REVIEW: Contribution of Genosystematics to Current Concepts of Phylogeny and Classification of Bryophytes
A. V. Troitsky1*, M. S. Ignatov2, V. K. Bobrova1, and I. A. Milyutina1
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: (495) 939-3181; E-mail: tav@genebee.msu.su2Main Botanic Garden of the Russian Academy of Sciences, ul. Botanicheskaya 4, 127276 Moscow, Russia; E-mail: misha_ignatov@list.ru
* To whom correspondence should be addressed.
Received September 17, 2007
This paper is a survey of the current state of molecular studies on bryophyte phylogeny. Molecular data have greatly contributed to developing a phylogeny and classification of bryophytes. The previous traditional systems of classification based on morphological data are being significantly revised. New data of the authors are presented on phylogeny of Hypnales pleurocarpous mosses inferred from nucleotide sequence data of the nuclear DNA internal transcribed spacers ITS1-2 and the trnL-F region of the chloroplast genome.
KEY WORDS: molecular phylogenetics, genosystematics, hepatics, liverworts, hornworts, pleurocarpous ITS1-2, trnL-FDOI: 10.1134/S0006297907120115
The bryophytes comprise a group of the most simply organized embryophytes. They are characterized by the dominance of a gametophyte in their life cycle; furthermore, they lack a developed vascular system. The almost only function of the diploid sporophyte is spore production, as it is a parasite on the autotrophic gametophyte. In many bryophytes the vegetative propagation is predominant, by means of fragile parts of plants, such as caducous leaves, or specialized organs, such as brood bodies and buds. In this case, sporophytes are very rare and only found in confined areas of the species ranges, and in some species they are completely unknown.
Many bryophyte species demonstrate wide geographical distribution, grow under diverse ecological conditions, and often are pioneers in extreme habitats. Undoubtedly, those species played an important biota-forming role in the settlement of land by plants.
Three main phyla of modern bryophytes--hornworts, liverworts, and mosses--include about 100, 5000, and 10,000 species, respectively (estimated by one of the authors, M. S. Ignatov, on the basis of existing catalogs and databases).
The time of origin of bryophytes and their phyla is not well determined from paleontological data [1]. The cause is a bad preservation of these plants in sediments and rarity of findings of sporophytes, on which the modern systematics is greatly based. Most of such well-identified forms are <60 million years old. However, the age of fossil spores, which can be classified as bryophytes, is 440-450 million years, which is in good agreement with the most reliable molecular-genetic chronology of the origin of land plants (425-490 million years ago) [2].
This article is largely a survey of recent achievements in bryophyte systematics, which are mostly provided by widespread introduction of genosystematics methods. The molecular-genetic study of bryophyte phylogeny began later than that on phylogeny of seed plants, particularly flowering plants, and is not so intensive. Rare publications concerning macrosystematics of bryophytes or their particular groups and usually based on analysis of no more than ten to twenty samples began to appear in second half of the 1990s. A critical point is the year 2000, when seven papers concerning various bryophyte groups were published in the second volume of the journal Bryologist. The current state of the problem is reflected in [3-5]. As of September 2007, there are 337 entries for hornworts, 5517 for liverworts, and 17,412 for mosses in the GenBank database.
BRYOPHYTE ORIGIN AND MACROSYSTEMATICS
Long before the appearance of molecular phylogenetics, it was generally recognized that bryophytes are not a uniform group, and this division was subdivided into two classes, namely, leafy mosses or simply mosses (Bryopsida) and liverworts (Hepaticopsida) comprising both thalloid and leafy forms. Later, a perception that hornworts essentially differ from liverworts led to segregation of these thalloid bryophytes into an individual class, Anthocerotopsida. In modern classifications these three groups have the rank of divisions: Bryophyta, Marchantiophyta, and Anthocerotophyta, or even superdivisions [6]. Many hypotheses have been proposed on bryophyte origin. Bryophytes were thought to derive from red, brown, or green algae as an independent lineage of land plant evolution, considered as an intermediate group of evolution from charophyceans to vascular plants (tracheophytes), or supposed to be a result of reduced organization of more advanced embryophytes [6].
The morphological cladistic analysis has revealed monophyly of land plants, whose closest ancestors are charophyceans, and paraphyly of three monophyletic bryophyte divisions, which form a grade preceding the thracheophytes. However, relationships between the bryophyte divisions remained ambiguous [7-11]. Resolution of this problem was complicated because of small data sets and indefinite interpretation of main morphological signs as homologous. Cladistic analysis of ultrastructure and ontogenesis of male gametes has demonstrated monophyly rather than paraphyly of bryophytes, which are, together with Selaginella (lycophytes) sister to other lycophytes and other vascular plants [12].
Molecular data--short sequences of 5S rRNA--were first applied to phenetic analysis of four bryophyte species [13, 14] that revealed monophyly of bryophytes. However, a comparison of longer sequences of 18S and 26S rRNA of eight bryophyte species has shown again a paraphyly of bryophytes, with liverworts in the basal group and mosses and hornworts sister to vascular plants [15]. Phylogenetic analysis of the chloroplast rbcL gene sequence of six bryophytes has led to conclusion on polyphyly of both bryophytes and vascular plants [16]. Analysis of 26S rRNA along with analysis of small ribosomal subunit RNA genes from all three genetic compartments of the cell and the rbcL gene has shown hornworts as a basal group of embryophytes and moss-liverwort clade as sister to vascular plants [17, 18].
Thus, these early molecular studies of evolution have led to conflicting conclusions on relationships between three bryophyte groups and tracheophytes. However, small numbers of bryophyte species were analyzed in all three above-mentioned studies, whereas significantly large sampling of species in the studied taxa provides the only reliable phylogenetic reconstructions. Analysis of longer sequences by means of combining the data on many genes cannot compensate possible effect of homoplasy and difference in the molecular evolution rates in different lineages. In this context, it is most likely to get a single well-supported tree that does not reflect the genuine phylogeny. (This kind of mistake is considered in the paper of Soltis and coworkers [19]).
So, one has to look with a critical eye at phylogenetic conclusions deduced from analysis of small species sampling, even if it includes long sequences. Those are conclusions of Nishiyama et al. [20] on monophyly of bryophytes, which were made from trees constructed on the basis of 51 gene sequences from whole chloroplast genomes of 20 higher plant and algal species. However, only three bryophyte and two vascular plant (psilotum and fern) species were taken into analysis. In another study [21], in which a significant part (57) of chloroplast genomes from 17 plant species including single species of mosses, liverworts, and hornworts was compared, the monophyly of bryophytes was also established, with hornworts as the most primitive ones.
In our laboratory, the phylogenetic trees for 38 species of bryophytes, seven species of lycophytes, and two species of algae were constructed from sequences of inner transcribed spacers of chloroplast rRNA genes: ITS2, 3, and 4 using three different methods [22]. According to the phylogenetic reconstruction, hornworts (four species were used in analysis) are sister to vascular plants, liverworts comprise a basal group of land plants, whereas mosses occupy an intermediate position. (Earlier, the “liverworts-basal” topology was established from analysis of ITS2-4 from smaller species sampling [23].) These conclusions were afterwards confirmed by scale reconstruction of phylogeny of about 200 green plant species by the sequences of six genes [24].
Some signs of chloroplast and mitochondrial genome organization are also indicative of hornwort position sister to vascular plants. In particular, the loss of gene ycf66 in Anthoceros formosae chloroplast DNA [25] and character of mitochondrial gene nad5 splicing [26] are synapomorphies integrating hornworts with vascular plants. Paraphyly of bryophytes, which form a grade with liverworts in basal position and hornworts sister to vascular plants, is confirmed by analysis of intron gains and losses in other mitochondrial genes and by highly supported reconstruction of phylogeny of 11 bryophyte species based on sequences of the mitochondrial nad5 gene (~4700 positions) [26].
Nonetheless, there are other conclusions: in a phylogenetic tree of 37 hornwort, six liverwort, and four moss species reconstructed from sequence analysis of rbcL, nad5, and 18S rRNA genes, hornworts are the most ancient, and mosses and liverworts form a clade sister to tracheophytes [27]. At the same time, insufficient representation of mosses and liverworts is obvious.
LIVERWORTS (MARCHANTIOPHYTA)
The classical systematics [28, 29] traditionally subdivides liverworts into two big groups: marchantioid or complex thalloid liverworts and jungermannioid liverworts, including the leafy and simple thalloid taxa. Paraphyly of liverworts expressed as affiliation of jungermannioid liverworts with mosses on the analysis of 18S rRNA [30] and rbcL [31] was not confirmed by analysis of chloroplast ITS2-4 DNA [23] and some other sequences.
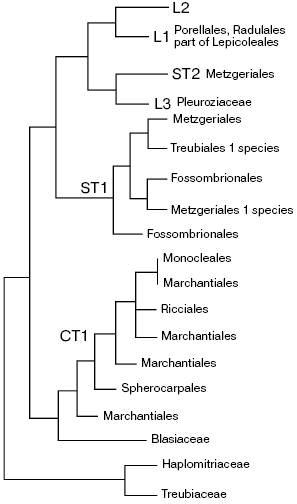
The most common liverwort systems of Schuster [28] and Grolle [32], widely used until recently have undergone major changes as a result of incorporation of molecular data. New systems were quite recently developed [33, 34] on the basis of multigene analysis of extended sampling of liverworts [35, 36]. The datasets used for phylogenetic reconstructions included sequences from all three genetic compartments of the cell: chloroplast trnL-F, rps4, rbcL, atpB, and psbA; mitochondrial nad5; and nuclear rRNA genes of 50-173 liverwort species. Some authors supplemented molecular data with anatomical and physiological characters. So reconstructed phylogenies and systems deduced from them in the above-mentioned papers are not completely identical, but their “backbones” are about the same. One can suppose that the main drawn conclusions will no longer be seriously revised. Figure 1 shows the “backbone” tree of liverwort phylogeny in accordance with the data of Forrest et al. [34], and the Scheme - new system of liverworts, which is put forward by He-Nygren et al. [37]. A whole series of taxa which were thought to be monophyletic turn out to be para- or even polyphyletic, including Mezgeriidae commonly considered as a basal group of Jungermanniidae.
Fig. 1. Phylogenetic tree of liverworts according to Forrest et al. [34] with some modifications. L1, L2, and L3 are leafy liverworts; ST1 and ST2 are simple thalloids, and CT1 is complex thalloids.

System of liverworts proposed by He-Nygren et al. [37]
Scheme
In general, the classical subdivision into marchantioids and jungermannioids remains valid, but this dichotomy is not first on the phylogenetic tree of liverworts--the Treubia plus Haplomitrium clade separates still earlier. Basal position of Haplomitrium relatively to other liverworts is also supported by presence (unlike other liverworts) of active mitochondrial gene nad7 [38]. Haplomitrioids were often considered as a primitive, basal group of jungermannioids, but always referred to leafy groups and never compared with morphologically different thalloid treubioids. However, phylogenetic reconstructions supporting affinity of Haplomitrium and Treubia seem to be reliable. A particular position of treubioids was noted by Schuster [28], who put them into the basis for a system of thalloid jungermannioids. The revelation of a particular position of treubioids in the system provoked studies of their anatomy, which has found unique characteristics (particularly, very peculiar mycorrhiza), which make Treubia kin with Haplomitrium [5].
The second completely surprising conclusion is attribution of the genus Blasia, which all bryologists earlier unanimously included in jungermannioids, to the group of marchantioid liverworts, in which it was elevated to a new order or, in some other interpretations, to a separate class. Such rating enables a new look at a series of morphological structures of Blasia that have no obvious homology within jungermannioids.
HORNWORTS (ANTHOCEROTOPHYTA)
Until quite recently, hornworts remained relatively unexplored, although its modern diversity hardly achieves one hundred species. Their system was not actually developed, although a phylum Notothylas with shortened sporophytes was sometimes segregated in a distinct family.
Studies of hornworts by methods of molecular phylogenetics [27, 39] allow a new system for them. On a phylogenetic tree reconstructed from sequences of rbcL, nad5, and 18S rRNA genes, Anthoceros, Folioceros, and Sphaerosporoceros form a separated clade (subclass Anthocerotidae) sister to most other hornworts affiliated to the subclass Notothylatidae [27]. Thus, it has been found that the genus Notothylas is closer to the wide-spread genus Phaeoceros, than the latter is to the second wide-spread genus Anthoceros (both latter have never been considered to be so very different and often placed to one genus).
Yet the main discovery from molecular studies of evolution is associated with a small plant from Central America, Leiosporoceros. This genus is found to be not only sister to other hornworts, but highly differs from them, so that it is currently segregated to the second autonomous class of Anthocerotophyta, Leiosporocerotopsida. Naturally, this isolated position determined its active investigation. Despite small difference in appearance from other hornworts, Leiosporoceros demonstrates very peculiar traits associated with its symbionts. It is known that all hornworts live in symbiosis with cyanobacteria populating the mucilage cavities. However, in Leiosporoceros these cavities grow with growth of a thallus to form strands penetrating, like a midrib, each branch of the thallus. No other plant has such conformation of cavities [7]. The low level of RNA editing is also a peculiarity of Leiosporoceros. In this feature, it differs from all other hornworts. Analysis of phylogenetic relationships of Leiosporoceros and extended sampling of other hornworts based on molecular data and effect of character of RNA editing on conclusions is reported by Duff and coworkers [27].
MOSSES (BRYOPHYTA)
Various DNA regions were determined in all moss families and in the great majority of moss genera. The latest system, which has assimilated the genosystematics data and is largely based on them, divides mosses into classes Takakiopsida, Sphagnopsida, Andreaeopsida, Andreaeobryopsida, Oedipodiopsida, Politrichopsida, Tetraphidopsida, and the largest one, Bryopsida [40]. Except for the former five classes occupying basal position on the phylogenetic trees, other mosses have peristome--a specialized and having no analogs in other plants structure controlling dispersal of spores from the sporophyte capsule.
The new system has radically altered the position and relationships of many taxa (Fig. 2). The most significant conclusions made by molecular phylogenetics are: although the general idea on primitiveness of sphagnous and andreaeid mosses has been confirmed, some disagreements with systems adopted in the XX century are found, concerning, firstly, two or three basal groups; secondly, interrelation of main groups distinguished for structure of peristome; and thirdly, pleurocarpous mosses, for which the former system turned out to be absolutely inadequate to new concepts.
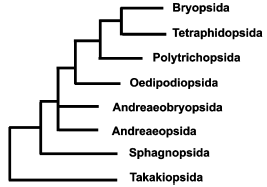
Basal groups of mosses. Relative positions of basal groups on phylogenetic trees reconstructed from analysis of various genes or their sets are not quite in agreement for various samplings and methods [5, 40-43]. In our opinion, the most reasoned are the evolutionary relationships shown in Fig. 3.Fig. 2. Scheme of interrelations between moss systems of Brotherus [55] and Goffinet and Buck [40]. A, acrocarpous mosses; P, pleurocarpous mosses.
Takakia (Takakiopsida). One of two presently known representatives of this enigmatic genus was first collected in 1951, but only described in 1958, because it was quite unclear, what plant group it should be compared with, the more especially as only male plants were found, and sporophyte was not. Till the end of the 1980s, virtually all systematists ranged Takakia in liverworts (according to preferential comparison with Haplomitrium), although already in 1988 Murray [44] noted morphological similarities that may link Takakia with Andreaeobryum.Fig. 3. Phylogenetic relationships between basal groups of mosses based on molecular data.
Another opinion was expressed concerning the systematic position of Takakia: Crandall-Stotler and coworkers [45, 46] investigated the ultrastructure of the meristem area and concluded that neither mosses nor liverworts have such apical cells, which, however, are found in hornworts and especially in lycophytes, so that they propose to range Takakia in a special division, Takakiophyta.
In 1990, at last the Takakia sporophytes were discovered [47, 48], which allowed ranging of Takakia in one of the basal moss groups according to morphological traits. Soon after the data of molecular analysis appeared, according to which Takakia groups with Andreaea, taking off from the main stem of the moss tree after Sphagnum, which occupies the basal position [49], or consolidating with it [24, 41, 42].
On the other hand, we have found in our laboratory [22] the 27-bp insertion in chloroplast ITS3 of Takakia, which is absent in mosses including Andreaea and Sphagnum, but present in all other land plants. Thus, deletion of this segment should be considered as a synapomorphy, and Takakia--as the most ancient living moss. The same conclusion ensues from characteristics of chloroplast RNA editing, which draw Takakia together with hornworts rather than mosses [50].
Andreaeobryum (Andreaeobryopsida). This peculiar monotypic genus was described in 1976. It somewhat resembles Andreaea (Andreaeopsida), but so different from it that Murray proposed to range Andreaeobryum in a separate order [44]. The molecular data [40, 42] are in good agreement with morphological ones and provide reason to elevate this high enough rank to class. Andreaeobryopsida and Andreaeopsida form a clade or grade on phylogenetic trees reconstructed from molecular data.
Oedipodium. Unlike Takakia and Andreaeobryum, Oedipodium has been known from the beginning of XIXth century. In the XXth century its position in the system altered insignificantly, it was placed either in Splachnaceae or in its own family that is separate but in one way or another near Splachnaceae and Funariaceae. The absence of peristome hampered their rating to these groups, but peristome reduction is a frequent phenomenon in many moss groups, so there was no presumption to suspect whatever but reduction. Nevertheless, the absence of peristome in Oedipodium, may well be primary ancestral: molecular analysis on five genes resolved Oedipodium position on the phylogenetic tree between the basal genera (Takakia, Sphagnum, and Andreaea) and Polytrichaceae (the first group in which peristome appears although in this case it is constructed fundamentally differently than peristome in all other mosses); interestingly, the most primitive Polytrichaceae are also peristome-less [43]. On the trees constructed from four-gene analysis, Oedipodium, with strong bootstrap support, is sister to peristomate mosses [41, 51].
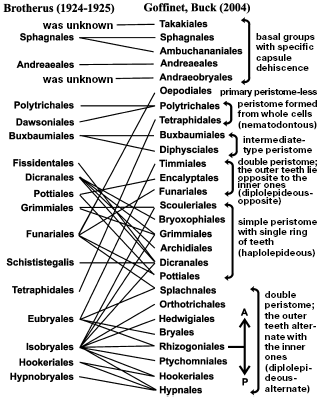
Main groups distinguished by peristome architecture. The very primitive groups have peristome composed of entire cells, whereas more complexly built ones of coalescent remnants of cell walls, which allows it to execute multivarious hygroscopic movements. Molecular data have confirmed such a general evolutionary trend. At the same time, complex peristome is also morphologically differentiated: in different taxa it can possess one ring of 16 teeth or two rings with either alternately or oppositely arranged teeth. Traditional morphology considered the chain of evolutionary transformations of peristome, so that the peristome consisting of one ring of teeth was regarded as the primitive type. However, molecular data have shown [40, 52] that more primitive is a double peristome with opposite elements, and both the double-alternate and single peristomes are its derivatives. This seemingly small refinement has awakened close attention to the character of collocation of peristome teeth in various taxa, gave an impulse to revision of peristome structure in whole families (such as Timmiaceae), and significantly altered the system of peristomate mosses (Fig. 2).
Pleurocarpous mosses. This group includes approximately 50% of all present-day moss species. Unlike acrocarpous mosses which grow orthotropically, the pleurocarps commonly spread along a substrate to give them the ability to form rapidly extensive coverings on soils, rocks, or trees. Ecological diversity of habitats determines high morphological plasticity of pleurocarpous mosses making them difficult to classify.
The newest system of pleurocarpous mosses integrates morphological and molecular data and is largely based on the latter. This group combines subclass Hypnidae - orders Hypnales (4400 species), Hookeriales (750 species), and Ptychomniales (100 species) sister to them, as well as a series of families of the order Rhizogonianae arranged in Bryidae. Rhizogonianae are basal to Hypnidae [40, 53, 54]. Rhizogonianae seem to be non-monophyletic.
In the system of Brotherus [55] that dominated through almost all of the XXth century, pleurocarpous mosses were subdivided into three groups: Hookeriales (this group substantially persists in the new system) and two orders, each combining two-three tens of families and about two thousand species: Hypnobryales (= Hypnales) characterized by complete development of peristome and Isobryales (= Leucodontales) characterized by highly deviating peristome architecture.
Molecular phylogenetic data have shown that separation into Isobryales and Hypnobryales is not supported [56-60] or requires significant changes in conceptions of about a quarter of families with complete change of diagnostic characteristics [61, 62]. The peristome architecture (so stable in other parts of the moss system) in pleurocarpous mosses according to a phylogeny reconstructed by molecular data was proved to be prone to frequent reduction changes. In many cases, molecular data enforce displacement of species and genera into distant families [62].
The “truthfulness” of molecular phylogeny is supported by resemblance in some other characteristics (often in gametophyte structure), which earlier were not regarded as important and, hence, were not considered, and by the fact that in complexes of close species reduced peristome is characteristic of species inhabiting chiefly tree trunks. The latter fact explains much: epiphytical ecotope type strongly determines a whole complex of morphological characteristics including those of peristome architecture [63], which earlier were considered as conservative and on which the system was based. And since such transition to the epiphytical way of life is observed in many families, most of the characteristics used in classification proved to be improper for ascertaining relationships.
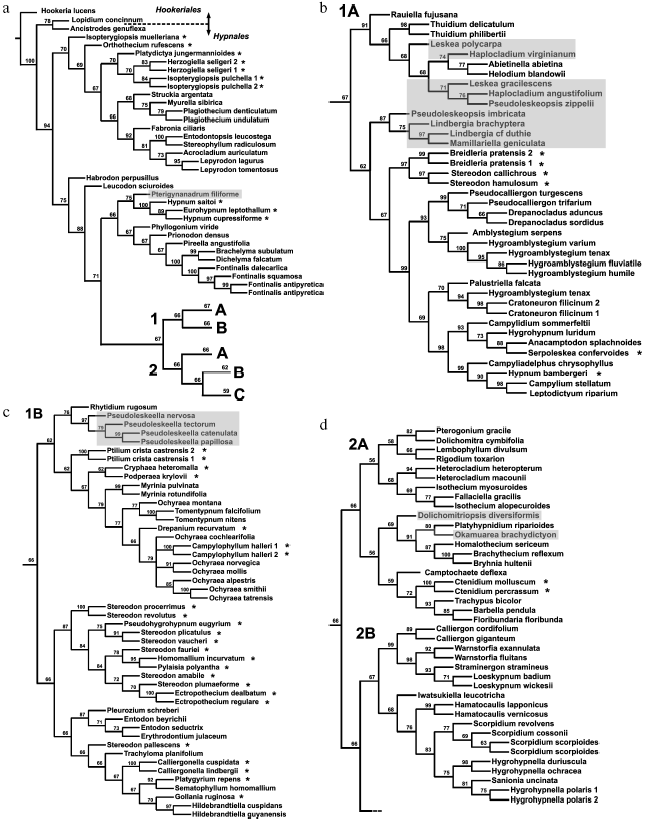
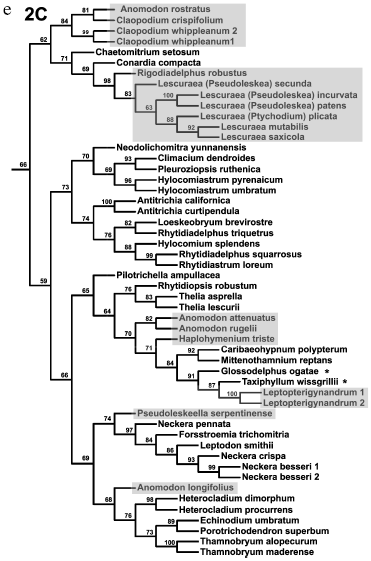
Analysis of distinct regions of the chloroplast genome (sometimes in combination with mitochondrial genes) led to the conclusion that diversification of pleurocarpous mosses occurred very rapidly. A consideration was put forward on this basis that any construction of their reliable supported phylogeny is impossible [60]. However, such skepticism seems exaggerated. Inclusion of nuclear ITS into the analysis has enabled increased resolution and more reliably supported phylogenies with completely rational (although not always recognized) groups [61, 62]. Figure 4 shows one of the recent phylogenetic trees for 218 samples of pleurocarpous mosses reconstructed in our works from the analysis of nuclear ITS1-2 sequences and trnL-F of chloroplast genome. Hypnales representatives separated into two large clades generally coincided with two groups, 1 and 2, revealed from the results of exclusively morphological (but very accurate and all-embracing) analysis fulfilled by Hedenäs during the pre-molecular epoch [64]. Two clades, in turn, are divided into subclades 1 (A, B) and 2 (A, B, C). The branches leading to these main groups are substantially shorter than those at the distal part of the tree and their support is not high (62-70%). Therefore, interrelationships between them cannot be definitely stated. A number of taxa in a traditional system (genus Hypnum, families Hypnaceae, Leskeaceae, Amblystegiaceae, etc.) turned out to be non-monophyletic. Volumes of many families require reconsideration. Some new groups correlate with particular anatomical and morphological features.
It may seem that the above-represented conclusions of gene systematics on phylogeny and systematics of bryophytes are too categorical. This remark is right in part, because we did not make a detailed analysis of existing contradictions and their probable causes. Very rapid explosive evolution, like in pleurocarpous mosses, and extinction of a substantial part of representatives of ancient groups are serious obstacles for the reconstruction of phylogenies. Sufficiently large sampling of the analyzed taxons is one of the most important factors providing confidence of phylogenetic reconstructions.Fig. 4. a-e) Consensual phylogenetic tree constructed using the maximum parsimony method by nuclear ITS1-2 and chloroplast trnL-F sequences for 218 samples of pleurocarpous mosses. Jackknife support values are indicated for nodes. Shadowing denotes representatives of genera traditionally assigned to Leskeaceae, and asterisks denote representatives traditionally assigned to Hypnaceae.
On the other hand, the elongation of analyzed sequences by grouping the data by a number of genes provides lower significance. So, for instance, the phylogenetic tree for 48 thalloid liverworts built by five chloroplast genes was not substantially changed after the addition of two nuclear and one mitochondrial genes [36]. The result of new analysis for the same species differs from the former one in two alterations of bootstrap support values by 1-2% and in a solution of one formerly non-resolved node in frames of one clade. There are many examples like this.
The choice of genome loci used for the analysis plays an important role as well. Unfortunately, at present the set of sequences applicable for these aims is rather limited. Nuclear genome ITSs are most variable in bryophytes among popular loci in plant gene systematics. These sequences are used most frequently for studies at the species level, and sometimes at generic and populational level. However, as we have shown, it can be successfully employed at higher taxonomic level in bryophytes.
Not only construction of trees by sequences, but also analysis of structural genome rearrangements and localization of introns may play very important roles in revealing of phylogenetic interrelations. These genome characteristics are subjected to homoplasies to a lesser extent and their regarding can be determining in solution of problem situations.
Molecular phylogenetics has introduced a determining contribution to the solution of phylogenetic and systematic problems in bryophytes and opened wide perspectives for populational and microevolutional studies. It has provided a strong impulse to the search for new morphological and anatomical markers of phylogenesis and reconceptualization of morphological evolution pathways. Revelation of key groups in evolution has put forward new tasks for comparative anatomy (it was previously focused on “typical representatives”, whereas evolutionally important subjects were avoided). Now the latter become the subjects for the closest attention, that is, the molecular phylogenetics determines the tasks for investigations on evolution of distinct systems: genetic, biochemical, physiological, anatomic, and morphological. Of course, many taxonomic problems are not solved by traditional botany. It is clearly important to solve the problems, and this task primarily concerns botanists themselves.
This study was supported by the Russian Foundation for Basic Research (grants 06-04-49493 and 07-04-00013).
REFERENCES
1.Meyen, S. V. (1987) Fundamentals of Palaeobotany
[Russian translation], Nedra, Moscow.
2.Sanderson, M. J. (2003) Am. J. Bot.,
90, 954-956.
3.Goffinet, B., Hollowell, V., and Magill, R. (eds.)
(2004) Molecular Systematics of Bryophytes, Missouri Botanical
Garden Press, St. Louis.
4.Newton, A. E., and Tagney, R. S. (eds.) (2007)
Pleurocarpous Mosses: Systematics and Evolution, CRC Press, Boca
Raton.
5.Renzaglia, K. S., Schuette, S., Duff, R. J.,
Ligrone, R., Shaw, A. J., Mishler, B. D., and Duckett, J. G. (2007)
Bryologist, 110, 179-213.
6.Kenrick, P., and Crane, P. R. (1997) The Origin
and Early Diversification of Land Plants, Smithsonian Institution
Press, Washington.
7.Sluiman, H. J. (1983) Protoplasma,
115, 160-175.
8.Bremer, K., Humphries, C. J., Mishler, B. D., and
Churchill, S. P. (1987) Taxon, 36, 339-349.
9.Mishler, B. D., and Churchill, S. P. (1985)
Brittonia, 37, 282-285.
10.Whittemore, A. T. (1987) Cladistics,
3, 60-65.
11.Theriot, E. (1988) Taxon, 37,
913-919.
12.Garbary, D. J., Renzaglia, K. S., and Duckett, J.
G. (1993) Plant Syst. Evol., 188, 237-269.
13.Hori, H., Lim, B.-L., and Osawa, S. (1985)
Proc. Natl. Acad. Sci. USA, 82, 820-823.
14.Van de Peer, Y., de Baere, R., Cauwenberghs, J.,
and de Wachter, R. (1990) Plant Syst. Evol., 170,
85-96.
15.Waters, D. A., Buchheim, M. A., Dewey, R. A., and
Chapman, R. L. (1992) Amer. J. Bot., 79, 459-466.
16.Manhart, J. R. (1994) Mol. Phylogenet.
Evol., 3, 114-127.
17.Hedderson, T. A., and Chapman, R. L. (1996)
Plant Syst. Evol., 200, 213-224.
18.Nickrent, D. L., Parkinson, C. L., Palmer, J. D.,
and Duff, R. J. (2000) Mol. Biol. Evol., 17,
1885-1895.
19.Soltis, D. E., Albert, V. A., Savolainen, V.,
Hilu, K., Qiu, Y.-L., Chase, M. W., Farris, J. S., Stefanovic, S.,
Rice, D. W., Palmer, J. D., and Soltis, P. S. (2004) Trends Plant
Sci., 9, 477-483.
20.Nishiyama, T., Wolf, P. G., Kugita, M., Sinclair,
R. B., Sugita, M., Sugiura, C., Wakasugi, T., Yamada, K., Yoshinaga,
K., Yamaguchi, K., Ueda, K., and Hasebe, M. (2004) Mol. Biol.
Evol., 21, 1813-1819.
21.Goremykin, V. V., and Hellwig, F. H. (2005)
Plant Syst. Evol., 254, 93-103.
22.Samigullin, T. Kh., Yacentyuk, S. P.,
Degtyaryeva, G. V., Valiejo-Roman, K. M., Bobrova, V. K., Capesius, I.,
Martin, W. F., Troitsky, A. V., Filin, V. R., and Antonov, A. S. (2002)
Arctoa, 11, 31-43.
23.Samigullin, T. H., Valiejo-Roman, K. M.,
Troitsky, A. V., Bobrova, V. K., Filin, V. R., Martin, W., and Antonov,
A. S. (1998) FEBS Lett., 422, 47-51.
24.Qiu, Y. L., Li, L. B., Wang, B., Chen, Z. D.,
Knoop, V., Groth-Malonek, M., Dombrovska, O., Lee, J., Kent, L., Rest,
J., Estabrook, G. F., Hendry, T. A., Taylor, D. W., Testa, C. M.,
Ambros, M., Crandall-Stotler, B., Duff, R. J., Stech, M., Frey, W.,
Quandt, D., and Davis, C. C. (2006) Proc. Natl. Acad. Sci. USA,
103, 15511-15516.
25.Kugita, M., Kaneko, A., Yamamoto, Y., Takeya, T.,
Matsumoto, T., and Yoshinaga, K. (2003) Nucleic Acids Res.,
31, 716-721.
26.Groth-Malonek, M., Pruchner, D., Grewe, F., and
Knopp, V. (2005) Mol. Biol. Evol., 22, 117-125.
27.Duff, R. J., Villarreal, J. C., Cargill, D. C.,
and Renzaglia, K. S. (2007) Bryologist, 110, 214-243.
28.Schuster, R. M. (1966) The Hepaticae and
Anthocerotae of North America, East of the Hundredth
Meridian, Columbia University Press, New York.
29.Shlyakov, R. N. (1975) The Liverworts.
Morphology, Phylogeny, and Classification [in
Russian], Nauka, Leningrad.
30.Capesius, I., and Bopp, M. (1997) Plant Syst.
Evol., 207, 87-97.
31.Lewis, L. A., Mishler, B. D., and Vilgalys, R.
(1997) Mol. Phylogenet. Evol., 7, 377-393.
32.Grolle, R. (1983) Acta Bot. Fennica,
121, 1-62.
33.Frey, W., and Stech, M. (2005) Nova
Hedwigia, 81, 55-78.
34.Forrest, L. L., Davis, E. C., Long, D. G.,
Crandall-Stotler, B. J., Clark, A., and Hollingsworth, M. L. (2006)
Bryologist, 109, 303-334.
35.Forrest, L. L., and Crandall-Stotler, B. J.
(2005) J. Hattori Bot. Lab., 97, 127-159.
36.Crandall-Stotler, B. J., Forrest, L. L., and
Stotler, R. E. (2005) Taxon, 54, 299-316.
37.He-Nygren, X., Juslen, A., Ahonen, I., Glenny,
D., and Piippo, S. (2006) Cladistics, 22, 1-31.
38.Groth-Malonek, M., Wahrmund, U., Polsakiewicz,
M., and Knoop, V. (2007) Mol. Biol. Evol., 24,
1068-1074.
39.Duff, R. J., Cargill, D. C., Villarreal, J. C.,
and Renzaglia, K. S. (2004) in Molecular Systematics of Bryophytes
(Goffinet, B., Hollowell, V., and Magill, R., eds.) Missouri
Botanical Garden Press, St. Louis, pp. 41-58.
40.Goffinet, B., and Buck, W. R. (2004) in
Molecular Systematics of Bryophytes (Goffinet, B., Hollowell,
V., and Magill, R., eds.) Missouri Botanical Garden Press, St. Louis,
pp. 205-239.
41.Newton, A. E., Cox, C. J., Duckett, J. G.,
Wheeler, J. A., Goffinet, B., Hedderson, T. A. J., and Mishler, B. D.
(2000) Bryologist, 103, 187-211.
42.Cox, C. J., Goffinet, B., Shaw, A. J., and Boles,
S. B. (2004) Syst. Bot., 29, 234-250.
43.Hyvönen, J., Koskinen, S., Merrill, G. L.
S., Hedderson, T. A., and Stenroos, S. (2004) Mol. Phylogenet.
Evol., 31, 915-928.
44.Murray, B. M. (1988) Nova Hedwigia,
90, 289-336.
45.Crandall-Stotler, B. (1986) J. Bryol.,
14, 1-23.
46.Crandall-Stotler, B., and Bozzola, J. J. (1988)
J. Hattori Bot. Lab., 64, 197-218.
47.Smith, D. K., and Davison, P. G. (1993) J.
Hattori Bot. Lab., 73, 263-271.
48.Higuchi, M., and Da-cheng, Z. (1998) J.
Hattori Bot. Lab., 84, 57-69.
49.Hedderson, T. A., Chapman, R., and Cox, C. J.
(1998) in Bryology for the Twenty-First Century (Bates, J. W.,
Ashton, N. W., and Duckett, J. G., eds.) The British Bryological
Society and Maney Publishing, Leeds, pp. 65-77.
50.Sugita, M., Miyata, Y., Maruyama, K., Sugiura,
C., Arikawa, T., and Higuchi, M. (2006) Biosci. Biotechnol.
Biochem., 70, 2268-2274.
51.Magombo, Z. L. K. (2003) Syst. Bot.,
28, 24-38.
52.Ignatov, M. S., and Ignatova, E. A. (2003)
Moss Flora of the Middle European Russia [in Russian], KMK
Scientific Press Ltd., Moscow.
53.Bell, N. E., and Newton, A. E. (2004) in
Molecular Systematics of Bryophytes (Goffinet, B., Hollowell,
V., and Magill, R., eds.) Missouri Botanical Garden Press, St. Louis,
pp. 270-290.
54.O'Brein, T. J. (2007) in Pleurocarpous Mosses:
Systematics and Evolution (Newton, A. E., and Tagney, R. S., eds.)
CRC Press, Boca Raton, pp. 19-49.
55.Brotherus, V. F. (1925) Musci. Die Naturlichen
Pflanzenfamilien (Engler, A., and Prantl, K., eds.) Vol. 11, W.
Engelmann, Leipzig, pp. 1-522.
56.Tsubota, H., Arikawa, T., Akiyama, H., de Luna,
E., Gonzales, D., Higuchi, M., and Deguchi, H. (2002) Hikobia,
13, 645-665.
57.Tsubota, H., de Luna, E., Gonzales, D., Ignatov,
M., and Deguchi, H. (2004) Hikobia, 14, 149-170.
58.Buck, W. R., Goffinet, B., and Shaw, A. J. (2000)
Mol. Phylogenet. Evol., 16, 180-198.
59.Goffinet, B., Cox, C. J., Shaw, A. J., and
Hedderson, T. J. (2001) Ann. Bot., 87, 191-208.
60.Shaw, A. J., Cox, C. J., Goffinet, B., Buck, W.
R., and Boles, S. B. (2003) Evolution, 57, 2226-2241.
61.Gardiner, A., Ignatov, M., Huttunen, S., and
Troitsky, A. (2005) Taxon, 54, 651-663.
62.Ignatov, M. S., Gardiner, A. A., Bobrova, V. K.,
Milyutina, I. A., Huttunen, S., and Troitsky, A. V. (2007) in
Pleurocarpous Mosses: Systematics and Evolution (Newton, A. E.,
and Tagney, R. S., eds.) CRC Press, Boca Raton, pp. 177-213.
63.Huttunen, S., Ignatov, M. S., Müller, K.,
and Quandt, D. (2004) in Molecular Systematics of Bryophytes
(Goffinet, B., Hollowell, V., and Magill, R., eds.) Missouri
Botanical Garden Press, St. Louis, pp. 328-361.
64.Hedenas, L. (1995) J. Bryol., 18,
723-781.