Transcription Factor GATA-3 Is Involved in Repression of Promoter Activity of the Human Interleukin-4 Gene
G. T. F. Schwenger1, V. A. Mordvinov2*, and C. J. Sanderson1
1Curtin University School of Biomedical Sciences and the Western Australian Institute of Medical Research, Molecular Immunology Group, Level 5 Rear 50 Murray Street, Perth 6000, Western Australia; fax: (618) 9266-1650; E-mail: c.sanderson@curtin.edu.au2Institute of Cytology and Genetics, Siberian Division, Russian Academy of Sciences, pr. Lavrentieva 10, 630090 Novosibirsk, Russia; fax: (7-3833) 331-278; E-mail: mordvin@bionet.nsc.ru
* To whom correspondence should be addressed.
Received March 31, 2005; Revision received May 16, 2005
GATA-3 was shown to bind to two sites of the IL-4 gene promoter in human T-cell lines PER-117 and Jurkat. A motif located in the region of position -860 and responsible for GATA-3 binding was detected for the first time. Mutation or deletion of this site increased the promoter activity. The findings suggest a direct involvement of GATA-3 in regulation of the human IL-4 gene transcription as a repressor of the promoter activity.
KEY WORDS: T-cells, transcription factor GATA-3, interleukin-4, promoter, repressor
Abbreviations: PMA) phorbol 12-myristate 13-acetate; CaI) calcium ionophore; alphaCD28) antibodies to CD28 (alpha-Leu-CD28).
GATA-3 is one of the most important transcription factors providing for
maturation and activity of T-cell-related immunity. This factor is
necessary for development of thymocytes, expansion and proliferation of
T-cell precursors, realizing of mechanisms responsible for T-cell
differentiation, and regulation of gene transcription of cytokines
produced by T-cells [1-3].
GATA-3 is most actively expressed in T-helpers 2 (Th2). In these cells,
GATA-3 interacts with an intergene regulatory region of the gene
cluster of Th2 cytokines and induces transcriptional competence of the
chromosome locus, which includes the genes of interleukin-4 (IL-4),
IL-5, and IL-13 [3, 4].
Moreover, GATA-3 is directly involved in regulation of expression of
the genes IL-13 and IL-5 [5-7].
However, the role of this factor in the regulation of activity of the
human gene IL-4 promoter is still obscure.
IL-4 is among the key cytokines involved in formation and regulation of the body's protective functions. In addition to B-lymphocytes and mast cells, this immunomodulator has naive T-cells as targets. IL-4 activates in these cells the transcription factor STAT6, which in turn provides for a high level of GATA-3 expression. Thus, IL-4 induces the expression of GATA-3, which is required for effective production of Th2 cytokines. This mechanism is essential for triggering the T-cell polarization and production of the Th2 subpopulation [3]. There is a regulatory relationship between IL-4 and GATA-3; therefore, it seems probable that GATA-3 can control IL-4 expression not only on the level of regulation of the Th2 cytokine locus transcription, but also on the level of regulation of the gene promoter activity.
It seems that only one of three possible GATA elements of the mouse IL-4 gene promoter can interact with GATA-3 and has regulatory features [8]. This site, similarly to the two functionally inactive motifs of GATA, is located in the limits of the first 300 nucleotides of the IL-4 promoter sequence. We have not found data on the presence of GATA elements more distantly in the regulatory regions of human and mouse IL-4 genes.
The purpose of the present work was to study the role of GATA-3 in the regulation of activity of the human gene IL-4 promoter. The study was performed according to the routine scheme of analysis of regulatory regions, which included the computer-aided search for GATA elements in the gene sequence, determination of the binding efficiency of the detected elements to GATA-3, and their functional testing with a reporter system. In experiments, the human T-cell lines PER-117 and Jurkat were used, which are traditional cell models for studies on regulatory mechanisms of the Th2 cytokine gene transcription [9, 10].
MATERIALS AND METHODS
Computer-aided analysis of the gene IL-4 sequence was performed with the TFsearch program [11].
PER-117 and Jurkat cells were grown in RPMI 1640 medium containing 10% fetal calf serum (Trace Scientific, VIC, Australia), 2 mM L-glutamine and 75 µM monothioglycerol (Sigma, USA), 10 mM HEPES (pH 7.3) and 1 mM sodium pyruvate (Life Technologies, Inc.). The cells were activated with PMA (10 ng/ml) and 1 mM cAMP (Sigma), 0.25 µM CaI (A23187; Sigma), and alpha-Leu-CD28 (0.2 µg/ml; Becton Dickinson).
Labeling of oligonucleotides, isolation of nuclear proteins, and studies on their binding to the oligonucleotide samples were performed as described in [12]. Primary sequences of the oligonucleotides used in this work are presented in the table. All oligonucleotides were synthesized by Sigma Genosys (Australia).
Oligonucleotides used in experiments on determination of the
protein-binding activity of possible regulatory elements of GATA and on
performing the addressed mutagenesis
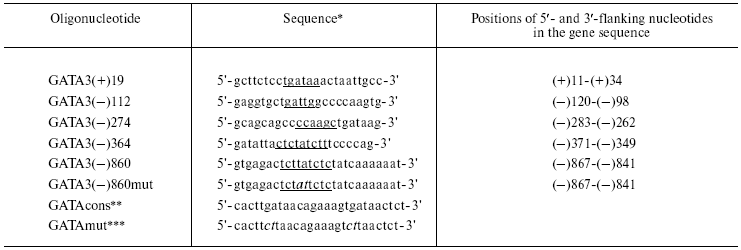
*Sequences of only the upper chain are presented. GATA motifs
are underlined; mutations are italicized.
**The oligonucleotide that includes a consensus motif for
binding the GATA-3 factor.
***The oligonucleotide that includes a mutation in the consensus
motif for binding the GATA-3 factor.
To perform addressed mutagenesis, a fragment of the human IL-4 gene located between positions -1075 and +60 was cloned inside the pALTER vector (Promega, USA). The mutagenesis was realized using an Altered Sites II in vitro Mutagenesis System kit (Promega) and the oligonucleotide GATA3(-)860mut (table).
Reporter constructions pIL4bk and pIL4mut were prepared by transfer of the initial and changed fragments of the IL-4 gene from the vector pALTER into the plasmid pGL3 Basic (Promega). The pIL4del construction was produced by removal of the fragment Acc65I/PstI of the IL-4 gene from pIL4bk.
The cells PER-117 and Jurkat were transfected by electroporation as described in [12]. In all experiments the cells were transfected in 3-5 repeats. Upon the electroporation, the cells were incubated for 6 h at 37°C in the presence of 5% CO2, activated, and placed into a CO2 incubator for 16 h. After the incubation, the cells were subjected to lysis in a solution to determine the activity of luciferase. This solution contained 50 mM Tris-HCl (pH 7.8), 15 mM MgSO4, 33.3 mM DTT, 0.1 mM EDTA, 250 µM LiCoA (Sigma), 500 µM sodium luciferin (Molecular Probes, USA), and 0.5% Triton X-100. The luciferase activity was determined with a Victor 1420 multifunctional analyzer (Wallac).
RESULTS AND DISCUSSION
Computer-aided analysis of the human gene IL-4 sequence located between positions -1075 and +66 revealed five possible sites for binding the transcription factor GATA-3. In the primary sequence of the gene, these sites are located between positions +19 and +24, -112 and -107, -274 and -269, -364 and -356, and -860 and -852. In the following text these sites are nominated according to position in the gene sequence of the 5´-flanking base of the corresponding site: GATA3(+)19, GATA3(-)112, GATA3(-)274, GATA3(-)364, and GATA3(-)860.
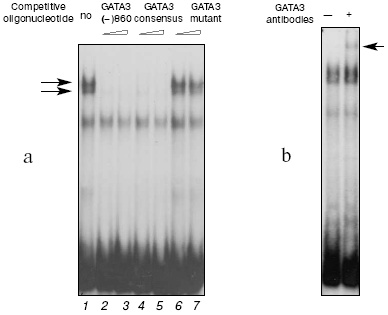
The protein-binding activities of the GATA motifs were determined using a set of oligonucleotides that contained the sequences of all possible GATA elements of the human IL-4 gene. The probe GATA3(-)860 and nuclear extracts from the PER-117 cells produced two complexes with very similar electrophoretic mobilities (Fig. 1a, lane 1). Activation of the cells did not markedly change the intensity of complexing, and the data for extracts from the intact and activated cells were similar. The oligonucleotide GATA3(-)860 and the oligonucleotide which contained the consensus motif for binding the GATA-3 factor completely inhibited the complexing (lanes 2-5). However, an oligonucleotide with mutation in the sequence of the GATA-3 consensus motif did not affect the efficiency of the interaction of the sample with the nuclear proteins (lanes 6 and 7).
Addition of antibodies to GATA-3 (Santa Cruz Biotechnology, Inc., USA) during the binding of the GATA3(-)860 probe with the nuclear extracts from the PER-117 cells resulted in formation of an additional complex with a low electrophoretic mobility (Fig. 1b). Antibodies to other members of the GATA family did not affect production of the main complexes and failed to induce generation of additional ones. These data suggested that the GATA(-)860 complexes with the nuclear extracts from the PER-117 cells should contain the GATA-3 factor. Experiments with nuclear extracts from the Jurkat cells gave similar results.Fig. 1. Binding of nuclear proteins from non-activated PER-117 cells with synthetic double-stranded oligonucleotide GATA3(-)860 that included the sequence between positions -867 and -841 in the human gene IL-4 promoter. a) During the binding reaction 50- and 100-fold molar excess of competitive oligonucleotides was added (specific DNA-protein complexes from the GATA3(-)860 sample and nuclear extracts are shown by the arrows); b) during the binding reaction, antibodies to GATA-3 were added (the complex generated on the addition of antibodies during the binding reaction is shown by the arrow).
Determination of the protein-binding activities of the other GATA motifs indicated that the probe GATA3(-)274 and T-cell nuclear extract also produced specific complexes which included the factor GATA-3. But the probes corresponding to the sequences GATA3(+)19, GATA3(-)112, and GATA3(-)364 lacked this ability.
Thus, of five possible GATA elements detected by us in the human IL-4 gene sequence, only GATA3(-)274 and GATA3(-)860 bound the transcription factor GATA-3. Data on the interaction of GATA3(-)274 with GATA-3 and also data on the lack of specific protein-binding activities in GATA3(+)19 and GATA3(-)112 are in agreement with findings for similar sites of the mouse IL-4 gene [8, 13]. Therefore, it is suggested that the functional role of GATA3(-)274 should not significantly differ from the functional role of the corresponding GATA element of the mouse gene. Conversely to this site, the motif GATA3(-)860 was for the first time detected in the sequence of the IL-4 promoter, and that is why we subsequently concentrated on studies of functional features of this possible regulatory element.
To study functional features of the site GATA3(-)860, a set of plasmids was created which expressed the luciferase gene under the control of the IL-4 promoter. The basic reporter construction denoted as pIL4bk was prepared by cloning inside the vector pGL3 Basic of a fragment of the human IL-4 gene located between the positions -1075 and +60. By addressed mutagenesis, the construction pIL4mut was prepared, with substitutions in the motif GATA3(-)860. It was shown in preliminary experiments that the probe corresponding to the altered sequence GATA3(-)860 (the oligonucleotide GATA3(-)860mut) lacked the specific protein-binding activity. This indicated that the altered sequence GATA3(-)860 had no sites interacting with transcription factors capable of affecting the activity of the promoter.
Another reporter construction, pIL4del, was prepared as a result of deletion from the promoter IL-4 region pIL4bk of an area carrying the motif GATA3(-)860. As the promoter of the luciferase gene, pIL4del included the gene IL-4 fragment located between the positions -744 and +60.
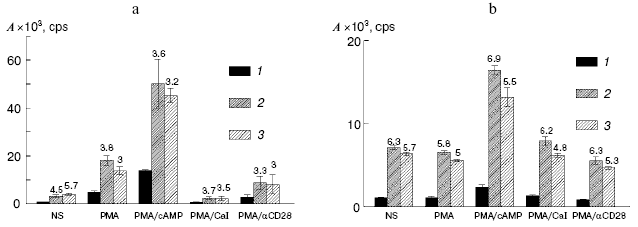
The PER-117 and Jurkat cells were transfected with these constructions, and their expressions were assessed by activity of luciferase. The mutation in the GATA3(-)860 sequence and deletion of the DNA fragment which included this element pronouncedly increased the expressions of the reporter constructions (Fig. 2). The activity of luciferase in the cells transfected with the constructions pIL4mut and pIL4del was 3-7 times higher than the activity of the reporter protein in the cells transfected with pIL4bk. Note, that the mutation in GATA3(-)860 and deletion of this element increased the activity of the reporter plasmids not only in the stimulated but also in the non-stimulated cultures of PER-117 and Jurkat. Consequently, the element GATA3(-)860 was involved in repression of both the basal and induced activities of the IL-4 promoter. Because GATA3(-)860 interacted with the transcription factor GATA-3 in intact cultures and activation of the cells had a slight effect on this interaction, this factor was concluded to be a component of the mechanism restraining the basal activity of the IL-4 promoter and suppressing the induced transcription of the gene.
GATA-3 has a similar function in the regulation of the human IL-5 gene expression. The element for GATA-3 binding located in the region of the -400 position of the IL-5 sequence is involved in repression of the induced and basal activities of the promoter [7]. Thus, GATA-3 seems to act as a suppressor of IL-4 and IL-5 transcription. But this contradicts data on the positive role of GATA-3 in initiation of transcription of Th2 cytokine genes in total and during activation of the IL-5 expression [3, 6]. This contradiction may be solved with a model that considers GATA-3 as an architectural factor responsible for a specific conformation of DNA. In this case, constant contact of GATA-3 with the corresponding elements of the gene is required for creating a functionally active spacial structure of the promoter. Destruction of the GATA-3 binding site will partially change this structure, and this will be manifested by changes in the efficiency of transcription activators or repressors acting through both regulatory elements of the promoter and sequences located in other regions of the gene cluster of Th2 cytokines.Fig. 2. Activities (A) of luciferase in the cell cultures transfected with the reporter constructions pIL4bk (1), pIL4mut (2), and pIL4del (3). The transfected PER-117 (a) and Jurkat (b) cells were not stimulated (NS) or were treated with various combinations of activators; 16 h later the cells were subjected to lysis and the activity of luciferase was determined. Mean values of four independent experiments are presented with the confidence interval.
The unique role of GATA-3 in T-cell differentiation and activation of expression of Th2 cytokines suggests this transcription factor to be a possible target for anti-allergic pharmaceuticals. In fact, an increased content of this protein was recorded in the lungs of asthma patients [14]. In an asthma model in mice suppression of the GATA-3 expression resulted in disappearance of the allergic reaction and prevented development of hypersensitivity of the respiratory tract [15, 16].
Elaboration of anti-allergic drugs needs careful studies on mechanisms that promote GATA-3 to control both transcription of the gene locus of Th2 cytokines as a whole and expression of individual genes of Th2 cytokines. In this context, data on the role of GATA-3 in regulation of activity of the gene IL-4 promoter significantly supplement the general picture of the role of this factor in formation of T-lymphocyte subpopulations and activation of Th2 cytokine expression. Our work is the first to show the involvement of the motif for GATA-3 binding in the repression of the IL-4 promoter activity. Our findings are in good correlation with data on the role of GATA-3 in the regulation of IL-5 expression [7] and allow us to consider GATA-3 to be an architectural factor responsible for the functionally active spacial structure of the promoter. Such a model of the action of GATA-3 completely corresponds to universally accepted ideas about the involvement of this protein in initiation of chromatin reconstruction to produce the transcriptional profile of Th2 and regulate the expression of Th2 cytokine genes.
This work was supported by the Program of Basic Research of the Asthma Foundation of Western Australia and Interdisciplinary Integration Project of Basic Research of the Siberian Branch, Russian Academy of Sciences, on Gene Networks: Theoretical Analysis, Computerized Modeling, and Experimental Construction, No. 119.
REFERENCES
1.Ting, C. N., Olson, M. C., Barton, K. P., and
Leiden, J. M. (1996) Nature, 384, 474-478.
2.Hendriks, R. W., Nawijn, M. C., Engel, J. D., van
Doorninck, H., Grosveld, F., and Karis, A. (1999) Eur. J.
Immunol., 29, 1912-1918.
3.Murphy, K. M., and Reiner, S. L. (2002) Nat.
Rev. Immunol., 2, 933-944.
4.Takemoto, N., Kamogawa, Y., Jun Lee, H., Kurata,
H., Arai, K. I., O'Garra, A., Arai, N., and Miyatake, S. (2000) J.
Immunol., 165, 6687-6691.
5.Corry, D. B. (1999) Curr. Opin. Immunol.,
11, 610-614.
6.Siegel, M. D., Zhang, D. H., Ray, P., and Ray, A.
(1995) J. Biol. Chem., 270, 24548-24555.
7.Schwenger, G. T. F., Fournier, R., Kok, C. C.,
Mordvinov, V. A., Yeoman, D., and Sanderson, C. J. (2001) J. Biol.
Chem., 276, 48502-48509.
8.Ranganath, S., Ouyang, W., Bhattarcharya, D., Sha,
W. C., Grupe, A., Peltz, G., Murphy, K. M. (1998) J. Immunol.,
161, 3822-3826.
9.Staynov, D. Z., Cousins, D. J., and Lee, T. H.
(1995) Int. Arch. Allergy Immunol., 107, 217-219.
10.Mordvinov, V. A., and Sanderson, C. J. (2001)
Arch. Immunol. Ther. Exp. (Warsz.), 49, 345-351.
11.Heinemeyer, T., Wingender, E., Reuter, I.,
Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A.,
Podkolodnaya, O. A., Kolpakov, F. A., Podkolodny, N. L., and Kolchanov,
N. A. (1998) Nucleic Acids Res., 26, 362-367.
12.Mordvinov, V. A., Schwenger, G. T., Fournier, R.,
De Boer, M. L., Peroni, S. E., Singh, A. D., Karlen, S., Holland, J.
W., and Sanderson, C. J. (1999) J. Allergy Clin. Immunol.,
103, 1125-1135.
13.Szabo, S. J., Gold, J. S., Murphy, T. L., and
Murphy, K. M. (1993) Mol. Cell Biol., 13, 4793-4805;
erratum in: (1993) Mol. Cell. Biol., 13, 5928.
14.Nakamura, Y., Ghaffar, O., Olivenstein, R., Taha,
R. A., Soussi-Gounni, A., Zhang, D. H., Ray, A., and Hamid, Q. (1999)
J. Allergy Clin. Immunol., 103, 215-222.
15.Zhang, D. H., Yang, L., Cohn, L., Parkyn, L.,
Homer, R., Ray, P., and Ray, A. (1999) Immunity, 11,
473-482.
16.Finotto, S., De Sanctis, G. T., Lehr, H. A.,
Herz, U., Buerke, M., Schipp, M., Bartsch, B., Atreya, R., Schmitt, E.,
Galle, P. R., Renz, H., and Neurath, M. F. (2001) J. Exp. Med.,
193, 1247-1260.