Analysis of Proteins Interacting with TRIP8b Adapter
N. V. Popova1*, A. N. Plotnikov2, R. Kh. Ziganshin1, I. E. Deyev1, and A. G. Petrenko1,2
1Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, 117991 Moscow, Russia; fax: (495) 335-7103; E-mail: petrea01@med.nyu.edu; petrenko@ibch.ru; popova@ibch.ru2New York University, 10016 New York, USA
* To whom correspondence should be addressed.
Received November 12, 2007; Revision received February 13, 2008
Calcium-independent receptor of latrotoxin (CIRL) is an orphan heptahelical receptor implicated in regulation of exocytosis. To characterize molecular mechanisms of CIRL functioning, we searched for its intracellular partners using the yeast two-hybrid SR system with the cytoplasmic C-terminal fragment of CIRL as bait. One of the interacting proteins was identified as TRIP8b, a putative cytosolic adapter protein with multiple tetratricopeptide repeats. To understand functional significance of CIRL-TRIP8b interaction, we further isolated TRIP8b-interacting proteins by affinity chromatography of brain extracts on immobilized recombinant TRIP8b. Sixteen proteins were identified by mass spectrometry in the purified preparations. Clathrin and subunits of AP2 complex appeared to be the major TRIP8b-interacting proteins. Our data suggest a role of TRIP8b in receptor-mediated endocytosis.
KEY WORDS: CIRL/latrophilin, TRIP8b, endocytosis, clathrinDOI: 10.1134/S0006297908060035
Abbreviations: CIRL) calcium-independent receptor of alpha-latrotoxin; GPCR) G-protein coupled receptor; GPS) GPCR proteolysis site; FERM) RhoGEF and pleckstrin domain-containing protein 2; GST) glutathione S-transferase; HCN channel) hyperpolarization-activated cyclic nucleotide-regulated channel; IPTG) isopropyl-beta-D-thiogalactopyranoside; Ndufa9) NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 9; NIPSNAP) 4-nitrophenylphosphatase domain and non-neuronal SNAP-25-like protein; PDZ) domain name composed of first letters of names of three homologous proteins (post synaptic density protein, PSD-95; Drosophila disc large protein, Dlg; zonula occludens-1, ZO-1); PMSF) phenylmethylsulfonyl fluoride; PTS1) peroxisome-targeting signal 1; Rab) Ras-related proteins in brain; SOS) Son of sevenless; SR system) SOS recruitment system; TRIP8b) TPR-containing Rab8b interacting protein; TPR) tetratricopeptide repeat.
Calcium-independent receptor of alpha-latrotoxin or latrophilin
(CIRL) [1, 2] belongs to the
GPS family of G-protein-coupled receptors (GPCR) with large N-terminal
ectodomains, that include structural motives characteristic of cell
adhesion proteins [3]. These heptahelical receptors
are of interest because of their potential to couple extracellular
adhesion interactions with G-protein mediated intracellular signaling
[4].
CIRL consists of two noncovalently bound subunits p120 and p85. p120 is a hydrophilic protein oriented into the extracellular space, while p85 is an integral membrane protein with seven transmembrane helices. Both subunits are encoded by the same gene, and the two-subunit CIRL structure is a result of endogenous proteolysis of a receptor precursor protein [1, 3].
It is known that CIRL is one of the receptors of alpha-latrotoxin [5, 6], a presynaptic toxin causing massive release of neurotransmitter by stimulation of synaptic vesicles exocytosis [7]. alpha-Latrotoxin stimulates release of various neurotransmitters including acetylcholine, glutamate, and gamma-aminobutyric acid. Moreover, alpha-latrotoxin stimulates exocytosis in secretory cells such as chromaffin and pancreatic cells [8-10], and exogenous CIRL expression in chromaffin cells increases their sensitivity to alpha-latrotoxin [9].
Natural agonistic CIRL ligands are still unknown, although some proteins interacting with the extended C-terminal CIRL fragment have been identified. These soluble multidomain scaffold proteins with PDZ domains belong to the ProSAP/SSTRIP/Shank family [11, 12]; its representatives play a key role in the formation of multimolecular complexes of membrane and cytoskeletal proteins in synapses. Their interaction with CIRL supposedly targets receptors to the zones of exocytosis.
Using the yeast two-hybrid SR system, we detected interaction of CIRL cytoplasmic part with TRIP8b protein that was originally discovered as a partner of Rab8b small GTPase and also of HCN membrane channel protein [13, 14]. The analysis of TRIP8b interacting proteins suggests that this protein regulates CIRL transport from the cytoplasmic membrane to endosomes.
MATERIALS AND METHODS
The following reagents were used in this study: NaCl and Triton X-100 from Amresco (USA); Tris and Hepes from Merck (Germany); Tween-20 and dried milk from BioRad (USA); restriction endonucleases from Fermentas (Lithuania); SuperSignal West Pico chemiluminescence detection kit from Pierce (USA). We also used nitrocellulose membrane from GE Healthcare (USA), Ni-NTA from Qiagen (USA), BrCN-activated Sepharose from Sigma (USA), trypsin from Promega (USA), HRP-conjugated anti-rabbit and anti-mouse IgG from Jackson ImmunoResearch Laboratories (USA). Antibodies against clathrin light chain were kindly provided by Dr. Thomas Südhof, antibodies against Caspr by Dr. James Salzer, and yeast two-hybrid SR system (SOS recruitment system) including rat brain cDNA library by Dr. Michael Karin.
Yeast two-hybrid SR system. For screening, we used rat brain cDNA library, in which cDNA was cloned in the polylinker of yeast expression vector pYes2 following the nucleotide sequence encoding myristoylation signal [15]. The sequence encoding the C-terminal part of CIRL-1 p85 subunit (bait) linked with shortened human SOS protein (hSOS) was cloned in pADNS vector [16].
Screening was performed as described in [15]. Yeasts were grown on minimal medium with glucose (the medium contained the necessary amino acids, 2% glucose, 0.5% (NH4)2SO4, 0.17% yeast extract, and 4% agar) or on the galactose medium containing 3% galactose, 2% raffinose, and 2% glycerol instead of glucose. Yeast cdc25-2 strain cells were co-transfected with 3 µg of pADNS-CIRL1-CT plasmid and 3 µg of pYes2-cDNA plasmid library and grown for 4 days at 25şC on the minimal glucose plates. Then colonies were placed onto minimal galactose plates and grown for 2-3 days at 36şC. Plasmids isolated from 17 colonies were transformed in Escherichia coli and used for DNA insertion sequencing.
Isolation of proteins and preparation of sorbents for affinity chromatography. DNA encoding full-sized TRIP8b protein was isolated as a result of screening and cloned in pET-32a(+) vector via NcoI/EcoRV sites. The construction thus obtained encodes TRIP8b bearing Trx-tag (thioredoxin), His-tag (sequence of six histidine residues), and S-tag (S-peptide sequence bonded to S-protein) at the N-end. Escherichia coli BL21(DE3)pUBS520 strain cells [17] were transformed by pET-32a-TRIP8b plasmids or pET-32a (control), and protein synthesis was induced by addition of isopropyl-beta-D-thiogalactopyranoside (IPTG) to final concentration 0.5 mM. Proteins were purified by chromatography on nickel-containing resin (Ni-NTA) under native conditions (Tag is control protein containing Trx-tag, His-tag, and S-tag sequences) or under denaturing conditions with urea (TRIP8b) according to the producer's protocol. In case of TRIP8b obtained under denaturing conditions, stepwise dialysis and refolding were performed according to Qiagen's recommendations. Protein purity was monitored by SDS-PAGE (10% polyacrylamide gel) according to Laemmli.
To obtain TRIP8b-Sepharose and Tag-Sepharose (control), proteins were linked with BrCN-activated Sepharose so that final protein concentrations were 2 mg/ml of resin.
Preparation of antibodies against TRIP8b. To obtain antibodies against TRIP8b, the DNA sequence of its C-terminus (207-602 aa) was cloned into pET28a(+) vector via NdeI/XhoI sites. The hybrid recombinant protein with six additional histidine residues at the N-end was purified on Ni-containing resin according to the producer's protocol. Rabbits were immunized by subdermal injections of the recombinant protein mixture with Freund's complete or incomplete adjuvant.
Analysis of interaction between CIRL and TRIP8b in vitro. Escherichia coli BL21(DE3)pUBS520 strain cells were transformed by pGEX-kg-CT2 plasmid encoding CIRL-1 C-terminal part (1269-1471 aa) linked with glutathione S-transferase (GST-CT2). Vector pGEX-kg encoding glutathione S-transferase (GST) was used as control. Protein synthesis was induced by addition of IPTG to final concentration 0.5 mM. Cell lysate was applied to glutathione-Sepharose and washed with phosphate-buffered saline (PBS). Then an aliquot of resin (50 µl) was incubated with COS cell lysate transfected by pcDNA3.1_TRIP8b plasmid encoding full-sized TRIP8b.
The resin was washed with buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF); resin-bound proteins were eluted with buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM glutathione. After elution, proteins were separated by SDS-PAGE (10% gel) and applied onto a nitrocellulose membrane. TRIP8b was detected using rabbit antibodies specific to its C-terminal part.
Affinity chromatography of rat brain proteins. Rat brains (20 g) were homogenized in 20 ml of buffer A (20 mM Tris-HCl or Hepes-KOH, pH 7.3, 10 mM EDTA, 1 mM PMSF) and centrifuged at 15,000g for 30 min (all procedures were performed at 4şC). NaCl was added to the supernatant to the final concentration 150 mM, and the mixture was centrifuged at 100,000g for 1 h. The supernatant consisting of soluble rat brain proteins was applied to TRIP8b- or Tag-Sepharose (resin volume 7 ml) at the rate 5 ml/h. The sorbent was washed with 150 ml of buffer B (20 mM Tris-HCl or Hepes-KOH, pH 7.3, 150 mM NaCl, 1 mM EDTA, 0.05% Triton X-100) and bound proteins were eluted with buffer C (20 mM Tris-HCl or Hepes-KOH, pH 7.3, 1 M NaCl, 1 mM EDTA).
To obtain rat brain membrane extract, the pellet after low-speed centrifugation was homogenized in buffer D (20 mM Tris-HCl, pH 7.3, 1 mM EDTA, 1.5% Triton X-100, 1 mM PMSF), incubated for 40 min on ice, and centrifuged at 100,000g for 1 h. NaCl was added to the supernatant to the final concentration 150 mM, and the mixture was used for chromatography as described above, with buffers C and D additionally containing 0.2% Triton X-100.
Eluate proteins were concentrated and separated by SDS-PAGE (8-20% gradient gel). After the electrophoresis, gels were stained with Coomassie R-250.
Western blot. Proteins were separated by SDS-PAGE (10% gel) and blotted onto nitrocellulose membrane. For blocking, the membranes were incubated in TBS-T buffer (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.1% Tween-20) containing 5% fat-free milk overnight at 4şC. To detect proteins, the membranes were incubated for 90 min at room temperature with the primary antibodies (anti-TRIP8b, 1 : 5000; anti-clathrin light chains, 1 : 750; anti-Caspr, 1 : 5000 v/v), washed with TBS-T buffer, and incubated with secondary antibodies (anti-mouse IgG and anti-rabbit IgG conjugated with horseradish peroxidase, 1 : 10,000 v/v) for 1 h. The blots were developed with the SuperSignal West Pico chemoluminescence detection kit.
Protein mass spectrometry. Samples were prepared according to [18] with some modifications. After SDS-PAGE (8-20% gradient gel), the gel was stained with Coomassie R-250 and protein bands were excised and cut into 1 × 1-mm pieces. A piece of gel was washed twice for 5 min in 70 µl of 50% 200 mM NH4HCO3-50% acetonitrile mixture (v/v) and placed in 70 µl of acetonitrile for 15 min, then acetonitrile was removed, and the gel was dried on SpeedVac for 20 min. The dried gel piece was put in 3 µl of trypsin solution (10 ng/µl) in 50 mM NH4HCO3, kept on ice for 30 min, and then incubated overnight at 37şC. The next day 6 µl of 50% acetonitrile in 0.1% trifluoroacetic acid (TFA) was added to the gel, and the gel was incubated for 2 h. After incubation, 2 µl of the above-gel solution was taken and mixed with template solution (10 mg/ml 2,5-dihydroxybenzoic acid in 50% acetonitrile-0.1% TFA, v/v) and applied on a mass-spectrometric target. Mass spectra were recorded using an Ultraflex TOF/TOF mass spectrometer from Bruker in reflectory mode in the m/z range 600-6000 daltons.
The data were processed using Bruker Daltonics Flex Analysis 2.2 software, and peaks of trypsin contained in samples were used for calibration. The following search parameters were used: accuracy of mass determination 100 m.f., SwissProt protein sequence Database, Rodentia taxon (rodent), one missed cleavage, possible methionine oxidation. If the value of Score parameter calculated for each protein exceeded 70, identification was accepted as reliable. The method for calculation of the Score parameter is described at site http://www.matrixscience.com/help/scoring_help.html.
RESULTS
The search for intracellular molecular partners of CIRL was performed using the yeast two-hybrid SOS recruitment (SR) system [15]. Use of this system to search for proteins interacting with membrane proteins is more justified compared with the conventional two-hybrid system, because in this case proteins interact not in the nucleus but on the membrane.
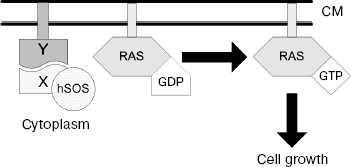
The main principle of this method is as follows: being positioned near the membrane, C-terminally truncated hSOS (human SOS protein) fused to protein of interest (X “bait”) is able to complement temperature-sensitive mutation of yeast factor (cdc25-2 protein) of GDP/GTP exchange on Ras protein. In the SR system, hSOS translocation to membrane occurs in case of X “bait” interaction with unknown protein Y, which may be an integral membrane protein or soluble protein anchored in membrane by myristoylation signal (Fig. 1).
The C-terminal part of CIRL-1 p85 subunit linked with the truncated hSOS was used as “bait”. As a result of screening of rat brain cDNA library, 17 possibly positive clones were revealed in yeast expressing vector. Sequencing of their DNA showed that seven of them encode C-terminal fragments of TRIP8b protein (AAK38580). Then a full-sized cDNA of this protein was cloned using a rat brain cDNA phage library as described in [1].Fig. 1. Principle of yeast SR system functioning. Protein X fused to C-terminally truncated hSOS and protein Y localized on cytoplasmic membrane are co-expressed in the yeast cell. Interaction between X and Y proteins recruits hSOS to the plasma membrane where it stimulates GDP/GTP exchange on Ras protein; as a result, the cell gains the capacity of growing at 36şC. CM, cytoplasmic membrane.
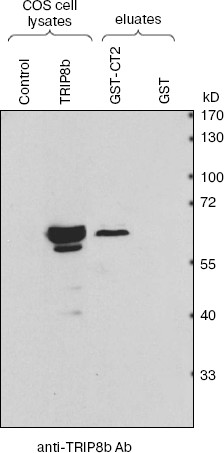
To confirm interaction between CIRL and TRIP8b, we used precipitation of their fragments obtained by artificial expression. The CIRL-1 C-terminal part fused to glutathione-S-transferase sequence at the N-end (GST-CT2) was obtained by expression in bacterial cells and then sorbed on glutathione-Sepharose. The lysate of COS cells transfected by plasmid encoding full-sized TRIP8b was incubated with GST-CT2-Sepharose or with control GST-Sepharose. After elution, proteins were separated by SDS-PAGE and blotted onto nitrocellulose membrane, and then TRIP8b was detected with antibodies. As shown in Fig. 2, TRIP8b is absent from the eluate from control resin, but is stained in the eluate from resin on which CIRL-1 C-terminal fragment (GST-CT2) was sorbed.
TPR-containing Rab8b interacting protein (TRIP8b) was first identified as a protein interacting with Rab8b protein [13]. It was also shown that TRIP8b interacts with a protein forming HCN channel (hyperpolarization-activated cyclic nucleotide-regulated channel) [14]. However, the physiological role and detailed mechanisms of TRIP8b functioning are still unclear.Fig. 2. Checking for interaction between TRIP8b and CIRL-1 (immunoblot analysis). Glutathione-Sepharose on which CIRL-1 cytoplasmic part (202 aa) fused with GST (GST-CT2) or GST (control) was sorbed, was incubated with COS cell lysate containing TRIP8b. Bound proteins were eluted with glutathione-containing buffer. Eluate aliquots were separated by SDS-PAGE (10% gel), blotted onto nitrocellulose membrane, and then TRIP8b was detected by antibodies.
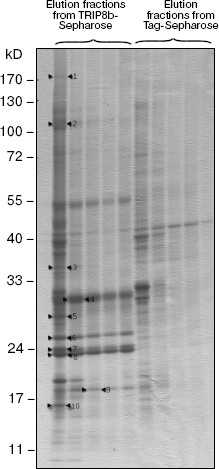
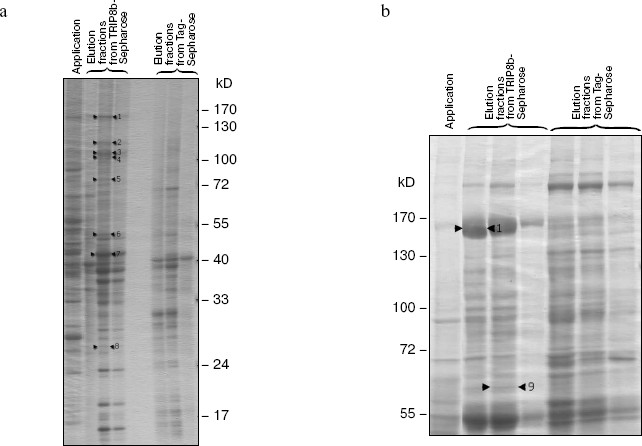
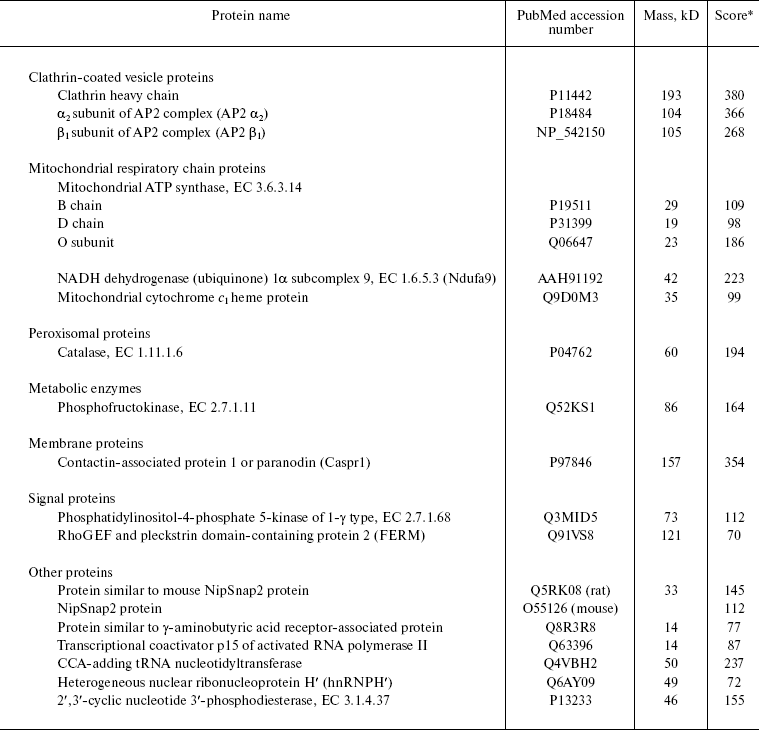
To obtain additional information about this protein, we searched for TRIP8b-interacting proteins, using affinity chromatography on resin with immobilized TRIP8b bearing Trx-tag, His-tag, and S-tag at the N-terminus. Sorbent with immobilized Trx-, His-, and S-tags was used as control resin (Tag-Sepharose). The fraction of soluble rat brain proteins or detergent extracts of crude membrane fraction, which contained nuclei and mitochondria along with cytoplasmic membranes, was applied to the resin. The resin was washed with initial buffer, and bound proteins were eluted with high-salt buffer and separated by SDS-PAGE (Figs. 3 and 4). Protein bands present only in the eluates from the TRIP8b-Sepharose, but not in the eluates from the control resin were excised from the gel, and these samples were used for mass spectrometry. Proteins reliably identified by mass spectrometry are presented in the table and also shown by arrows in Figs. 3 and 4. It should be noted that in the presence of Tris-containing buffer efficiency of clathrin binding to TRIP8b-Sepharose markedly decreased (Fig. 4, a and b); this possibly indicates that Tris-buffer causes disassembly of clathrin forming clathrin vesicles [19].
Fig. 3. Results of chromatography of proteins of rat brain membrane solubilizate on TRIP8b-Sepharose (8-20% gradient SDS-polyacrylamide gel). Proteins of rat brain membrane solubilizate were applied on TRIP8b- or Tag-Sepharose, the resin was washed with initial buffer, then bound proteins were eluted with high-salt buffer, separated by gradient SDS-PAGE, and detected by staining with Coomassie R-250. Marked by arrows: 1) Caspr; 2) alpha2 subunit of AP2 complex; 3) Ndufa9; 4) cytochrome c1; 5) protein similar to NipSnap2 protein; 6) ATP synthase B chain; 7) ATP synthase D chain; 8) ATP synthase O subunit; 9) protein similar to GABA receptor-associated protein; 10) transcriptional coactivator p15 of activated RNA polymerase II.
Results of mass spectrometry of proteins isolated on TRIP8b-SepharoseFig. 4. Results of chromatography of soluble rat brain proteins on TRIP8b-Sepharose (8-20% gradient SDS-polyacrylamide gel). Soluble rat brain proteins were applied on TRIP8b- or Tag-Sepharose in the initial buffer with Tris-HCl (a) or with Hepes-KOH (b), the resin was washed with initial buffer, then bound proteins were eluted with high-salt buffer, separated by gradient SDS-PAGE, and detected by staining with Coomassie R-250. Marked by arrows: 1) clathrin heavy chain; 2) FERM; 3) beta1 subunit of AP2 complex; 4) alpha2 subunit of AP2 complex; 5) phosphofructokinase; 6) heterogeneous nuclear ribonucleoprotein H´; 7) mixture of tRNA nucleotidyltransferase and 2´,3´-cyclic nucleotide 3´-phosphodiesterase; 8) protein similar to NipSnap2 protein; 9) catalase.
*Score is parameter characterizing identification reliability of a certain protein. In general, at score value > 60-70, identification may be considered as reliable.
Thus, we identified 16 proteins and protein complexes specifically sorbed on TRIP8b-Sepharose. As can be seen, proteins of cytoplasmic membrane (Caspr) and mitochondria (ATP synthase subunits, cytochrome c1, NADH dehydrogenase) as well as proteins of clathrin-coated vesicles (clathrin, AP2 complex), peroxisomes (catalase), cytoplasmic proteins (e.g. phosphofructokinase, phosphatidylinositol-4-phosphate 5-kinase), nuclear proteins (transcriptional coactivator p15 of activated RNA polymerase II), proteins participating in tRNA processing (tRNA nucleotidyltransferase), and some other proteins interact with the affinity resin. Interaction of some proteins with TRIP8b is possibly mediated; nonspecific sorption of certain proteins also cannot be excluded.
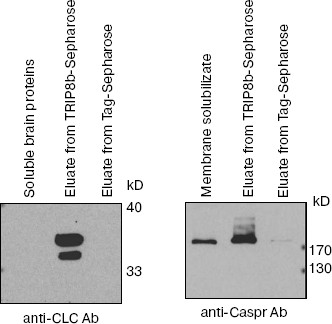
To confirm the results of mass-spectrometry and to test the specificity of the interaction of detected proteins with TRIP8b, proteins of elution fractions were analyzed by Western blotting with antibodies against clathrin light chains or against Caspr (Fig. 5). As can be seen on the immunoblot, antibodies detect clathrin and Caspr, which are present only in elution fractions from TRIP8b-Sepharose but not from Tag-Sepharose.
Fig. 5. Protein identification by antibodies (immunoblot analysis). Proteins of fractions mentioned in the figure were separated by SDS-PAGE (10% gel) and blotted onto nitrocellulose membrane; then clathrin light chains (CLC) or Caspr were detected by antibodies.
DISCUSSION
TRIP8b protein interacting with cytoplasmic domain CIRL-1 was found using the yeast two-hybrid SR system. We confirmed this interaction by coprecipitation of full-sized TRIP8b with CIRL-1 C-terminal part (Fig. 2). We also showed that proteins involved in endocytosis, signal proteins, components of mitochondrial respiratory chain, and some other proteins interact with TRIP8b-Sepharose (table).
TRIP8b is characterized by the presence of six tetratricopeptide repeats (TPR). This structural motif, present in many proteins, mediates protein-protein interactions and provides assembly of multisubunit complexes. TPR domain may consist of 3-16 sequential repeats of 34 amino acid residues, although certain TPR motifs may be dispersed through the protein sequence. TPR motifs have been identified in various organisms from bacteria to humans. Proteins containing TPR domains are involved in various biological processes such as cell cycle regulation, transcription control, transport in mitochondria and peroxisomes, neurogenesis, and protein folding [20].
Pex5p protein (or PTS1 receptor) is the nearest homolog of TRIP8b. TRIP8b is 40% identical to Pex5p protein, and their C-terminal regions bearing TPR domains are 57% identical (another name of TRIP8b is protein relative to Pex5p (Pex5Rp)) [21]. Pex5p participates in transport of proteins bearing C-terminal sequence SerLysLeu (PTS1) in peroxisomes [22].
As mentioned above, TRIP8b was originally detected as a protein interacting with Rab8b [13]. It appeared that it is the TPR domain of TRIP8b that is necessary for its interaction with Rab8b. Rab8b, an intracellular small GTPase, is located in cell contact areas and possibly regulates dynamics of intercellular membrane complexes as well as endo/exocytosis [23]. Enhanced expression of TRIP8b resulted in increased release of adrenocorticotropin by AtT20 cells [13].
TRIP8b also directly interacts with protein forming hyperpolarization-activated cyclic nucleotide-regulated channel (HCN channel) [14]. Coexpression of these proteins in mammalian cells results in decreased HCN concentration on the membrane and its accumulation in early endosomes.
Isolation of clathrin, one of the major proteins in elution fractions from TRIP8b-Sepharose (Fig. 4b), and adapter complex AP2 (alpha and beta_subunits) suggests that TRIP8b participates in endocytosis, since it is known that receptor endocytosis in most cases proceeds via a clathrin-dependent mechanism [24]. Together with clathrin, various adapter proteins, arrestins, and protein kinases are involved in this process [25].
Phosphofructokinase and 2´,3´-cyclic nucleotide 3´-phosphodiesterase (table) were also found in clathrin-coated vesicles [26].
It is not surprising that we isolated catalase on TRIP8b-Sepharose (table), because TRIP8b is known to recognize a sequence that peroxisome proteins bear at the C-end (PTS1). Moreover, it was shown that TRIP8b in vitro interacts with peroxisomal proteins, in particular, with catalase, but does not participate in peroxisomal import [21]. This fact confirms the resin specificity, but this interaction is not likely to have any functional importance in the cell.
The interaction of TRIP8b with such proteins as ATP synthase, Ndufa9, and cytochrome c1 is also functionally unclear (these proteins are components of mitochondrial respiratory chain and participate in oxidative phosphorylation); the same can be said about the interaction of TRIP8b with tRNA-nuleotidyltransferase, which adds conservative CCA sequence to the 3´-end of tRNA during its maturation [27]. Since the detailed intracellular distribution of TRIP8b is unknown, specificity and functionality of the TRIP8b interaction with these proteins is not clear. Another explanation could be possible artifacts resulting from solubilization of membrane proteins with detergent and specific conditions for affinity chromatography.
The potential interaction of TRIP8b with NipSnap2 protein is of interest; this protein belongs to NIPSNAP family and although the function of these proteins is still unclear, it was suggested that proteins of this family participate in vesicle transport. This hypothesis was based on the fact that in C. elegans, the nipsnap gene is located in the operon that encodes proteins participating in this process [28].
Interaction of TRIP8b with three cytoplasmic membrane proteins (HCN, CIRL-1, and Caspr) has been found. On the other side, clathrin and other components of the endocytotic mechanism were the major proteins present in eluates from TRIP8b-Sepharose. Interaction of the C-terminal fragment of CIRL with TRIP8b suggests that TRIP8b, possibly together with Rab8b, defines the specificity of CIRL transport from the cytoplasmic membrane to endosomes, thus regulating receptor concentration on the cell surface. Further studies using other experimental techniques would allow to test this hypothesis, and to determine the mechanisms of CIRL intracellular transport, as well as to determine the role of TRIP8b in endocytosis.
This work was financially supported by the Russian Foundation for Basic Research (grant No. 06-04-49706a), Fundamental Sciences for Medicine and Molecular and Cellular Biology Programs of Presidium of the Russian Academy of Sciences, and NIH Fogarty grant 5 RO3 TW007210.
REFERENCES
1.Krasnoperov, V. G., Bittner, M. A., Beavis, R.,
Kuang, Y., Salnikow, K. V., Chepurny, O. G., Little, A. R., Plotnikov,
A. N., Wu, D., Holz, R. W., and Petrenko, A. G. (1997) Neuron,
18, 925-937.
2.Lelianova, V. G., Davletov, B. A., Sterling, A.,
Rahman, M. A., Grishin, E. V., Totty, N. F., and Ushkaryov, Y. A.
(1997) J. Biol. Chem., 272, 21504-21508.
3.Krasnoperov, V., Lu, Y., Buryanovsky, L., Neubert,
T. A., Ichtchenko, K., and Petrenko, A. G. (2002) J. Biol.
Chem., 277, 46518-46526.
4.Hayflick, J. S. (2000) J. Recept. Signal
Transduct. Res., 20, 119-131.
5.Davletov, B. A., Shamotienko, O. G., Lelianova, V.
G., Grishin, E. V., and Ushkaryov, Y. A. (1996) J. Biol. Chem.,
271, 23239-23245.
6.Krasnoperov, V. G., Beavis, R., Chepurny, O. G.,
Little, A. R., Plotnikov, A. N., and Petrenko, A. G. (1996) Biochem.
Biophys. Res. Commun., 227, 868-875.
7.Rosenthal, L., and Meldolesi, J. (1989)
Pharmacol. Ther., 42, 115-134.
8.Bittner, M. A., and Holz, R. W. (2000) J. Biol.
Chem., 275, 25351-25357.
9.Bittner, M. A., Krasnoperov, V. G., Stuenkel, E.
L., Petrenko, A. G., and Holz, R. W. (1998) J. Neurosci.,
18, 2914-2922.
10.Lang, J., Ushkaryov, Y., Grasso, A., and
Wollheim, C. B. (1998) EMBO J., 17, 648-657.
11.Tobaben, S., Sudhof, T. C., and Stahl, B. (2000)
J. Biol. Chem., 275, 36204-36210.
12.Kreienkamp, H. J., Soltau, M., Richter, D., and
Bockers, T. (2002) Biochem. Soc. Trans., 30, 464-468.
13.Chen, S., Liang, M. C., Chia, J. N., Ngsee, J.
K., and Ting, A. E. (2001) J. Biol. Chem., 276,
13209-13216.
14.Santoro, B., Wainger, B. J., and Siegelbaum, S.
A. (2004) J. Neurosci., 24, 10750-10762.
15.Aronheim, A., Zandi, E., Hennemann, H., Elledge,
S. J., and Karin, M. (1997) Mol. Cell. Biol., 17,
3094-3102.
16.Colicelli, J., Birchmeier, C., Michaeli, T.,
O'Neill, K., Riggs, M., and Wigler, M. (1989) Proc. Natl. Acad. Sci.
USA, 86, 3599-3603.
17.Brinkmann, U., Mattes, R. E., and Buckel, P.
(1989) Gene, 85, 109-114.
18.Terry, D. E., Umstot, E., and Desiderio, D. M.
(2004) J. Am. Soc. Mass Spectrom., 15, 784-794.
19.Pearse, B. M., and Robinson, M. S. (1984) EMBO
J., 3, 1951-1957.
20.D'Andrea, L. D., and Regan, L. (2003) Trends
Biochem. Sci., 28, 655-662.
21.Amery, L., Sano, H., Mannaerts, G. P., Snider,
J., van Looy, J., Fransen, M., and van Veldhoven, P. P. (2001)
Biochem. J., 357, 635-646.
22.Subramani, S. (1998) Physiol. Rev.,
78, 171-188.
23.Lau, A. S., and Mruk, D. D. (2003)
Endocrinology, 144, 1549-1563.
24.Brodin, L., Low, P., and Shupliakov, O. (2000)
Curr. Opin. Neurobiol., 10, 312-320.
25.Reiter, E., and Lefkowitz, R. J. (2006) Trends
Endocrinol. Metab., 17, 159-165.
26.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak,
S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V.,
Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J.,
and McPherson, P. S. (2004) Proc. Natl. Acad. Sci. USA,
101, 3833-3838.
27.Xiong, Y., and Steitz, T. A. (2006) Curr.
Opin. Struct. Biol., 16, 12-17.
28.Seroussi, E., Pan, H. Q., Kedra, D., Roe, B. A.,
and Dumanski, J. P. (1998) Gene, 212, 13-20.