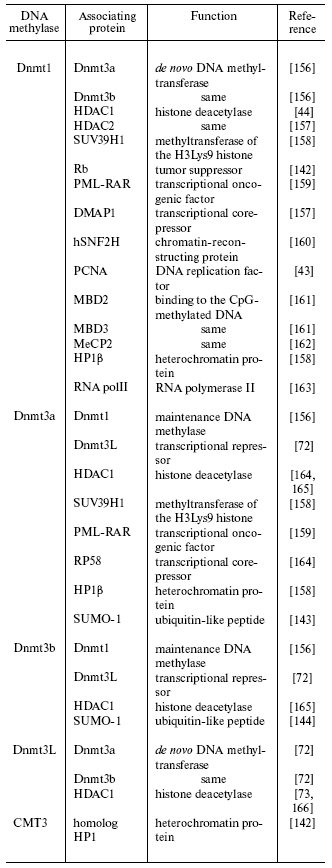
The N-terminal domain of DNA methylase Dnmt1 contains varied specific functional sequences, such as the nuclear localization signal (NLS), the cysteine-enriched zinc-binding motif, and a special sequence directing the methylase into the area of DNA replication (Fig. 1). The enzyme is associated with the replication foci during S-phase and is diffused in nucleoplasm of the cells off the S-phase [41]. The human DNMT1 gene cDNA has also been cloned and characterized [37, 42]. The structure of this methylase, including the N-terminal domain, resembles the structure of the corresponding mouse methylase. The human and animal Dnmt1 methylase is a component of the replicative complex. In particular, this is confirmed by finding the enzyme in the complex with the nuclear antigen of human proliferating cells [43]. The N-terminal regulatory and C-terminal catalytic domains of the Dnmt1 molecule are bound through GK amino acid repeats [34, 37]. Note that the DNA methylase Dnmt1 contains in its N-terminal domain amino acid sequences homologous to the transcriptional repressor HRX through which the enzyme is in vivo associated with histone deacetylase [44]. Some proteins capable of association with eukaryotic DNA methylases are presented in the table.
Proteins associating with DNA methyltransferasesFig. 1. Structure of the Dnmt1 family of DNA methyltransferases. The N- and C-terminal domains are separated by GK repeats. Roman figures denote the major conservative motifs of the C-terminal catalytic domain; TRD is the region responsible for recognition of the specific methylated sequence. In the N-terminal regulatory domain, functional sequences are marked which determine binding the proliferating cell nuclear antigen (PCNA), nuclear localization signal (NLS), protein targeting to DNA replication foci (TRF), binding with Zn2+ (Cys-rich), and homology with polybromo-1-protein (PBHD). Met(s) and Met(o) are the N-terminal positions of the somatic and oocyte specific protein forms, respectively.
Although Dnmt1 methylates mainly half-methylated sites, this enzyme can also methylate unusual substrates with varied structural anomalies [45, 46]. The mouse DNA methylase can methylate single-stranded DNA if the latter contains m5C in its chain. The enzyme is suggested to recognize m5C in the single-stranded DNA chain as a signal for methylation of unmodified cytosine residues in the CpG sequences and to operate on the loop areas of the single-stranded chain [47]. Similarly, the Eco dam bacterial DNA methylase involved in DNA replication and repair can modify single-stranded DNAs [48, 49]. And m5C can similarly function as a signal for induction of methylation on half-methylated and fully unmethylated CpG sequences in duplex substrates, and the rate of de novo methylation of such half-methylated substrates is several times higher than the rate of methylation of unmodified substrates by the enzyme [50]. The human Dnmt1 can selectively recognize and modify half-methylated asymmetric duplex substrates consisting of the “purpose target” CpG in one chain and the paired methylated cytidine residue in the complementary chain. But the neighborhood of this methylated cytidine with guanosine in the same chain is not necessary: any nucleoside or its derivative can be a neighbor [51] (Fig. 2). Substrates optimal for the de novo methylation contain other sequences of 13-17 nucleotides between unmodified CpG dinucleotides [50, 52, 53]. Note that the mouse DNA methylase can de novo methylate cytosine residues in sequences different from CpG, and this ability is more pronounced on single-stranded DNA [50]. Thus, it is not excluded that asymmetric methylation of cytosine residues in DNA observed in the cells of various eukaryotes can be realized by known DNA methylases, with involvement of specific regulatory factors modulating the specificity of recognition of the sequence to be methylated.
Inactivation of the mouse methylase DNMT1 gene resulted in a significant (up to 70%) decrease in the genome methylation and to death of developing embryos [54-56]. The remaining 30% level of DNA methylation and the ability of embryonal stem cells deprived of the Dnmt1 methylase for de novo methylation of retroviral DNA suggest that these functions were performed by other DNA methylases [56]. Such methylases were searched for in animals, and new enzymes of the Dnmt2 and Dnmt3 families were found.Fig. 2. Substrate specificity of DNA methyltransferases of the Dnmt1 family [51]. A half-methylated duplex sequence determines methylation of cytosine (*C) in the CpG sequence of the lower chain. The pairing of *C with guanine of the upper chain is not necessary. The asymmetric recognition site is in the frame.
The Dnmt2 family. The DNA methylase Dnmt2 consists of 415 amino acid residues, has no N-terminal domain, and seems to have no methyltransferase activity [57, 58]. But later the activity of this enzyme was found in human [59] and drosophila [60] cells. Inactivation by homologous knockout of the DNMT2 gene in mouse embryonal stem cells did not change the maintenance and de novo methylation of DNA, thus, the function of this enzyme remains unclear [61]. Genes of “short” DNA methylases were also detected in plants [62] and fungi [63].
The Dnmt3 family. The DNA methylases Dnmt3a and Dnmt3b are responsible for de novo methylation of DNA [61, 64]. The human Dnmt3a and Dnmt3b consist of 908 and 859 amino acid residues, respectively, and the DNMT3B gene can encode some shorter polypeptides by alternative splicing [64]. The cDNA of the mouse genes DNMT3A and DNMT3B are highly homologous to the corresponding human cDNAs [61, 64]. The de novo methylation of DNA is indicated by similar efficiency of modification by these enzymes of the CpG sequences in half-methylated and unmethylated native and synthetic substrates and also by their markedly decreased activities in mature somatic tissues [61]. The genes DNMT3A and DNMT3B are highly expressed in nondifferentiated embryonal stem cells where the methylases encoded by these genes are important not only for establishing but also for maintaining the general pattern of DNA methylation [65]. However, in differentiating cells and in somatic tissues of the mature organism the expression of these genes is extremely low [61]. On inactivation of the genes DNMT3A and DNMT3B in embryonal stem cells by homologous knockout, these cells lost their ability for de novo methylation of retroviral DNA. These genes are also required for normal post-embryonal development of mice: deficient animals died at about four weeks of age [66].
Although the enzymes Dnmt3a and Dnmt3b perform similar or overlapping functions, they also display specific differences. Thus, the Dnmt3b methylase is responsible for methylation of centromeric linker satellite repeats [66], and mutations in the human DNMT3B gene result in the ICF-syndrome (immunodeficiency centromeric instability, facial anomalies) [67]. The ICF-syndrome is a rare autosomal recessive disease, which is characterized by various immunological defects and abnormal structure of the face. This syndrome is associated with instability of centromeric heterochromatin [67]. In the ICF-syndrome, satellite DNA II and III (the major components of the constitutive heterochromatin) are hypomethylated [68].
A special protein of the Dnmt3 family denoted as Dnmt3L is also present in mammalian cells [69, 70]. This protein has no methyltransferase activity because some key amino acid motifs in it are absent or short. However, the Dnmt3L protein stimulates in the cell activities of the DNA methylases Dnmt3a and Dnmt3b and interacts with them [71, 72]. All proteins of the Dnmt3 family are colocated in the nucleus, and Dnmt3L is necessary for regulation of the de novo DNA methylation and establishing gene imprinting [70]. Note that Dnmt3L in association with histone deacetylase acts also as a transcriptional repressor [73].
The CpA sequences in DNA of mouse embryonal stem cells are significantly methylated, whereas the CpT sequences are methylated markedly less (15-20% of the total methylation of cytosine) [74]. Such a “non-CpG” methylation is found in both symmetric Cp(N)npG sequences and asymmetric sites. In embryonal stem cells with inactivated gene of the DNA methylase Dnmt1, the fraction of “non-CpG” methylation increased to 45% of the total methylation of DNA, and this suggested the association of this DNA methylase activity with the DNMT3A and DNM3B genes. Indeed, DNA of transgenic drosophila cells with the expressed DNMT3A gene, in addition to the CpG sequence, contained methylated cytosine also in the CpA sequence [74]. The Dnmt3 methylases have a significantly shorter N-terminal domain as compared to the Dnmt1 methylase, but this domain contains areas responsible for binding to various site-specific transcriptional repressors (table). Owing to this feature, all methylases of the Dnmt3 family can act as transcriptional repressors, and this function does not depend on their catalytic function of DNA methyltransferase [73, 75].
DNA METHYLTRANSFERASES OF PLANTS
The METI family. Three families of DNA methylases are found in plants, one of which is similar in structure and functions to the DNMT1 family of mammals. Thus, genes of DNA methylases encoding the proteins METI and METII are found in Arabidopsis thaliana [76]. The structure of DNA methylases METI and METII is similar to that of the mouse enzyme DNMT1, but these plant methylases are different in the structure of their N-terminal domains [76]. Two genes of DNA methylases are found in carrot and rice, which are homologous to DNA methylases METI of Arabidopsis [77, 78]. Transformed Arabidopsis plants with the anti-sense form of the gene of the DNA methylase METI are characterized by a decreased level of methylation and various anomalies in development [79]. Genes of the first class DNA methylases also include the recently identified gene NtMETI of tobacco, which encodes a 175-kD protein of 1556 amino acid residues [80]. Transcripts of this gene are found only in the cells of actively proliferating plant meristem and can be detected only during the S-phase in a synchronized suspension culture of tobacco cells. DNA methylase NtMETI is 59% homologous to the METI enzyme from Arabidopsis, and the anti-sense form of the NtMETI gene affects the morphogenesis of tobacco plants [80].
A single DNA methylase gene of the METI family is identified in pea cells [81]. The gene encodes a 174-kD protein of 1554 amino acid residues and is 65% homologous to the metI gene of DNA methylase from Arabidopsis. These proteins are 61% homologous on the level of amino acid sequence. DNA methylase of pea has the regulatory N-terminal domain and the catalytic C-terminal domain which includes eight of ten conservative regions of prokaryotic cytosine-(C5)-DNA methyltransferases. The catalytic domains of DNA methylases from pea and Arabidopsis are 78 and 52% homologous to the corresponding human domain. The gene of DNA methylase from pea is mainly expressed in the apical meristem and embryonal tissue with rapidly proliferating cells and is not expressed in leaves, hypocotyl, and mature roots. In the baculovirus expression system cDNA of the pea DNA methylase gene determines a 182-kD protein which displays the methyltransferase activity on pea DNA and duplex synthetic oligonucleotides containing the CG sequence and the half-methylated CWG sequence (where W = A or T) [81].
Thus, the single pea DNA methylase displays both the maintenance and de novo activity, as well as the methylation types CpG and CpNpG. At first glance, these properties do not correspond to isolation of two DNA methylases from pea cells, one of which (with molecular weight of 150 kD) has the CpG type of methylation and the other (of 140 kD) methylates the CpA/TpG sequence [82]. However, the variable TRD domain of the pea DNA methylase, which is responsible for methylation of the specific nucleotide sequence, noticeably differs in size from the TRD of the mouse and human enzymes. Tandem TRDs responsible for separate methylation of the CpG and CpNpG sequences can also be located in this region [81]. Clusters of basic amino acids, which are possible targets of proteinases, are found in different regions of the N-terminal domain of rice and pea DNA methylases. However, these sequences are absent in the C-terminal domain [78, 81]. Possibly, proteolytic processing leads to the appearance of the pea DNA methylase with molecular weights of 140, 110, 100, and 53 kD, which retain the intact catalytic domain. In fact, low-molecular-weight DNA methylases with the CpG methylation type were isolated from nuclei of various plants (pea [82], wheat [83, 84], and rice [85]). Thus, in plants the same gene of DNA methylase can encode proteins performing either two types of CpG and CpNpG methylation or one of these types. But the CpNpG sequences and, possibly, asymmetric sites in plant DNA can be also methylated by DNA methylases of a special family of chromomethylases (CMT) (Fig. 3).
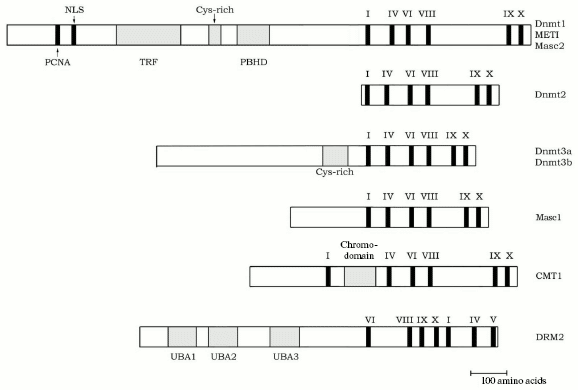
The chromomethylase family. Polymorphic DNA methylases of this family were first discovered in Arabidopsis. These methylases contain between blocks I and IV a chromodomain of 80 amino acid residues, which provides for their interaction with specific chromatin proteins and the nuclear membrane [86]. In Arabidopsis cells, CMT3 is involved in the maintenance hypermethylation in CpNpG and asymmetric DNA sites, and the lack of methylation in these sequences reactivates endogenous transposons [87]. The chromomethylase Zmet2 from maize performs a similar function [88]. So far chromomethylases have been found only in plants.Fig. 3. Scheme of structures of different families of DNA methyltransferases. The functional sequences in the N-terminal domain of methylases of the Dnmt1 and Dnmt3 families are denoted as in Fig. 1. Roman numerals indicate the major conservative motifs of the C-terminal catalytic domain; UBA are areas of potential ubiquitination of the DRM2 family methylases.
The DRM family. Plants possess another class of DNA methylases, domain rearranged methylases (DRM), which have conservative motifs in the catalytic domain disposed otherwise: VI - IX - X - I - II - III - IV - V (Fig. 3) [89]. Genes of these enzymes have been found in Arabidopsis, maize, and, possibly, in soya. Although the functionally important amino acid blocks are rearranged, the three-dimensional packing of the catalytic domain in these molecules is similar to the structure of the bacterial cytosine DNA methylase HhaI. The N-terminal regulatory region of DRM contains ubiquitin-binding sequences, and this suggests a possibility of ubiquitination of these DNA methylases. These proteins are most similar in functions with DNA methylases of the Dnmt3 family. It is suggested that DRM can de novo methylate DNA in asymmetric sequences and maintain this modification of cytosine during inactivation of transposons and transgenic silencing [89].
Plants are likely to have also other DNA methylases. Thus, in Arabidopsis the gene METIII of DNA methylase has been detected which encodes an enzyme deprived of the N-terminal domain. This gene cannot hybridize with the genes of METI and METII DNA methylases [62]. Unlike the situation in animals, decreased methylation of the plant genome is not lethal but causes anomalies in development and appearance of new phenotypes [76, 90-92].
TOTAL METHYLATION OF CYTOSINE IN SEQUENCES OF PLANT DNA AND RNA
INTERFERENCE
Gene silencing occurs at about 30% probability among independent genetic plant transformants and is also described for other transgenic organisms [93]. Gene silencing depends on the existence of repeating copies of transgenes or the presence in the transgenes and inherent genes of homologous sequences. In particular, gene silencing is observed at the repeating transformation of transgenic plants caused by homologous genetic constructions [94]. A coordinated suppression (co-suppression) of homologous transgenes and the inherent genes is also found in transformed plants [95, 96]. A similar process called quelling is also reported for Neurospora crassa. Gene silencing can be associated with the cell defense against invasion of foreign genetic elements [97]. Gene silencing is accompanied by their intense methylation, including asymmetric sequences. Such a specific picture of DNA methylation in plant cells was first observed in the transgenic plants Petunia hybrida which expressed the maize gene of dihydroflavonol reductase and showed unexpected changes in flower color [98].
The cDNA of potato spindle tuber virus integrated into the tobacco genome is intensely hypermethylated in both chains (90-100% of all cytosine residues) only if the transgenic plants are infected with this virus and it replicates in the plant cells. Involvement of RNA is necessary for the intense de novo methylation of DNA in symmetric and asymmetric sequences [99, 100].
There are two types of gene silencing that depend on the presence of homologous sequences: transcriptional and post-transcriptional gene silencing (TGS and PTGS, respectively). In TGS, there is no transcription of transgenes, whereas PTGS is characterized by prolonged transcription, but the half-life of the RNA is lowered so sharply that it is difficult to detect polyadenylated transcripts [101]. Both TGS and PTGS in plants are accompanied by hypermethylation of the DNA that is extended onto the majority of cytosine residues [102].
Both TGS and PTGS associated with hypermethylation of the turned off genes are directed by specific RNA molecules [103]. This RNA is double-stranded, which is unusual for normal cells but characteristic for replicating viruses and viroids. The antiviral resistance of plants and silencing of genes can be obtained by concurrent expression in the cell of both the sense and anti-sense RNA [104]. Note, that the double-stranded RNA corresponding to the sense and anti-sense sequences of endogenous mRNA induces gene silencing in various organisms, from trypanosomes to mammals and plants [105].
In the cells of transgenic plants with PTGS-induced silencing of foreign genes the sense and anti-sense 21-25-meric oligoribonucleotides are present complementary to the appropriate mRNA. Such oligoribonucleotides are absent in the cells of transgenic plants with normally expressed foreign genes [106]. In Drosophila, similar 21-23-meric duplex short interfering RNAs (siRNA), which induced the mRNA degradation, were found a little later [107, 108]. This phenomenon was called RNA interference. It is established that PTGS in plants, quelling in Neurospora crassa, and RNA interference in mammals are functionally equivalent and have in common the same mechanism of generation of siRNAs which induce degradation of mRNAs. In plants, the RNA-directed methylation modifies the majority of cytosine residues inside the area of RNA-DNA homology and results in methylation of the gene-encoding regions during PTGS and of the promotor areas during TGS [109]. RNA interference occurs in organisms with poorly or incompletely methylated DNA, e.g., in Drosophila and in the nematode Caenorhabditis elegans. However, the mechanism of siRNA generation is the same in all organisms, and RNA interference triggers similar processes of degradation of mRNAs, histone methylation, and production of the untranscribable heterochromatin [107, 108].
Thus, DNA methylation in plants, and possibly in mammals, does not determine RNA interference but during this process seems to perform some very specific functions, which may be absent in other eukaryotes. The mechanism of the RNA-directed intense DNA methylation during gene silencing has not been studied. The type of DNA methylase involved is unknown. Another problem is associated with the mechanism of maintaining the de novo established asymmetric DNA methylation in transformed plant cells.
Information about asymmetric DNA methylation in plant cells is mainly obtained for foreign genes and concerns de novo DNA methylation. However, endogenous sequences of plant DNA can be methylated similarly. Thus, in mutant Arabidopsis plants with decreased DNA methylation (DDM) and in transgenic plants with the anti-sense form of the DNA methylase METI gene the single-copy genes SUP (SUPERMAN) and AG (AGAMOUS) that determine the flower morphology are hypermethylated [110, 111]. The cytosine methylation in these genes is the highest in asymmetric pyrimidine-enriched sequences, lower in symmetric CpNpG sequences, and minimal in the CpG sequence. These genes are hypermethylated on the background of general hypomethylation of DNA and disordered development of the plants, and this picture is similar to the DNA methylation in tumor cells and expression of genes, which are normally repressed [112].
Hypermethylation in plants can occur under stress conditions. Thus, in the presence of antibiotics massive DNA hypermethylation of cytosine in tobacco plants mainly occurs in the CpG sequence [113]. It seems that with hypermethylation asymmetric DNA methylation begins on the palindrome sequences CpG or CpNpG and then extends onto other cytosine residues in a rather long DNA area.
TOTAL METHYLATION OF FUNGAL DNA SEQUENCES
The specific recognition and modification of repeating nucleotide sequences was first detected in the fungus Neurospora crassa [114, 115] and was designated repeat-induced point mutation (RIP). A similar process, methylation-induced premeiotically (MIP), was discovered in the fungus Ascobolus immersus [116, 117]. It seems that both processes initially protected the genome against transposons [118]. In the cells of fungi, these finely regulated processes occur during the sexual cycle in the time interval between fertilization and karyogamy when haploid nuclei of both crossing cells are present in the joint cytoplasm. If the haploid genome contains two or more copies of sequences of more than 300 n.p., they undergo modifications. During RIP in N. crassa duplicated sequences concurrently undergo mutational transitions G-C →_A-T and methylation, i.e., are changed genetically and epigenetically [118]. In A. immersus, duplicated sequences are only methylated during MIP [119]. Large repeating sequences, which escape RIP or MIP, are only tandem repeats of ribosomal RNAs, and this seems to be associated with their location in the nucleolus [119]. However, the tandem-organized genes of ribosomal RNAs of N. crassa are likely to belong to a small number of genes which are ordinarily methylated independently of RIP [120, 121]. Both tandem duplicated sequences and unconnected duplications are subjected to MIP. The MIP efficiency depends on the duplication size of these sequences [122-124] and is higher for tandem duplications [123]. Tandem repeats with the size of >630 n.p. are always subjected to MIP. The decrease in their size from 630 to 317 n.p. is associated with a dramatic decrease in the MIP rate. Unlike the case of tandem duplications, the methylation efficiency of unconnected duplications smoothly correlates with their size [119]. Methylation of cytosine residues inside the repeating genes results in their reversible silencing which can be, in particular, removed with the demethylating agent 5-azacytidine [117, 122]. Methylation of fungal genes during MIP and RIP decreases the content of corresponding mRNAs or fully eliminates them [123]. Methylation during MIP in Ascobolus does not affect initiation of transcription but inhibits its elongation [123]. In Neurospora, DNA methylation during RIP seems also to inhibit the elongation of transcription [118]. This feature discriminates the above-mentioned fungi from mammals and plants, in which DNA methylation prevents the initiation of transcription.
DNA methylation in Neurospora [125] and Ascobolus [126] is not limited to only symmetric sequences CpG and CpNpG but engages the whole duplicated DNA sequence. The question arises about the mechanism of inheritance and maintaining of this asymmetric methylation during every cycle of DNA replication in vegetative cells of Ascobolus mycelium. Possibly, the maintenance methylation of cytosine in asymmetric sequences depends on methylation of the adjacent symmetric sites. During MIP in Ascobolus in the duplicated DNA sequences with size of ~1000 n.p. all cytosine residues are methylated, whereas in the shorter sequences only CpG dinucleotides are methylated [124]. Thus, it is suggested that in the A. immersus cells there are two different mechanisms of the maintenance DNA methylation. The first mechanism controls the methylation of CpG dinucleotides, with involvement of a DNA methylase similar to the Dnmt1 enzyme of mammals, whereas the other mechanism determines the methylation of asymmetric sequences. The efficiency of the second mechanism depends on the size of the repeating DNA region. The methylation only of CpG dinucleotides in the short repeats suggests the interrelation between the two postulated mechanisms of DNA methylation [124]. In fact, two methylase genes of this ascomycete are known, MASC1 and MASC2.
The Masc1 family. The MASC1 gene encodes a 61.5-kD protein of 537 amino acid residues. The protein contains all 10 conservative motifs of cytosine(C5)-DNA methyltransferases, but is specified in the short TRD region between conservative motifs VIII and IX and the small size of the N-terminal domain nonhomologous to the N-terminal domain of the Dnmt1 family methylases [127]. The protein has no DNA methyltransferase activity in vitro. Homologous knockout of this gene fails to affect ascomycete viability and the maintenance methylation in its vegetative cells, but forbids the de novo methylation of repeating DNA sequences during MIP and results in sterility of the strains homozygous in this mutation [127].
The other methylase gene MASC2 of Ascobolus encodes a protein that has both the conservative catalytic domain and the large N-terminal domain and belongs to the Dnmt1 methylase family [128]. Mutation of the MASC2 gene does not influence the ability of Ascobolus for MIP, maintenance methylation, and de novo methylation in vegetative cells. Double mutations in the MASC1 and MASC2 genes also have no effect on the maintenance methylation of various genes [129]. Thus, these data suggest the presence in Ascobolus of at least the third gene responsible for the maintenance and de novo methylation in vegetative cells.
However, N. crassa has only the DNA methylase responsible for methylation of the whole genome [130]. This enzyme is encoded by the DIM-2 (defective in methylation) gene, and mutation in it leads to the full demethylation of DNA in symmetric and asymmetric sequences. The dim-2 protein consists of 1454 amino acid residues organized in the N- and C-terminal domains without the GK connection between them and is a member of the Dnmt1 family. However, it is the most unlike this family of proteins, especially in the structure of the N-terminal domain. This enzyme performs in the cell the maintenance and de novo methylation, but it is not involved in RIP, and its inactivity does not cause growth anomalies [130].
REGULATION OF EXPRESSION AND MODULATION OF ACTIVITY OF DNA
METHYLTRANSFERASES
DNA methylation is involved in the change-over of different genetic programs of the cell. Therefore, mechanisms capable of regulating the expression and activity modulation of DNA methylases themselves must exist. Studies on these mechanisms have been initiated only recently, and the regulation of expression of the appropriate genes is studied better. Transcription of DNA methylase genes of the Dnmt1, Dnmt3a, and Dnmt3b families is coordinated in normal human tissues, but this coordination is disturbed in tumors. Along with a moderately increased expression of the DNMT1 and DNMT3A genes, expression of the DNMT3B gene is increased significantly [131]. The complicated structure of eukaryotic genes of DNA methylases and of the encoded proteins suggests that they have varied regulatory elements. In particular, alternative splicing and transcription from different promotors can regulate on the gene level the expression of DNA methylase genes. Thus, the mouse DNMT1 gene (>56 t.n.p.) consists of 39 exons from 32 to 352 n.p. in size [39]. The long isoform of the Dnmt1 DNA methylase is translated from the third ATG codon of the first exon [132] and is present in embryonal stem cells and somatic tissues, whereas the short isoform is translated from the fourth ATG codon of the fourth exon and is found in oocytes and pre-implanted embryos [133].
Different isoforms of the human Dnmt3b DNA methylase are tissue specific [131]. Two sex-specific exons of the DNMT1 gene control its expression in mammalian oocytes [134]. Different isoforms of DNA methylases might be different in substrate specificity. The human DNMT1 gene can be transcribed from one major and three minor initiation sites and is regulated by independent promotors and enhancers [135], and this correlates with the existence of different isoforms of this enzyme in embryonal and somatic cells [134, 136]. The P1 site of the major promotor of the DNMT1 gene has a high content of CG sequences, which is characteristic for house-keeping genes, whereas these sequences are deficient in the P2-P4 sites of the minor promotors.
Thus, methylation of the DNA methylase gene can regulate its expression. Indeed, in mouse embryonal stem cells with highly expressed DNMT3L gene none of CpG dinucleotides in its promotor region are methylated, whereas they are methylated in the differentiated cells and tissues [137]. Between the P1 and P2-P4 promotors, three enhancers are located which are activated by the protooncogenic signal Ras-c-Jun and repressed by the tumor Rb-suppressor [135]. Thus, regulation of the DNMT1 gene transcription is essential for normal and oncogenic programs of the cell. The structure of the mouse DNMT1 gene includes the regulatory element AP-1 activated via the Ras-Jun protooncogenic signal pathway [138, 139]. The AP-1 regulatory area contains 29 CpG dinucleotides, which act as sensors of the genome methylation [140]. The DNMT1 gene expression is supposed to be regulated by the feedback principle. According to this hypothesis, the end product of methylation, i.e., methylated DNA, regulates the DNMT1 gene expression in the cis-position [140]. This hypothesis explains the paradoxical coexistence in tumor cells of generally insufficient DNA methylation and the high DNA methylase activity.
In the plant cell nucleus DNA methylation is controlled by phytohormones. Addition of gibberellin, 6-benzylaminopurine, and fusicoccin to nuclear extracts of wheat seedlings increased 30-65% the level of wheat DNA methylation. However, these phytohormones failed to stimulate the activity of partially purified DNA methylases, which suggests that their effect is mediated through nuclear proteins [84, 141]. It has been mentioned that the N-terminal domain of most eukaryotic DNA methylases contains varied functionally important sequences that determine their association with nuclear proteins (table). Therefore, the activities of DNA methylases are supposed to be regulated on the level of protein-protein interactions. Thus, the tumor Rb-suppressor can modulate the activity of human Dnmt1 methylase [142]. Proteolytic processing and enzymatic covalent modifications can also modulate activities of DNA methylases. In particular, modification of the Dnmt3a methylase by the ubiquitin-like peptide SUMO-1 modulates its interaction with histone deacetylases and the function of the transcriptional repressor [143]. Dnmt3b methylase is also modified by the peptide SUMO-1 [144]. At present, the place of DNA methylation is studied in the hierarchy of development of the epigenetic status of chromatin. Thus, DNA methylation in the CpNpG sequences in Arabidopsis plants is controlled by the primary methylation of the histone H3. This control is realized through interaction of the chromomethylase CMT3 with a homolog of the heterochromatin protein HP1 which, in turn, interacts with methylated lysine 9 of the H3 histone (H3Lys9) modified by the specific lysine histone H3 methyltransferase [145]. In N. crassa, expression of the DNA methylase dim-2 is also controlled by the histone H3Lys9 methyltransferase [146]. Methylation of Lys9 in histone H3 can, in turn, depend on the primary methylation of the CpG sequences in DNA [147].
It was recently established that in most eukaryotes the genome is methylated by numerous DNA methylases specifically involved in different genetic processes, sometimes without manifesting their catalytic methyltransferase function. Even in cells of lower eukaryotes possessing only one DNA methylase, this enzyme can perform multiple functions and modify cytosine in varied specific DNA sequences with involvement of yet unknown modulating factors.
Methylation of DNA can be also controlled on the level of DNA-protein interactions. Thus, the transcriptional factor Sp1 is often associated with unmethylated CpG islands in promotors of the house-keeping genes forbidding the de novo methylation and supporting the constitutive expression of these genes [148]. Deletion of the promotor area of the APRT gene with GC boxes or mutagenesis in them of the Sp1-recognizable sequences with the CpG sites resulted in de novo methylation of the CpG island of this gene [149].
However, proteins that bind to methylated DNA can retain its methylated state. Therefore, the existence should be noted of a special class of the DNA(m5CpG)-binding proteins [150]. It is possible that just these proteins can determine the strong feedback between the CpG- and CpNpG-types of methylation of specific genetic regions and their non-alternative CpG-type hypermethylation in some tumors [151]. Possibly, a shielding of one of the DNA chains by specific proteins, along with modulation of the DNA methylase activities, results in massive differential methylation of cytosine in one of the DNA chains of the centromeric region in chromosomes of plant seedlings [152].
In the cell, DNA methylases do not act on the level of simple DNA-protein complexes, but on DNA inside a complicated chromatin structure. The interrelations between the genome modification and numerous epigenetic modifications of chromatin proteins are fully realized on the level of chromatin. At present, it is important to elucidate the cause-effect consequence of DNA methylation and enzymatic modifications of nuclear proteins in the formation of specific chromatin structures.
It is also important to determine the role of different site-specific types of methylation in the normal genome and in disease. This information would be useful for diagnosis and prediction of diseases associated with abnormal DNA methylation and for treatment of such diseases. Therefore, it is necessary to elaborate approaches for monitoring individual structural-functional types of methylation of the total genome and subgenomic fractions and searching for abnormally methylated marker DNA sequences. Even now there are significant achievements in obtaining the genomic pattern of abnormal methylation of the CpG islands [153], some approaches have been developed for analysis of methylation of the CpNpG sequences in the total genome and subgenomic fractions [154], and some markers of abnormal DNA methylation have been found for some forms of carcinogenesis [155]. Further studies on the structural-functional picture of the eukaryotic genome methylation and pathways of its regulation are very important for understanding molecular principles of epigenetic processes.
The work was supported by the Russian Foundation for Basic Research (the project No. 04-04-48582).
REFERENCES
1.Vanyushin, B. F., and Belozersky, A. N. (1959)
Dokl. Akad. Nauk SSSR, 129, 944-946.
2.Marinova, E. I., Vkadychenskaya, N. S., and
Antonov, A. S. (1969) Dokl. Akad. Nauk SSSR, 185,
945-948.
3.Vanyushin, B. F., Tkacheva, S. G., and Belozersky,
A. N. (1970) Nature, 225, 948-949.
4.Vanyushin, B. F., Mazin, A. L., Vasilyev, V. K.,
and Belozersky, A. N. (1973) Biochim. Biophys. Acta, 299,
397-403.
5.Sulimova, G. E., Mazin, A. L., Vanyushin, B. F.,
and Belozersky, A. N. (1970) Dokl. Akad. Nauk SSSR, 193,
1422-1425.
6.Berdyshev, T. D., Korotaev, T. K., Boyarskikh, G.
V., and Vanyushin, B. F. (1967) Biokhimiya, 32,
988-993.
7.Drozhdenyuk, A. P., Sulimova, G. E., and Vanyushin,
B. F. (1976) Mol. Biol. (Moscow), 10, 1378-1386.
8.Vanyushin, B. F., Nemirovsky, I. E., Klemenko, V.
V., Vasilyev, V. K., and Belozersky, A. N. (1973) Gerontologia,
19, 138-152.
9.Burtseva, N. N., Azizov, Yu. M., Itkin, V. Z., and
Vanyushin, B. F. (1977) Biokhimiya, 42, 1690-1696.
10.Vanyushin, B. F., and Romanenko, E. B. (1979)
Biokhimiya, 44, 78-85.
11.Romanov, G. A., and Vanyushin, B. F. (1981)
Biochim. Biophys. Acta, 653, 204-218.
12.Sinsheimer, R. L. (1954) J. Biol. Chem.,
208, 445-459.
13.Sinsheimer, R. L. (1955) J. Biol. Chem.,
215, 579-583.
14.Shapiro, H. S., and Chargaff, E. (1960)
Biochim. Biophys. Acta, 39, 68-82.
15.Doskocil, J., and Sorm, F. (1962) Biochim.
Biophys. Acta, 55, 953-959.
16.Grippo, P., Iaccarino, M., Parisi, E., and
Scarano, E. (1968) J. Mol. Biol., 36, 195-208.
17.Kirnos, M. D., Aleksandrushkina, N. I., and
Vanyushin, B. F. (1981) Biokhimiya, 46, 1458-1474.
18.Gruenbaum, Y., Naveh-Many, T., Cedar, H., and
Razin, A. (1981) Nature, 292, 860-862.
19.Bird, A. P. (1986) Nature, 321,
209-213.
20.Gardiner-Garden, M., and Frommer, M. (1987) J.
Mol. Biol., 196, 261-282.
21.Antequera, F., and Bird, A. (1993) Proc. Natl.
Acad. Sci. USA, 90, 11995-11999.
22.Antequera, F., and Bird, A. (1988) EMBO
J., 7, 2295-2299.
23.Clark, S. J., Harrison, J., and Frommer, M.
(1995) Nature Genet., 10, 20-28.
24.Selker, E. U., and Stevens, J. N. (1985) Proc.
Natl. Acad. Sci. USA, 82, 8114-8118.
25.Toth, M., Müller, U., and Doerfler, W.
(1990) J. Mol. Biol., 214, 673-683.
26.Meyer, P., Niedenhof, I., and ten Lohuis, M.
(1994) EMBO J., 13, 2084-2088.
27.Bestor, T. H. (1990) Philos. Trans. R. Soc.
Lond. B. Biol. Sci., 326, 179-187.
28.Sapienza, C., Peterson, A., Rossant, J., and
Balling, R. (1987) Nature, 328, 251-254.
29.Yoder, J. A., Walsh, C. P., and Bestor, T. H.
(1997) Trends Genet., 13, 335-340.
30.Bestor, T. H. (1998) Nature, 393,
311-312.
31.Lauster, R., Trautner, T. A., and Nouer-Weidner,
M. (1989) J. Mol. Biol., 206, 305-312.
32.Kumar, S., Cheng, X., Klimasauskas, S., Sha, M.,
Posfai, J., Roberts, R. J., and Wilson, G. G. (1994) Nucleic Acids
Res., 22, 1-10.
33.Klimasauskas, S., Kumar, S., Roberts, R. J., and
Cheng, X. (1994) Cell, 76, 357-369.
34.Bestor, T., Laudano, A., Mattaliano, R., and
Ingram, V. (1988) J. Mol. Biol., 203, 971-983.
35.Czank, A., Hauselmann, R., Page, A. W.,
Leonhardt, H., Bestor, T. H., Schaffner, W., and Hergersberg, M. (1991)
Gene, 109, 259-263.
36.Tollefsbol, T. O., and Hutchinson, C. A., III
(1995) J. Biol. Chem., 270, 18543-18550.
37.Yen, R. W., Vertino, P. M., Nelkin, B. D., Yu, J.
J., el-Deiry, W., Cumaraswamy, A., Lennon, G. G., Trask, B. J., Celano,
P., and Baylin, S. B. (1992) Nucleic Acids Res., 20,
2287-2291.
38.Bestor, T. H. (1992) EMBO J., 11,
2611-2617.
39.Margot, J. B., Aguirre-Arteta, A. M., Di Giacco,
B. V., Pradhan, S., Roberts, R. J., Cardoso, M. C., and Leonhardt, H.
(2000) J. Mol. Biol., 297, 293-300.
40.Zimermann, C., Guhl, E., and Graessmann, A.
(1997) Biol. Chem., 378, 393-405.
41.Leonhardt, H., Page, A. W., Weier, H.-U., and
Bestor, T. H. (1992) Cell, 71, 865-873.
42.Cumaraswamy, G., Lennon, G., Trask, B. J.,
Celano, B. J., and Baylin, S. B. (1992) Nucleic Acids Res.,
20, 2287-2291.
43.Chuang, L. S., Ian, H. I., Koh, T. W., Ng, H. H.,
Xu, G., and Li, B. F. (1997) Science, 277, 1996-2000.
44.Fuks, F., Burgers, W. A., Brehm, A.,
Hughes-Davies, L., and Kouzaridis, T. (2000) Nature Genet.,
24, 88-91.
45.Smith, S. S., Kan, J. L. C., Baker, D. J.,
Kaplan, B. E., and Dembek, P. (1991) J. Mol. Biol., 217,
39-51.
46.Smith, S. S., Kaplan, B. E., Sowers, L. C., and
Newman, E. M. (1992) Proc. Natl. Acad. Sci. USA, 89,
4744-4748.
47.Christman, J. K., Sheikhnejad, G., Marasco, C.
J., and Sufrin, J. R. (1995) Proc. Natl. Acad. Sci. USA,
92, 7347-7351.
48.Buryanov, Ya. I., Zinoviev, V. V., Vienozhinskis,
M. T., Malygin, E. G., Nesterenko, V. F., Popov, S. G., and Gorbunov,
Yu. A. (1984) FEBS Lett., 168, 166-168.
49.Buryanov, Ya. I., Zinoviev, V. V., Gorbunov, Yu.
A., Tuzikov, F. V., Rechkunova, N. I., Malygin, E. G., and Bayev, A. A.
(1988) Gene, 74, 67-69.
50.Tollefsbol, T. O., and Hutchinson, C. A., III
(1997) J. Mol. Biol., 269, 494-504.
51.Smith, S. S. (1994) Progr. Nucleic Acid. Res.
Mol. Biol., 49, 65-111.
52.Bolden, A. H., Nalin, C. M., Ward, C. A.,
Poonian, M. S., and Weissbach, A. (1985) Mol. Cell. Biol.,
6, 1135-1140.
53.Bolden, A., Ward, C., Nalin, C., and Weissbach,
A. (1986) Progr. Nucleic Acids Res. Mol. Biol., 33,
231-250.
54.Li, E., Bestor, T. H., and Jaenisch, R. (1992)
Cell, 69, 915-926.
55.Li, E., Beard, C., Forster, A. C., Bestor, T. H.,
and Jaenisch, R. (1993) Cold Spring Harbor Symp. Quant. Biol.,
58, 297-305.
56.Lei, H., Oh, S. P., Okano, M., Jüttermann,
R., Goss, K. A., Jaenisch, R., and Li, E. (1996) Development,
122, 3195-3205.
57.Yoder, J. A., and Bestor, T. H. (1998) Hum.
Mol. Genet., 7, 279-284.
58.Okano, M., Xie, S., and Li, E. (1998) Nucleic
Acids Res., 26, 2536-2540.
59.Herman, A., Schmitt, S., and Jeltsch, A. (2003)
J. Biol. Chem., 278, 31717-31721.
60.Lyko, F., Ramsahoye, B. H., and Jaenisch, R.
(2000) Nature, 408, 538-540.
61.Okano, M., Xie, S., and Li, E. (1998) Nature
Genet., 19, 219-220.
62.Finnegan, E. J., Genger, R. K., Peacock, W. J.,
and Dennis, E. S. (1998) Annu. Rev. Plant Physiol. Plant Mol.
Biol., 49, 223-247.
63.Wilkinson, C. R., Bartlett, R., Nurse, P., and
Bird, A. P. (1995) Nucleic Acids Res., 23, 203-210.
64.Xie, S., Wang, Z., Okano, M., Nogami, M., Li, Y.,
He, W. W., Okumura, K., and Li, E. (1999) Gene, 236,
87-95.
65.Chen, T., Ueda, Y., Dodge, J., Wang, Z., and Li,
E. (2003) Mol. Cell. Biol., 23, 5594-5605.
66.Okano, M., Bell, D. W., Haber, D. A., and Li, E.
(1999) Cell, 99, 247-257.
67.Xu, G.-L., Bestor, T. H., Bourc'his, D., Hsieh,
C.-L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J. J., and
Viegas-Pequignot, E. (1999) Nature, 402, 187-191.
68.Jeanpierre, M., Turleau, C., Aurias, A., Prieur,
M., Ledeist, F., Fischer, A., and Viegas-Pequignot, E. (1993) Hum.
Mol. Genet., 2, 731-735.
69.Bourc'his, D., Xu, G. L., Lin, C. S., Bollman,
B., and Bestor, T. H. (2001) Science, 294, 2536-2539.
70.Hata, K., Okano, M., Lei, H., and Li, E. (2002)
Development, 129, 1983-1993.
71.Chedin, F., Lieber, M. R., and Hsieh, C. L.
(2002) Proc. Natl. Acad. Sci. USA, 99, 16916-16921.
72.Suetake, I., Shinozaki, F., Miyagawa, J.,
Takeshima, H., and Tajima, S. (2004) J. Biol. Chem., 279,
27816-27823.
73.Aapola, U., Liv, I., and Peterson, P. (2002)
Nucleic Acids Res., 30, 3602-3608.
74.Ramsahoye, B. H., Biniskiewicz, D., Lyko, F.,
Clark, V., Bird, A. P., and Jaenish, R. (2000) Proc. Natl. Acad.
Sci. USA, 97, 5237-5242.
75.Bachman, K. E., Rountree, M. R., and Baylin, S.
B. (2001) J. Biol. Chem., 276, 32282-32287.
76.Finnegan, E. J., and Dennis, E. S. (1993)
Nucleic Acids Res., 21, 2383-2388.
77.Bernacchia, G., Primo, A., Giorgetti, L., Pitto,
L., and Cella, R. (1998) Plant J., 13, 317-329.
78.Teerawanichpan, P., Chandrasekharan, M. B.,
Jiang, Y., Narangajavana, J., and Hall, T. C. (2004) Planta,
218, 337-349.
79.Finnegan, E. J., Peacock, W. J., and Dennis, E.
S. (1996) Proc. Natl. Acad. Sci. USA, 93, 8449-8454.
80.Nakano, Y., Steward, N., Sekine, M., Kusano, T.,
and Sano, H. (2000) Plant Cell Physiol., 41, 448-457.
81.Pradhan, S., Cummings, M., Roberts, R. J., and
Adams, R. L. P. (1998) Nucleic Acids Res., 26,
1214-1222.
82.Pradhan, S., and Adams, R. L. P. (1995) Plant
J., 7, 471-481.
83.Theiss, G., Schleicher, R., Schimpff-Weil, R.,
and Follmann, H. (1987) Eur. J. Biochem., 167, 89-96.
84.Vlasova, T. I., Demidenko, Z. N., Kirnos, M. D.,
and Vanyushin, B. F. (1995) Gene, 157, 279-281.
85.Giordano, M., Mattachini, M. E., Cella, R., and
Pedrali-Noy, G. (1991) Biochem. Biophys. Res. Commun.,
177, 711-719.
86.Henikoff, S., and Comai, L. (1998)
Genetics, 149, 307-318.
87.Lindroth, A. M., Cao, X., Jackson, J. P.,
Zilberman, D., McCallum, C. M., Henikoff, S., and Jacobsen, S. E.
(2001) Science, 292, 2077-2080.
88.Papa, C. M., Springer, N. M., Muszynski, M. G.,
Meeley, R., and Kaeppler, S. M. (2001) Plant Cell, 13,
1919-1928.
89.Cao, X., Springer, N. M., Muszynski, M. G.,
Phillips, R. L., Kaeppler, S. M., and Jacobsen, S. E. (2000) Proc.
Natl. Acad. Sci. USA, 97, 4979-4984.
90.Buryanov, Ya. I., Zakharchenko, N. S., Shevchuk,
T. V., and Bogdarina, I. G. (1995) Gene, 157,
283-287.
91.Kakutani, T., Jeddeloh, J. A., Flowers, S. K.,
Munakata, K., and Richards, E. J. (1996) Proc. Natl. Acad. Sci.
USA, 93, 12406-12411.
92.Shevchuk, T. V., Zakharchenko, N. S., D'yachenko,
O. V., and Buryanov, Ya. I. (2001) Fiziol. Rast., 48,
556-561.
93.Vaucheret, H., Beclin, C., Elmayan, T.,
Fenerbach, F., Godon, C., Morel, J.-B., Mourrain, P., Palauqui, J.-C.,
and Vernettes, S. (1998) Plant J., 16, 651-659.
94.Matzke, M. A., Matzke, A. J. M., Primig, M., and
Trnousky, J. (1989) EMBO J., 8, 643-649.
95.Napoli, C., Lemieux, C., and Jorgensen, R. (1990)
Plant Cell, 2, 279-289.
96.Van der Krol, A. R., Mur, L. A., Beld, M., Mol,
J. N. M., and Stuitje, A. R. (1990) Plant Cell, 2,
291-299.
97.Ratcliff, F., Harrison, B. D., and Baulcombe, D.
C. (1997) Science, 276, 1558-1560.
98.Meyer, P., Heidmann, I., and Niedenhof, I. (1993)
Plant J., 4, 89-100.
99.Wassenegger, M., Heimes, S., Riedel, L., and
Sänger, H. L. (1994) Cell, 76, 567-576.
100.Pelissier, T., Thalmeir, S., Kempe, D.,
Sänger, H.-L., and Wassenegger, M. (1999) Nucleic Acids
Res., 27, 1625-1634.
101.De Carvalho, F., Gheysen, G., Kushnir, S., van
Montagu, M., Inze, D., and Castresana, C. (1992) EMBO J.,
11, 2595-2602.
102.Ingelbrecht, I., van Houdt, H., van Montagu,
M., and Depicker, A. (1994) Proc. Natl. Acad. Sci. USA,
91, 10502-10506.
103.Mette, M., van der Winden, J., Matzke, M. A.,
and Matzke, A. J. M. (1999) EMBO J., 18, 241-248.
104.Waterhause, P. M., Graham, M. W., and Wang,
M.-B. (1998) Proc. Natl. Acad. Sci. USA, 95,
13959-13964.
105.Fire, A. (1999) Trends Genet.,
15, 358-363.
106.Hamilton, A. J., and Baulcombe, D. C. (1999)
Science, 286, 950-952.
107.Zamore, P., Tuschl, T., Sharp, P., and Bartel,
D. (2000) Cell, 101, 25-33.
108.Zilberman, D., Cao, X., and Jacobsen, S. (2000)
Science, 299, 716-719.
109.Aufsatz, W., Mette, M. F., van der Winden, J.,
Matzke, A. J. M., and Matzke, M. (2002) Proc. Natl. Acad. Sci.
USA, 99, 16499-16506.
110.Jacobsen, S. E., and Meyerowitz, E. M. (1997)
Science, 277, 1100-1103.
111.Jacobsen, S. E., Sakai, H., Finnegan, E. J.,
Cao, X., and Meyerowitz, E. M. (2000) Curr. Biol., 10,
179-186.
112.Baylin, S. B., Herman, J. G., Graff, J. R.,
Vertino, P. M., and Issa, J.-P. (1998) Adv. Cancer Res.,
72, 141-196.
113.Schmitt, F., Oakeley, E. J., and Jost, J. P.
(1997) J. Biol. Chem., 272, 1534-1540.
114.Selker, E. U., Cambareri, E. B., Jensen, B. C.,
and Haack, K. R. (1987) Cell, 51, 741-752.
115.Cambareri, E. B., Jensen, B. C., Schabtach, E.,
and Selker, E. U. (1989) Science, 244, 1571-1575.
116.Rossignol, J.-L., and Faugeron, G. (1994)
Experientia, 50, 307-317.
117.Goyon, C., and Faugeron, G. (1989) Mol.
Cell. Biol., 9, 2818-2827.
118.Selker, E. U. (1997) Trends Genet.,
13, 296-301.
119.Rhounim, L., Rossignol, J.-L., and Faugeron, G.
(1992) EMBO J., 11, 4451-4457.
120.Perkins, D. D., Metzenberg, R. L., Raju, N. B.,
Selker, E. U., and Barry, E. G. (1986) Genetics, 114,
791-817.
121.Foss, H. M., Roberts, C. J., Claeys, K. M., and
Selker, E. U. (1993) Science, 262, 1737-1741.
122.Rhounim, L., Gregoire, A., Salama, S., and
Faugeron, G. (1994) Curr. Genet., 26, 344-351.
123.Barry, C., Faugeron, G., and Rosignol, J.-L.
(1993) Proc. Natl. Acad. Sci. USA, 90, 4557-4561.
124.Goyon, C., Barry, C., Gregoire, A., Faugeron,
G., and Rossignol, J.-L. (1996) Mol. Cell. Biol., 16,
3054-3065.
125.Selker, E. U., Fritz, D. Y., and Singer, M. J.
(1993) Science, 262, 1724-1728.
126.Goyon, C., Nogueira, T. I. V., and Faugeron, G.
(1994) J. Mol. Biol., 240, 42-51.
127.Malagnac, F., Wendel, B., Goyon, C., Faugeron,
G., Zickler, D., Rossignol, J.-L., Noyer-Weidner, M., Vollmayr, P.,
Trautner, T. S., and Walter, J. (1997) Cell, 91,
281-290.
128.Chernov, A. V., Vollmayr, P., Walter, J., and
Trautner, T. A. (1997) Biol. Chem., 378, 1467-1473.
129.Malagnac, F., Gregoire, A., Goyon, C.,
Rossignol, J.-L., and Faugeron, G. (1999) Mol. Microbiol.,
31, 331-338.
130.Kouzminova, E., and Selker, E. U. (2001)
EMBO J., 20, 4309-4323.
131.Robertson, K. D., Uzvolgyi, E., Liang, G.,
Talmadge, C., Sumegi, J., Gonzales, F. A., and Jones, P. A. (1999)
Nucleic Acids Res., 27, 2291-2298.
132.Glickman, J. F., Pavlovich, J. G., and Reich,
N. O. (1997) J. Biol. Chem., 272, 17851-17867.
133.Gaudet, F., Talbot, D., Leonhardt, H., and
Jaenisch, R. (1998) J. Biol. Chem., 273, 32725-32729.
134.Mertineit, C., Yoder, J. A., Taketo, T., Laird,
D. W., Trasler, J. M., and Bestor, T. H. (1998) Development,
125, 889-897.
135.Bigey, P., Ramchandani, S., Theberge, J.,
Araujo, F. D., and Szyf, M. (2000) Gene, 242,
407-418.
136.Deng, J., and Szyf, M. (1998) J. Biol.
Chem., 273, 22869-22872.
137.Aapola, U., Mäenpää, K., Kaipia,
A., and Petersen, P. (2004) Biochem. J., 380,
705-713.
138.MacLeod, A. R., Rouleau, J., and Szyf, M.
(1995) J. Biol. Chem., 270, 11327-11337.
139.Rouleau, J., MacLeod, A. R., and Szyf, M.
(1995) J. Biol. Chem., 270, 1595-1601.
140.Slack, A., Cervoni, N., Pinard, M., and Szyf,
M. (1999) Eur. J. Biochem., 264, 191-199.
141.Vlasova, T. I., Kirnos, M. D., Demidenko, Z.
N., and Vanyushin, B. F. (1994) Biochemistry (Moscow),
59, 1403-1410.
142.Pradhan, S., and Kim, G. D. (2002) EMBO
J., 21, 779-788.
143.Ling, Y., Sankpal, U. T., Robertson, A. K.,
McNally, J. G., Karpova, T., and Robertson, K. D. (2004) Nucleic
Acids Res., 32, 598-610.
144.Kang, E. S., Park, C. W., and Chung, J. H.
(2001) Biochem. Biophys. Res. Commun., 289, 862-868.
145.Jackson, J. P., Lingroth, A. M., Cao, X., and
Jacobsen, S. E. (2002) Nature, 416, 556-560.
146.Tamaru, H., and Selker, E. U. (2001)
Nature, 414, 277-283.
147.Soppe, W. J. J., Jasencakova, Z., Houben, A.,
Kakutani, T., Meister, A., Huang, M. S., Jacobsen, S. E., Schubert, I.,
and Fransz, P. F. (2002) EMBO J., 21, 6549-6559.
148.Hõller, M., Westin, G., Jiricny, J., and
Schaffner, W. (1988) Genes Dev., 2, 1127-1135.
149.MacLeod, D., Charlton, J., Mullins, J., and
Bird, A. P. (1994) Genes Dev., 8, 2282-2292.
150.Tatematsu, K. I., Yamazaki, T., and Ishikawa,
F. (2000) Genes Cells, 5, 677-688.
151.Marinitch, D. V., Vorobyev, I. A., Holmes, J.
A., Zakharchenko, N. S., Dyachenko, O. V., Buryanov, Ya. I., and
Shevchuk, T. V. (2004) Biochemistry (Moscow), 69,
340-349.
152.Luo, S., and Preuss, D. (2003) Proc. Natl.
Acad. Sci. USA, 100, 11133-11138.
153.Costello, J. F., Frühwald, M. C.,
Smiraglia, D. J., Rush, L. J., Robertson, G. P., Gao, X., Wright, F.
A., Feramisco, J. D., Peltomäki, P., Lang, J. C., Schuller, D. E.,
Yu, L., Bloomfield, C. D., Caligiuri, M. A., Yates, A., Nishikawa, R.,
Huang, H.-J. S., Petrelli, N. J., Zhang, X., O'Dorisio, M. S., Held, W.
A., Cavenee, W. K., and Plass, C. (2000) Nature Genet.,
25, 132-138.
154.Shevchuk, T. V., and Buryanov, Ya. I. (1999)
Bioorg. Khim., 25, 630-633.
155.Esteller, M., Corn, P. G., Baylin, S. B., and
Herman, J. G. (2001) Cancer Res., 61, 3225-3229.
156.Kim, G. D., Ni, J., Kelesoglu, N., Roberts, R.
J., and Pradhan, S. (2002) EMBO J., 21, 4183-4195.
157.Rountree, M. R., Bachman, K. E., and Baylin, S.
B. (2000) Nat. Genet., 25, 269-277.
158.Fuks, F., Hurd, P. J., Deplus, R., and
Kouzaridis, T. (2003) Nucleic Acids Res., 31,
2305-2312.
159.Di Croce, L., Raker, V. A., Corsaro, M., Fazi,
F., Fanelli, M., Faretta, M., Fuks, F., Coco, F. L., Kouzaridis, T.,
Nervi, C., Minucci, S., and Pelicci, P. G. (2002) Science,
295, 1079-1082.
160.Robertson, A. K., Geiman, T. M., Sankpal, U.
T., Hager, G. L., and Robertson, K. D. (2004) Biochem. Biophys. Res.
Commun., 322, 110-118.
161.Tatematsu, K. I., Yamazaki, T., and Ishikawa,
F. (2000) Genes Cells, 5, 677-688.
162.Kimura, H., and Shiota, K. (2003) J. Biol.
Chem., 278, 4806-4812.
163.Carty, S. M., and Greenleaf, A. L. (2002)
Mol. Cell. Proteomics, 1, 598-610.
164.Fuks, F., Burgers, W. A., Godin, N., Kasai, M.,
and Kouzaridis, T. (2001) EMBO J., 20, 2536-2544.
165.Bachman, K. E., Rountree, M. R., and Baylin, S.
B. (2001) J. Biol. Chem., 276, 32282-32287.
166.Deplus, R., Brenner, C., Burgers, W. A.,
Putmans, P., Kouzaridis, T., de Launoit, Y., and Fuks, F. (2002)
Nucleic Acids Res., 30, 3831-3838.