Effect of Hypermethylation of CCWGG Sequences in DNA of Mesembryanthemum crystallinum Plants on Their Adaptation to Salt Stress
O. V. Dyachenko1, N. S. Zakharchenko1, T. V. Shevchuk1, H. J. Bohnert2, J. C. Cushman3, and Ya. I. Buryanov1*
1Branch of Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, pr. Nauki 6, 142290 Pushchino, Moscow Region, Russia; fax: (8-27) 33-0527; E-mail: buryanov@fibkh.serpukhov.su2Department of Plant Biology, University of Illinois, 1201 West Gregory Drive, Urbana, IL 61801, USA; E-mail: bohnerth@life.uiuc.edu
3Department of Biochemistry, University of Nevada, Reno/MS 200, 1664 North Virginia Street, Reno, Nevada 89557, USA; E-mail: jcushman@unr.edu
* To whom correspondence should be addressed.
Received November 15, 2005
Under salt stress conditions, the level of CpNpG-methylation (N is any nucleoside) of the nuclear genome of the facultative halophyte Mesembryanthemum crystallinum in the CCWGG sequences (W = A or T) increases two-fold and is coupled with hypermethylation of satellite DNA on switching-over of C3-photosynthesis to the crassulacean acid metabolism (CAM) pathway of carbon dioxide assimilation. The methylation pattern of the CCWGG sequences is not changed in both the 5´-promoter region of the gene of phosphoenolpyruvate carboxylase, the key enzyme of C4-photosynthesis and CAM, and in the nuclear ribosomal DNA. Thus, a specific CpNpG-hypermethylation of satellite DNA has been found under conditions of expression of a new metabolic program. The functional role of the CpNpG-hypermethylation of satellite DNA is probably associated with formation of a specialized chromatin structure simultaneously regulating expression of a large number of genes in the cells of M. crystallinum plants on their adaptation to salt stress and switching-over to CAM metabolism.
KEY WORDS: Mesembryanthemum crystallinum, salt stress, adaptation, CAM metabolism, DNA, methylation, CpNpG sequences, PEP carboxylase and rRNA genes, satellite DNADOI: 10.1134/S000629790604016X
Abbreviations: CAM) crassulacean acid metabolism; N) any nucleoside; W) adenosine or thymidine; PEP) phosphoenolpyruvate.
Enzymatic methylation of DNA plays an important role in the regulation
of various cellular processes in plants, including differentiation and
morphogenesis, apoptosis, formation of the chromatin structure, and
gene expression [1-6]. Among
all eukaryotic organisms, plants are characterized by the highest level
of methylation of their genome because they have several types of
site-specific methylation of DNA. In addition to the symmetric CpG-type
of DNA methylation common for all eukaryotes, plants also have a
pronounced type of methylation of the symmetric CpNpG sequences (N is
any nucleoside) and an asymmetric NpCpN-type of methylation. All
functions of plant DNA methylation known to date have been established
by studies on the CpG-type of its methylation, whereas the functional
roles of other types of the site-specific DNA methylation are virtually
unknown. It is likely that these types of DNA methylation can execute
specific functions in the plant cells.
The facultative halophyte Mesembryanthemum crystallinum L. is a well-studied physiological model for investigation of plant adaptation to salination and water insufficiency. Under conditions of water shortage caused by drought or salination, this plant displays a specific ability to switch over its C3-photosynthesis to the CAM pathway of C4-photosynthesis (Crassulacean Acid Metabolism, or organic acid metabolism of the type of Crassulaceae) [7]. These two pathways of CO2 assimilation are significantly different biochemically and physiologically, and the CAM pathway itself is accompanied by an inverse circadian rhythm of stomata functioning: they are closed in daytime and open at night, which effectively protects CAM plants from water loss under conditions of its shortage.
To provide for a steady growth and development of plants, it is urgent to elucidate genetic and biochemical mechanisms of their adaptation to extreme conditions of the environment. Due to its physiological and biochemical features and a comparatively small size of the genome (about 350,000 kb), M. crystallinum is a suitable model for studies on adaptation of plants to conditions of salt stress and water insufficiency.
The CAM-specific form of phosphoenolpyruvate (PEP) carboxylase is a “diagnostic” marker of the CAM pathway functioning in M. crystallinum plants. The amounts of this PEP carboxylase and the corresponding mRNA sharply increase in the cells of these plants under conditions of water insufficiency or salination. Under these conditions, the transcription of the Ppc1 gene of the PEP carboxylase family is induced in the plants, as well as of other genes of the CAM pathway of CO2 assimilation [7]. But induction mechanisms of expression of these genes remain unclear. Enzymatic methylation of DNA is likely to play a specific role in these mechanisms. However, to date there is no information about specific features of DNA methylation in M. crystallinum plants under conditions of salt stress and its possible involvement in the regulation of CAM-specific genetic programs.
The purpose of the present work was to analyze the level of site-specific methylation of nuclear DNA and study the specificity of methylation of some genes of M. crystallinum plants grown under conditions of salt stress.
MATERIALS AND METHODS
Conditions of cultivation of M. crystallinum plants. The plants were grown from seeds to the age of 3-4 weeks in a soil culture under conditions of 12-h light day at the air temperature of 25°C in the daytime and 15°C at night. At the age of 3-4 weeks, the plants were transplanted into separate pots with 0.5 kg soil and cultivated under the same conditions to the age of 6-7 weeks. The plants most similar in appearance were chosen for study. These plants have well-developed three pairs and rapidly growing fourth pair of primary leaves of the main stalk. The fifth pair of primary leaves was in the initial stage of development. The experimental plants were watered once every two days with 0.5 M NaCl (50 ml per vessel), the control plants being watered with water.
Isolation of plant DNA. DNA was isolated from two-to-five pairs of the primary leaves of the experimental and control plants on the eighth day from the beginning of salination. DNA was isolated from the leaves finely powdered in liquid nitrogen by the standard method [8], which included extraction with phenol-chloroform mixture and treatment with RNase (RNase A from Sigma, USA).
The methylation level of cytosine in the CCWGG sequence was determined using DNA methyltransferases (DNA methylases) EcoRII and BstNI as described earlier in [9]. DNA methylase EcoRII from Escherichia coli B834(N3) cells and DNA methyltransferase BstNI from Bacillus stearothermophilus cells were isolated according to procedures presented in [10, 11]. The reaction mixture (20 µl) contained 40 mM Tris-HCl (pH 8.0), 2 mM EDTA, 7 mM 2-mercaptoethanol, 6.6 µM S-adenosyl-L-[methyl-3H]methionine (the specific activity 15 Ci/mmol; Amersham, England), 0.5 µg DNA, and DNA methylase (20 activity units) (1 activity unit is the enzyme amount catalyzing the transfer of 1 pmol methyl groups onto DNA of the phage lambda dcm- during 1 h at the optimum temperature). The reaction was performed for 2 h at 37°C with DNA methyltransferase EcoRII and at 60°C with DNA methyltransferase BstNI. A 19-µl aliquot of the reaction mixture was applied onto a DE-81 paper (Whatman, England) of 2 × 2 cm in size, washed successively with 0.2 M NaHCO3, H2O, and ethanol, and dried. Radioactivity of the samples was determined with a Beckman LS6800 scintillation counter. The level of in vivo methylation of DNA was determined by the formula:
![]()
Cleavage of DNA in different experiments was performed with restriction endonucleases HpaII, EcoRII, and MboI sensitive to methylation of inner cytosine in the sequences CCGG (HpaII), CCWGG (EcoRII), and GATC (MboI) and, respectively, with restriction endonucleases MspI, BstNI, and Sau3A insensitive to these types of methylation. All enzymes used in the present work were from New England Biolabs (USA) except for restrictases EcoRII and BstNI (isolated by us according to procedures presented in [11, 12]). DNA (8-12 µg) was hydrolyzed with restriction endonucleases (15-20 activity units per µg DNA) under conditions optimal for the activity of each enzyme. The reaction was performed for 16-18 h at 37°C with all enzymes except BstNI (60°C).
DNA hydrolysis products prepared with restrictases were subjected to electrophoresis in 0.8% agarose gel in TAE buffer (0.04 M Tris, 0.04 M CH3COOH, 2 mM EDTA (pH 8.0)) supplemented with ethidium bromide (0.5 µg/ml). Electrophoresis was performed at 3 V/cm. Separation of DNA fragments was monitored by UV at 310 nm.
The hydrolysis products were transferred onto a Hybond-N+ nylon membrane (Amersham, England) by electroblotting in a Mini Trans Blot apparatus (Bio-Rad, USA).
Blot-hybridization analysis of DNA. Southern-hybridization of DNA was performed by the standard scheme [13] with slight modifications. Prehybridization, hybridization with specific probes, and washing off the membrane were performed at 65°C according to recommendations of the membrane producer. The membranes were exposed to obtain radioautographs. DNA specimens were analyzed twice. To monitor the completeness of DNA cleavage with restrictases HpaII and EcoRII, DNA preparations were hydrolyzed with higher concentrations of these enzymes (25-30 activity units per µg DNA), and the hybridization pattern did not change.
DNA probe. For 32P-labeled hybridization probes, we used a fragment of the untranscribable 5´-regulatory promoter region of the ppc1 gene of PEP carboxylase of 1.5 kb in size, the plasmid pWRII containing the wheat rDNA genes [14], and the total DNA from the M. crystallinum plants not subjected to salt stress. The DNA probe was labeled by the random multiple primer approach (Multiprime DNA Labeling System Kit; Amersham) according to recommendations of the producer and using a domestic preparation [alpha-32P]dATP with the specific activity of 5000 Ci/mmol (Physico-Energetic Institute, Obninsk, Russia). The resulting probe had the specific activity of (0.5-1)*109 cpm per µg DNA.
RESULTS AND DISCUSSION
Methylation level of the CCWGG sequences in M. crystallinum. The methylation level of cytosine in the nucleotide sequences of DNA related to the CpNpG-methylated motif was determined using DNA methyltransferases EcoRII and BstNI [9]. Both enzymes recognize in DNA the same double-stranded symmetric sequence CCWGG (W = A or T) (with the central trinucleotide corresponding to the CpNpG sequence), where in each chain the inner cytosine residues are methylated but resulting in different products [10, 11]. DNA methylase EcoRII methylates cytosine residues by the C5-position of the pyrimidine ring with production of 5-methylcytosine. DNA methylase BstNI methylates the same cytosine residues by the exocyclic amino group producing N4-methylcytosine or N4,5-dimethylcytosine. Thus, the number of CH3-groups incorporated into DNA with enzymes EcoRII and BstNI represents, respectively, the number of the in vivo unmodified CCWGG sequences and the total number of these sequences in the DNA under study. Consequently, the difference between the number of methyl groups incorporated in the DNA under analysis with enzymes BstNI and EcoRII characterizes the in vivo methylation level of the CCWGG sequence of this DNA. The observed statistical regularities for the incidence of different nucleotide sequences in natural DNAs allowed us to use this approach not only to analyze the methylation level in DNA of the CCWGG sequences but also of the shorter CpNpG sequences.
By this enzymatic approach it has been found that under conditions of salination (0.5 M NaCl) the switching-over of the C3-photosynthesis to the CAM metabolism is associated with an increase in the methylation level of CCWGG sequences in the plant nuclear genome from 31.5 ± 3.8 to 67.2 ± 5.5% (mean arithmetic values and their standard deviations for three independent experiments performed in five repeats).
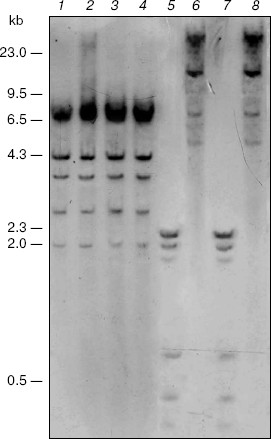
Search for targets of CpNpG-hypermethylation in the M. crystallinum genome. Blot-hybridization analysis of DNA from the control and salt stress-subjected M. crystallinum plants did not reveal difference in the pattern of CpNpG-methylation of the 5´-regulatory promoter region of the PEP carboxylase gene. However, this gene is CpNpG-methylated as it is shown by differences in the hydrolysis pattern of the gene with restrictase EcoRII (Fig. 1, lanes 2 and 4) and restrictase BstNI (Fig. 1, lanes 1 and 3). The differences in the hydrolysis pattern of the PEP carboxylase gene with restrictases MboI and Sau3A (Fig. 1, lanes 5-8) indicate the CpG-methylation of cytosine in the GATCG sequences of this gene. However, in the plants grown under normal physiological conditions and under conditions of salt stress the methylation pattern of these sequences in the PEP carboxylase gene is also unchanged. Thus, in the M. crystallinum plants the pattern of CpG- and CpNpG-methylation of specific cytosine residues in the 5´-regulatory promoter region of the PEP carboxylase gene does not change on the switching-over of the C3-photosynthesis to the CAM metabolism.
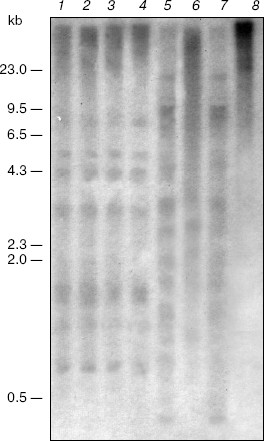
Also, salt stress does not affect methylation of the nuclear ribosomal DNA. The blot-hybridization analysis of DNA of the plants grown under normal conditions and under salination indicates the absence of differences in the methylation pattern of the CpNpG sequences in this region of the genome (Fig. 2, lanes 5-8). Note that under such different physiological conditions the plants lack methylation of the CpG sequences of the nuclear ribosomal DNA that is shown by similar patterns of its hydrolysis with restrictases HpaII and MspI (Fig. 2, lanes 1-4).Fig. 1. Blot-hybridization analysis of the PEP carboxylase gene of M. crystallinum plants. Lanes: 1, 3, 5, 7) DNA of the control plants; 2, 4, 6, 8) DNA of the plants cultivated under conditions of salt stress; 1, 3) hydrolysis with restrictase BstNI; 2, 4) hydrolysis with restrictase EcoRII; 5, 7) hydrolysis with restrictase MboI; 6, 8) hydrolysis with restrictase Sau3A. The dashes indicate positions of the marker fragments of the lambda/HindIII hydrolyzate and their size in kb.
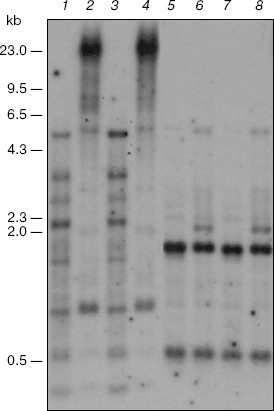
The fraction of satellite DNAs belongs to the CpNpG-hypermethylated targets of the genome of the M. crystallinum plants grown under salt stress conditions. This is shown by data of the blot-hybridization analysis of the M. crystallinum genomic DNA with the labeled total DNA of these plants used as a probe (Fig. 3). The presence of discrete DNA bands in the radioelectrophoregram suggests that the M. crystallinum plants have a specific fraction of satellite DNA. In the plants grown under different physiological conditions, satellite DNA is characterized by the absence of a noticeable methylation of the CCGG sequences (the CpG-methylation) that is confirmed by data on the DNA hydrolysis with restriction endonucleases MspI and HpaII (Fig. 3, lanes 1-4). However, analysis of DNA with restriction endonucleases BstNI and EcoRII revealed the CpNpG-type methylation of satellite DNA in the M. crystallinum plants grown under both normal and salt stress conditions (Fig. 3, lanes 5-8). These DNA sequences are hypermethylated in the plants grown under conditions of salination. This is indicated by a significant increase in the DNA resistance to cleavage with restriction endonuclease EcoRII that is demonstrated by the nearly complete absence of discrete DNA bands (Fig. 3, lane 8). Note that the CpNpG sequences in the genome of the M. crystallinum plants grown under the salination conditions were hypermethylated solely in satellite DNA.Fig. 2. Blot-hybridization analysis of ribosomal DNA of M. crystallinum plants. Lanes: 1, 3, 5, 7) DNA of the control plants; 2, 4, 6, 8) DNA of the plants cultivated under conditions of salt stress; 1, 3) hydrolysis with restrictase MspI; 2, 4) hydrolysis with restrictase HpaII; 5, 7) hydrolysis with restrictase BstNI; 6, 8) hydrolysis with restrictase EcoRII. The dash indications are the same as in Fig. 1.
Thus, this work is the first to reveal a specific CpNpG-hypermethylation of satellite DNA in the cells of M. crystallinum plants under conditions of switching on expression of a new metabolic program. The functional role of the CpNpG hypermethylation of satellite DNA seems to be associated with formation of a specialized chromatin structure concurrently regulating the expression of a large number of genes in the cells of these plants on their adaptation to salination and switching-over of C3-photosynthesis to the CAM metabolism.Fig. 3. Blot-hybridization analysis of satellite DNA of M. crystallinum plants. Lanes: 1, 3, 5, 7) DNA of the control plants; 2, 4, 6, 8) DNA of the plants cultivated under conditions of salt stress; 1, 3) hydrolysis with restrictase MspI; 2, 4) hydrolysis with restrictase HpaII; 5, 7) hydrolysis with restrictase BstNI; 6, 8) hydrolysis with restrictase EcoRII. The dash indications are the same as in Fig. 1.
The further development of this work includes isolation and characterization of the CpNpG-hypermethylated satellite DNA, establishment of its location on chromosomes of M. crystallinum plants, and investigation of the “CAM-specific” structure of chromatin.
This work was supported by the Basic Research Program of the Presidium of the Russian Academy of Sciences “Changes in Gene Funds of Plants, Animals, and Humans”.
REFERENCES
1.Meyer, P., Niedenhof, I., and ten Lohuis, M. (1994)
EMBO J., 13, 2084-2088.
2.Bestor, T. H. (1990) Philos. Trans. R. Soc.
Lond. B. Biol. Sci., 326, 179-187.
3.Sapienza, C., Peterson, A., Rossant, J., and
Balling, R. (1987) Nature, 328, 251-254.
4.Yoder, J. A., Walsh, C. P., and Bestor, T. H.
(1997) Trends Genet., 13, 335-340.
5.Bestor, T. H. (1998) Nature, 393,
311-312.
6.Finnegan, E. J., and Kovac, K. A. (2000) Plant
Mol. Biol., 43, 189-201.
7.Bohnert, H. J., Ostrem, J. A., Cushman, J. C.,
Michalowski, C. B., Rickers, J., Meyer, G., DeRocher, E. J., Vernon, D.
M., Krueger, M., Vazquez-Moreno, L., Velten, J., Hoefner, R., and
Schmitt, J. M. (1988) Plant Mol. Biol. Rep., 6,
10-28.
8.Dellaporta, S. L., Wood, J., and Hicks, J. B.
(1983) Plant Mol. Biol. Rep., 1, 19-21.
9.Shevchuk, T. B., and Buryanov, Ya. I. (1999)
Bioorg. Khim., 25, 630-633.
10.Buryanov, Ya. I., Bogdarina, I. G., and Bayev, A.
A. (1978) FEBS Lett., 88, 251-254.
11.Baryshev, M. M., Buryanov, Ya. I., Kosykh, V. G.,
and Baev, A. A. (1989) Biokhimiya, 54, 1894-1903.
12.Kosykh, V. G., Puntezhis, S. A., Buryanov, Ya.
I., and Baev, A. A. (1982) Biokhimiya, 47, 619-625.
13.Sambrook, J., Fritsch, E. F., and Maniatis, T.
(1989) in Molecular Cloning: A Laboratory Manual, 2nd Edn., Cold
Spring Harbor Press, Cold Spring Harbor Press. N. Y., 9.52.
14.Savel'ev, S. V., Ashapkin, V. V., and Vanyushin,
B. F. (1990) Mol. Biol. (Moscow), 24, 1042-1056.