Small RNAs in Human Brain Development and Disorders
E. I. Rogaev
Brudnick Neuropsychiatric Research Institute, Department of Psychiatry, University of Massachusetts Medical School, 303 Belmont Street, 01604 MA, USA; fax: 1-508-856-4004; E-mail: Evgeny.Rogaev@umassmed.eduLaboratory of Molecular Brain Genetics, Research Center of Mental Health, Russian Academy of Medical Sciences, Zagorodnoe Shosse 2, 113152 Moscow, Russia
Lomonosov Moscow State University, Faculty of Bioengineering and Bioinformatics, 119992 Moscow, Russia
Received November 3, 2005
Small RNA is a variable and abundant type of non-coding RNAs in brain. The function of these RNAs is mainly unknown. A specific class of small RNA, microRNA, is dynamically regulated in neurogenesis and in embryo brain development. The genes for synaptic formation and some mental retardation disorders are putative targets for microRNA predicted by computational algorithms. The molecular pathways for mental development, common forms of autisms, schizophrenia, and affective disorders have yet to be elucidated. The hypothesis proposed here is that small regulatory RNAs, specifically microRNAs, play a role in human brain development and pathogenesis of brain disorders, especially of neurodevelopmental conditions. Pilot tests using comprehensive arrays of microRNAs demonstrate that microRNAs derived from postmortem human brains are applicable for microRNA expression profiling. The abundant expression of many regulatory small RNAs in human brain implies their biological role that must be tested by functional assays in neurons and by genetic and comparative expression profiling.
KEY WORDS: schizophrenia genes, microRNA, brain development, microarray, gene expression
The brain is a rapidly evolving organ in Primates. The human brain is about three times larger than that of the chimpanzee. This enlargement is probably associated with alterations in regulation of divisions of cortical progenitor cells. The cerebral cortex and, especially, upper-cortical neurons are particularly developed in humans. Investigation of biochemical and genetic mechanisms underlying these specific brain developments is a challenging and exciting task. Only about 1.2% of the human genome belongs to genes encoding proteins. The number of mutations in encoding human genes in comparison to other higher primates is relatively low. Thus, it is conceivable that alterations in the regulation of the genes, rather than protein mutations, might be a driving force in recent evolution of brain in Hominoidea. There are common disorders of human brain representing impairments in conciseness, intellect, emotions, and memory. Among them are the group of neurodevelopmental diseases, such as mental retardation, autism, schizophrenia, and a group of progressive neurodegenerative diseases, e.g., dementias, Parkinson's disease, and ataxias. Recent developments from whole chromosome or whole genome tiling chip array analyses unexpectedly showed that a significant proportion of the genome may be transcribed. In contrast to the common view that non-coding junk RNAs (e.g., introns) are unstable and quickly eliminated, many non-coding RNAs (apart from housekeeping spliceosomal RNA, tRNA, and ribosomal RNAs) have a long half-life comparable to mRNA. These RNAs were found not only in the nucleus but also in the cytoplasm.
I propose the hypothesis, for the first time to my knowledge, that non-coding RNAs may play an important role in schizophrenia or other psychiatric disorders and specific normal and abnormal developments of the human brain.
Multiple small RNAs (<300-400 nt) have been found in brain. Many of these RNA species are brain-specific or enriched in brain. Their functions are not yet elucidated. Small nucleolar RNA (snoRNA) (~300 of known human snoRNAs of 60-300 nt in length) promote chemical modifications of other RNAs, mainly, non-protein-coding RNAs. However, some of the snoRNAs have complementary sequences in mRNA (e.g., to sequence harboring adenosite-to-ionosin RNA editing site of the serotonin 2C receptor gene) [1], and, therefore, may contribute to modification of mRNA of some neurogenes. Another abundant class of non-coding RNA are tiny RNAs (~19-25 nt) produced from longer double-stranded RNA. They can be classified as: 1) short interfering RNAs (siRNA) generated from exogenous or endogenous double-stranded RNA, and 2) microRNA derived from 60-90 nt precursor RNA forming imperfect hairpins. The tiny microRNAs may potentially transfer easily within and between nucleus and cytoplasm compartments, including transportations across the neuronal body, dendrites, and axons. In addition to microRNAs, varieties of other small non-coding RNAs are expressed in the brain with functions implicated in RNA and chromatin modifications. It is conceivable that other classes of small RNAs will be discovered in brain. Given the limited space, I will discuss the potential regulatory role of the microRNA type only.
MicroRNA. MicroRNAs (miRNAs) are a class of 19-22-nt regulatory RNAs that are products of small non-protein-coding genes found in animals and plants [2]. Several hundreds or, perhaps, thousands of evolutionarily conserved and non-conserved sequences for microRNA with a hairpin structure can be predicted in mammalian genomes by various computational prediction methods [3-5]. However, the experimental cloning of the microRNAs from human tissues has identified and validated to date 321 microRNA genes (http://microrna.sanger.ac.uk/sequences/). The microRNAs as an imperfect complement to one or several different mRNA targets may promote degradation of these mRNAs or, more often, suppress translation of the corresponding protein. Therefore, microRNAs can be considered as post-transcriptional suppressors. The functional role for any microRNA in mammals has not yet been clearly established using direct approaches, e.g., by identification of mutation associated with pathology or gene knock-out experiments in vivo. However, the evolutionary conservation of many microRNA genes and mutation analysis in invertebrates demonstrated their important regulatory role in development. Several observations can be used in support of the assumption that microRNA is a source of molecules that would be reasonable to test in pathogenesis of brain disorders.
1. The microRNAs and other small RNAs are abundant in brain; there are brain specific and brain enriched species of microRNAs (e.g., mi124a, mi9, mi218) [6].
2. In non-mammalian models, microRNAs are shown to be important for development of the nervous system and brain. The miR-430 rescued normal brain formation in a zebrafish mutant defective for the Dicer gene, the gene essential for processing pre-miRNA [7]. In the round worm Caenorhabditis elegans, lsy-6 microRNA mediates the bilateral asymmetry of chemosensory neurons [8].
3. The set of microRNAs changed expression upon induction of cultured cells to neurogenic status. MicroRNAs are temporally regulated during brain development (shown in rodents) [9].
4. Two most highly scored pathways for the predicted microRNA targeted genes [4] are: neurogenesis (may be related to abnormal neurodevelopment) and the ubiquitin-ligase protein regulation pathway (that may be related to neurodegeneration). There are many predicted targets for microRNAs of neurological interest, for example, BDNF (neuron-survival factor), MECP2 (Rett syndrome and X-linked mental retardation), and APP (Alzheimer's disease). The genes for synaptic proteins are commonly predicted targets. Intriguingly, under one of the prediction algorithms [4], two genes with the highest numbers of predicted sites for different multiple microRNA are synapsin 1 with putative sites for about 14 different microRNAs and FMRP1, with predicted sites for about 19 microRNAs. The FMRP1 (fragile X mental retardation protein) is supposed to be involved in repression of protein translation and synaptic plasticity. The inactivation of FMPR1 causes a common fragile X mental retardation. Remarkably, FMPR1 interacts with components of the microRNA processing machinery, Dicer and Argounate proteins [10]. Only a short stretch of complementary nucleotides in 5´-microRNA is critically required for targeting mRNA. Therefore, the number of predicted targets for any microRNA is broadly varied, depending on the computational methods applied for the prediction. The experimental validation will be required for any predicted microRNA/mRNA duo.
In summary, it is reasonable to suggest that non-coding genes for small RNAs are involved in pathogenesis of psychiatric illnesses and in particular, of the neurodevelopment pathway. Since no mutations or polymorphisms changing the protein structure in encoding genes have yet been found in schizophrenia, it is of interest to test this hypothesis in schizophrenia. The microRNAs, which are differentially expressed during early development-adolescence-adult stages, might be good candidate genes in this study. The mature microRNAs are small molecules (19-22 nt) near the minimal size required for detection by DNA/RNA hybridization or RT-PCR. Therefore, whether these forms of RNAs can be consistently detected and quantified in postmortem brain tissues (subjected to RNA degradation) has to be addressed. To elucidate expression of multiple microRNA genes in human brain specimens we have undertaken microRNA expression profiling using postmortem specimens of adult human neocortex. The total RNA was isolated from human parietal neocortex (Brodman area 7) from schizophrenic and normal individuals. The high quality of the isolated RNA was confirmed through an Agilent Bioanalyzer system (figure, panel (a)). The fractions of small RNA (<300 nt) from several unrelated individuals with no psychiatric illnesses were purified and separated from a higher molecular weight fraction of total RNA. The pooled probe was used for hybridization with recently designed miRNA Human Array (LC Science, USA). The Array contained probes for 312 mature microRNAs (Sanger v. 7.0, June 2005) with multiple controls including housekeeping small RNA genes (5S RNA and t-RNA species) and the oligonucleotide microRNA probes with single mutations (mismatch probe negative controls) (figure, panel (b)). Each probe for mature microRNA was spotted in 4-5 replications. Highly consistent results between spots for the same microRNA gene were found in most cases. An example of microRNA chip hybridization with a Cy5-labeled fraction of small RNA isolated from the human neocortex is demonstrated in the figure. Effective and specific hybridization was found for the microRNAs. According to the hybridization data, at least 35% of the tested microRNAs had detectable expression in the human brain. The highly expressed fraction included miR-125b, let-7a, miR-26a, let-7c, let-7f with the highest signals (the signal for these microRNAs was compatible with 5S RNA signal) and the second group of miR-9, miR-124a, let-7d, miR-15a, miR-9*, miR-23b, miR-128a, miR-128b. The results were confirmed by hybridization of independently pooled samples from normal and schizophrenia specimens with microRNA sets in several replicas. The stability of microRNAs and comparative level of expression was confirmed further for selected microRNAs by Northern blot hybridization (Burmistrova and Rogaev, unpublished). Our analysis of microRNA arrays imply that comparison of microRNA expression profiles in postmortem brain material may be a promising strategy to identify specific alterations in microRNA expression in neuropsychiatric disorders. The diseases of interest include early development pathologies of brain morphogenesis and function, e.g., macrocephaly and microcephaly, mental retardations, autism, schizophrenia, and affective and other adolescent or adult behavior disorders. Direct functional approaches can be applied by: (i) prediction of targets for the microRNA study, and (ii) direct testing in cultured neuronal cells or animal models whether the target is regulated by ectopic overexpression of the corresponding microRNA or down-regulation (a technically more challenging task) of endogenous microRNA.
I thank A. P. Grigorenko and O. A. Burmistrova for assistance in preparation of the manuscript. Postmortem brain tissue was donated by the Stanley Medical Research Institute Brain Collection courtesy of Drs. Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken.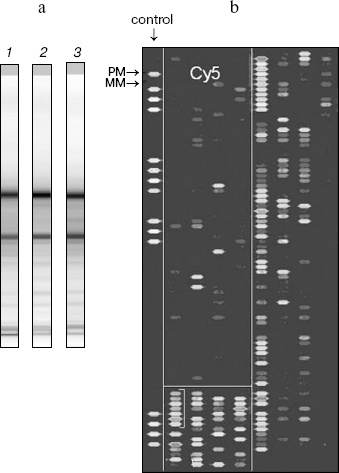
There are a few assumptions one may bear in mind. First, the level of protein (interaction in translational machinery) rather than the level of the mRNA may be affected by microRNA modulation. Second, the microRNAs are anticipated to be negative regulators. Thus, inactivation of microRNA will lead to upregulation of the protein target. Third, several microRNAs may regulate one gene with an additive effect and, vice versa, one microRNA may have pleiotropic effects on several gene targets. The role of non-coding small RNA, e.g., microRNA in the brain may be uncovered by: (i) searching for genetic variations contributing to individual differences in microRNA expression, e.g., via direct analysis of polymorphisms and mutations in precursor microRNA and nearby genomic regions, and (ii) comparative analysis of expression of the specific microRNA in psychiatric versus normal brain conditions. Finally, siRNA against specific viral nucleotide sequences may confer cellular immunity [11]. MicroRNA profiles are also changed globally in several human cancers [12]. Therefore, investigation of biological significance of small RNAs in response to infectious agents in brain and malignant cell states may be other perspective directions in brain research.
This work was supported in part by the Russian Foundation for Basic Research and the Stanley Research Foundation.
REFERENCES
1.Cavaille, J., Buiting, K., Kiefmann, M., Lalande,
M., Brannan, C. I., Horsthemke, B., Bachellerie, J. P., Brosius, J.,
and Huttenhofer, A. (2000) Proc. Natl. Acad. Sci. USA,
97, 14311-14316.
2.Ambros, V. (2001) Cell, 107,
823-826.
3.Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W.,
Bartel, D. P., and Burge, C. B. (2003) Cell, 115,
787-798.
4.John, B., Enright, A. J., Aravin, A., Tuschl, T.,
Sander, C., and Marks, D. S. (2004) PLoS. Biol., 2,
e363.
5.Berezikov, E., Guryev, V., van de Wienholds, E. B.
J., Plasterk, R. H., and Cuppen, E. (2005) Cell, 120,
21-24.
6.Sempere, L. F., Freemantle, S., Pitha-Rowe, I.,
Moss, E., Dmitrovsky, E., and Ambros, V. (2004) Genome Biol.,
5, R13.
7.Giraldez, A. J., Cinalli, R. M., Glasner, M. E.,
Enright, A. J., Thomson, J. M., Baskerville, S., Hammond, S. M.,
Bartel, D. P., and Schier, A. F. (2005) Science, 308,
833-838.
8.Johnston, R. J., and Hobert, O. (2003)
Nature, 426, 845-849.
9.Miska, E. A., Varez-Saavedra, E., Townsend, M.,
Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., and Horvitz,
H. R. (2004) Genome Biol., 5, R68.
10.Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto,
M., Mowrey, J., Jongens, T. A., Nelson, D. L., Moses, K., and Warren,
S. T. (2004) Nat. Neurosci., 7, 113-117.
11.Gitlin, L., Karelsky, S., and Andino, R. (2002)
Nature, 418, 430-434.
12.Lu, J., Getz, G., Miska, E. A., Varez-Saavedra,
E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H.,
Ferrando, A. A., Downing, J. R., Jacks, T., Horvitz, H. R., and Golub,
T. R. (2005) Nature, 435, 834-838.