REVIEW: Specific Features of 5S rRNA Structure - Its Interactions with Macromolecules and Possible Functions
A. V. Smirnov1,2*, N. S. Entelis2, I. A. Krasheninnikov1, R. Martin2, and I. A. Tarassov1
1Faculty of Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; E-mail: skard1@yandex.ru2Department of Molecular and Cellular Genetics, UMR 7156, Centre National de la Recherche Scientifique - Université Louis Pasteur, Strasbourg 67084, France
* To whom correspondence should be addressed.
Received November 19, 2007; Revision received February 18, 2008
Small non-coding RNAs are today a topic of great interest for molecular biologists because they can be regarded as relicts of a hypothetical “RNA world” which, apparently, preceded the modern stage of organic evolution on Earth. The small molecule of 5S rRNA (~120 nucleotides) is a component of large ribosomal subunits of all living beings (5S rRNAs are not found only in mitoribosomes of fungi and metazoans). This molecule interacts with various protein factors and 23S (28S) rRNA. This review contains the accumulated data to date concerning 5S rRNA structure, interactions with other biological macromolecules, intracellular traffic, and functions in the cell.
KEY WORDS: 5S rRNA, structure, RNA-protein interaction, ribosome, intracellular transportDOI: 10.1134/S000629790813004X
Small non-coding RNAs are now a topic of interest for researchers as relicts of a hypothetical “RNA world” that apparently preceded the modern stage of organic evolution on Earth. Among small RNAs, 5S rRNA is especially interesting because it is a component of ribosomes of all living beings, except those of mitochondria of fungi and metazoans, and interacts with various protein factors and with 23S (28S) rRNA. 5S rRNA is supposed to play an important role during protein synthesis on ribosomes. But, in spite of much data on 5S rRNA structure and interactions with other biological macromolecules, its function is still not clearly elucidated.
For more than 30 years of studies, 5S rRNA has been a classic model for investigations of structure and behavior of RNA in solutions and within complexes with proteins and also of RNA-protein recognition and binding. The discovery of eukaryotic 5S rRNA import into mitochondria of vertebrata was rather intriguing, although mechanism of this transfer is still unknown. The role of eukaryotic 5S rRNA in the prokaryotic system of mitochondria is also mysterious. The present review considers specific features of 5S rRNA structure in the context of its functions and interactions with various factors.
GENERAL INFORMATION ABOUT 5S rRNA
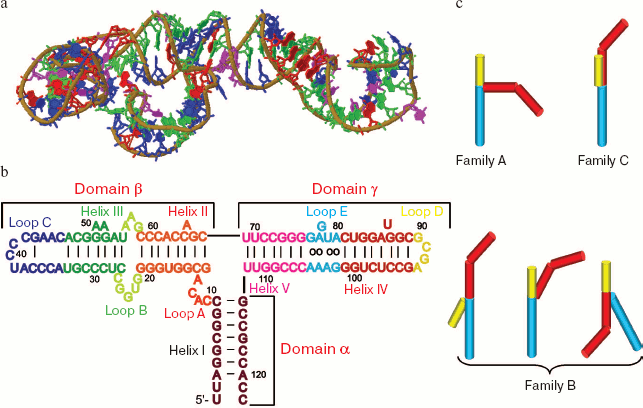
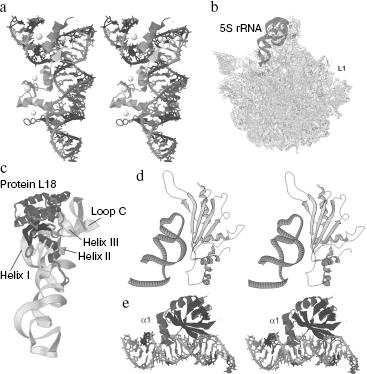
5S rRNA is a relatively small RNA molecule (~120 nucleotides) with highly conservative secondary and tertiary structures. However, its nucleotide sequence can vary even in limits of the same species (thus, 13 allelic variants of 5S rRNA are encoded in the human genome [1]). 5S rRNA molecules of both prokaryotes and eukaryotes have a three-domain Y-shaped organization called a “wishbone” [2-4]. Each “branch” of the molecule is a highly structured system of helices and terminal and inner loops with uninterrupted or nearly uninterrupted stacking (Fig. 1, a and b, see color insert). According to the modern nomenclature, the double-stranded regions of 5S rRNA are designated by Roman numerals and the inner and terminal loops by Latin letters. There are no routine designations for individual domains, but in some cases Greek letters are used for this purpose. Thus, the α-domain is formed by helix I, β-domain is formed by helices II and III and loops B and C, while the γ-domain includes helices IV and V and loops D and E.
Depending on specific features of every domain structure, and especially that of the γ-domain, all 5S rRNAs can be assigned to either bacterial or eukaryotic type. As a rule, 5S rRNAs of organelles have a classic bacterial organization, whereas 5S rRNAs of archaea are more like those of eukaryotes, and this also confirms the common origin of these organisms [5].Fig. 1. General information about 5S rRNA. a) Three-dimensional structure of 5S rRNA of the large ribosomal subunit from Haloarcula marismortui [26] (Jena Library). b) Secondary structure of the same 5S rRNA. c) Variants of geometry of the three-way junction exemplified by 5S rRNA-like molecules. The α-, β-, and γ-domains are shown, respectively, by yellow, red, and blue color (after [117]).
Spatial organizations of corresponding domains of different 5S rRNAs are usually rather similar and appear as more or less distorted helices. As to their arrangement, they are individual virtually independent structural units. Loop A is a special structural modulus that combines these 5S rRNAs into a united molecule. This loop plays a central role in the tertiary structure of 5S rRNA. As a three-way junction, loop A determines mutual locations of the molecular domains and acts as one of the major centers of tertiary interactions in 5S rRNAs. Loop A can be called the heart of the whole molecule. The nucleotide sequence in the loop is highly conservative. Mutations in this region usually lead to serious changes in the spatial structure of 5S rRNA and disorders in its interactions with protein factors.
On the basis of energy considerations, domains of apparently all 5S rRNAs display a tendency for producing a most elongated stacking system. This can be realized by several approaches, although there are only three fundamentally different types of geometry (Fig. 1c). All 5S rRNAs with an established three-dimensional structure within the large ribosomal subunit are characterized by geometry of the C family (when the β- and γ-domains are coaxial to each other), although theoretically such an organization of loop A can also have a conformation of the A family (coaxial α- and γ-domains).
Structures of other modules of the molecule are very complicated and strikingly diverse. Although 5S rRNA is relatively small, it is a “treasure-house” of very different non-canonical elements and motifs, which can be also detected in other RNAs but nowhere are so close to each other. Internal and terminal loops, bulges, continuous regions of non-canonical base pairing, and other structural features result in a unique relief of the 5S rRNA surface, where numerous widenings of the major groove expose multiple chemical groups capable of interactions with other macromolecules.
Similarly to any RNA within the cell, 5S rRNA is always bound with one or another protein factor. The protein component of the complex determines not only the stability of 5S rRNA but also the direction of its transfer inside the cell.
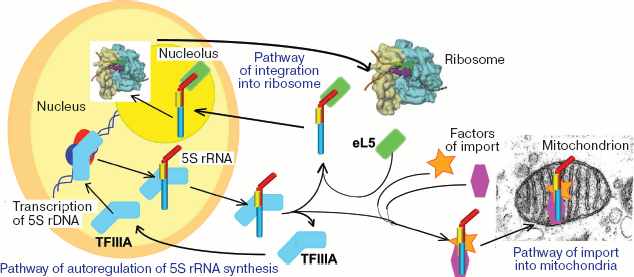
Among all known RNAs, 5S rRNA seems to have the most complicated transport pathways (Fig. 2, see color insert). The matter is that, as differentiated from genes of other rRNAs, transcription of the eukaryotic 5S rRNA genes is not associated with the nucleolus, and, thus, this component needs to be transported to the place of ribosomal subunit assembly. And the “delivery pathway” appears to be rather complicated [6-8]. First, newly synthesized 5S rRNA binds to transcription factor TFIIIA, which has earlier contributed to its generation (in addition to RNA polymerase III, transcription of the 5S rRNA genes requires the presence of three transcription factors: TFIIIA, TFIIIB, and TFIIIC). The resulting complex leaves the nucleus and comes into the cytosol where 5S rRNA can be accumulated for some time as the same complex (e.g. in oocytes of Xenopus laevis [9]) or within multicomponent RNPs, which include some proteins homologous to TFIIIA, some aminoacyl tRNA synthetases, and tRNAs [10]. But for penetrating into the nucleus, 5S rRNA has to bind with another partner, the ribosomal protein eL5. The eL5 protein, similarly to its prokaryotic homolog L18, is a major 5S rRNA-binding protein of the ribosome. Proteins of the eL5/L18 family are present in all living beings and are absolutely needed for 5S rRNA integration into the large subunit. In addition to L18, ribosomes of prokaryotes contain at least one 5S rRNA-binding protein, L5 (despite its name, it is not a relative of the eukaryotic L5). Finally, in many bacterial ribosomes 5S rRNA also interacts with the third protein factor, L25. Together 5S rRNA and ribosomal proteins associated with it form the main part of the central protuberance of the large subunit.
The functional role of 5S rRNA in the ribosome is still unclear. It is known that in the absence of 5S rRNA translation is impossible [11]. Nevertheless, numerous attempts to detect 5S rRNA in mitoribosomes of multicellular animals and fungi have been unsuccessful, and this makes doubtful its necessity and “indispensability” for biosynthesis of protein. At present, many authors agree that via multiple RNA-protein and RNA-RNA interactions, 5S rRNA works as a connecting link between functional centers of the ribosome and coordinates and harmonizes their activities [1, 12]. This viewpoint is supported by many theoretical and experimental data, but in general the functional role of 5S rRNA is still unclear.Fig. 2. Scheme of intracellular displacements of eukaryotic 5S rRNA.
SPECIAL STRUCTURAL FEATURES
Helix I
Helix I (9-10 bp in length) forms the α-domain and has a classic A-structure [13] that is usually rather stable due to high contents of G-C pairs (a high GC/AU ratio is specific for the whole 5S rRNA molecule and is never lower than 1). On one side of helix I there are 5′- and 3′-terminal regions of the molecule, which are often not paired on the ends or differ in the length (usually the 5′-end is located deeper). The other side of helix I is presented to loop A. In particular, helix I is specified by an increased (relatively to other modules of the molecule) number of G-U pairs, some of which are non-compensated (i.e. pyrimidine nucleotide is located in the 5′-direction from the guanine residue). Thus, in most cases the G7:U112 pair is non-compensated, whereas the G8:U111 pair is always compensated [14]. The presence of non-compensated G:U pairs results in local deformations of the helix because of ineffective stacking [15], and therefore this element of the secondary structure is potentially important for RNA-protein recognition. Note that water molecules are also involved in maintenance of the tertiary structure of this helix. They also contribute to stabilization of G-U pairs in the α-domain.
Helix I has rather considerable functional importance. Its specific secondary and tertiary structures are essential for stabilization of the molecule and its integration into the ribosome. Mutations that affect nucleotide pairing or interrupt stacking inside the helix decrease the rate of re-association of ribosomal subunits but do not influence translation quality [16].
Relatively recently the specific terminal structure of yeast 5S rRNA has been shown to be crucially significant for its stabilization, interaction with ribosomal protein eL5 and, as a result, for its integration into the large ribosomal subunit. 5S rRNA with properly formed 5′- and 3′-ends effectively interacts with eL5 protein and is highly resistant to the action of ribonucleases. But if the 5′-end of the molecule is even one nucleotide longer, it virtually loses its ability to interact with eL5 and can be easily cleaved by nucleases [17].
Intactness of helix I is also essential for the interaction of 5S rRNA with factor TFIIIA: deletions affecting its intact state virtually completely suppress binding [18]. This effect seems to be mediated, because helix I stability determines the structure of loop A, which is directly involved in the interaction with TFIIIA.
Helix II
Helix II is a part of domain β and in some traits differs from helix I. First, in all cases helix II is formed exclusively by canonical Watson-Crick pairs. Although the nucleotide sequence in this region is very variable, its secondary structure always remains unchanged. Second, helix II is specified by the presence of an unpaired nucleotide, but this does not disturb the general A-helical structure. The nucleotide type is not significant, although it is conservative within large taxons [5]. This nucleotide mainly promotes a local conformational shift which results in appearance of a binding site for protein factors (proteins of the eL5/L18 family, TFIIIA). Thus, on the binding of E. coli 5S rRNA binding with protein L18 this region (and also the 3′-side of loop B) acquires protection against cleavage by RNases. And bulged A-66 is supposed to be mainly responsible for the binding [19].
Studies on crystal structure of helix II from X. laevis 5S rRNA have shown that this structural element exists in two forms different only in the location of unpaired cytidine. In both cases, it is far beyond the helix limits, but in one form it is oriented towards loop A and just oppositely in the other form. By some parameters, this structure is unique. Cytidine displaced from the helix has a rare C3′-endo-trans-conformation. In one of the forms, the major groove is enlarged close to cytidine, and this promotes helix II binding with protein factors. Moreover, in the crystal this nucleotide is involved in a ternary interaction with nitrogen bases of adjacent molecules. It has been suggested that such interactions can occur on the in vivo binding with other RNAs, but this is still only a hypothesis [20].
B and C Loops
Single-stranded regions of 5S rRNA have an especially complicated organization. They are rather compact and well-structured elements with specific conformation and stability provided for by numerous non-canonical interactions, which especially characterize the A-, B-, and C-loops and correlate with the functional significance of these regions. Thus, loop B located between helices II and III not only fails to increase the inner mobility of domain β, but, in contrast, promotes an uninterrupted stacking of bases. In particular, this region is specified by a highly conservative pair C27:G58 inherent in the majority of prokaryotic molecules. Just this pair is responsible for integrity of the helical structure of the β-domain [2]. But although loop B plays a very important structural role, it seems to have no special function. It was mentioned above that this loop is a constituent of the 5S rRNA region protected by proteins of the eL5/L18-family but is not involved in formation of the binding site for these factors.
Terminal loop C plays a crucial role during interaction of 5S rRNA with ribosomal proteins of the eL5/L18-family and especially with bacterial protein L5. The spatial organization of loop C is very complicated. Some nucleotides seem to be involved in secondary and tertiary interactions, which maintain the structure of this loop. But analysis of mobility of guanine residues has shown that loop C conformation is half-open and permits a number of some bases to be available [21]. Together with a rigid “support” formed at the cost of secondary and tertiary contacts, loop C is an excellent site for specific binding of protein factors.
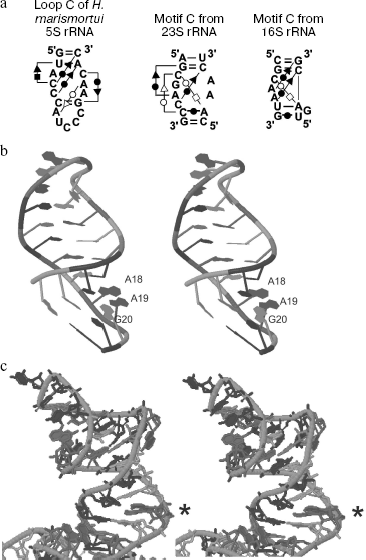
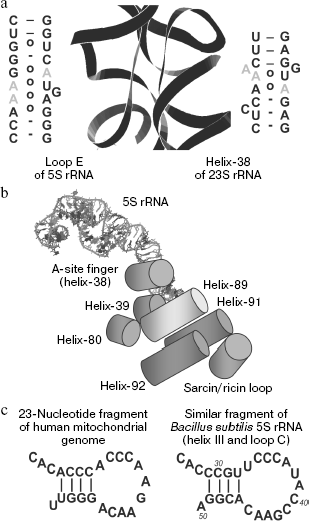
Analysis of isostericity matrices (an approach recently developed for comparison and identification of spatially similar RNA motifs) has shown that the C-loop of 5S rRNA belongs to so-called C-like motifs [22], which occur rather frequently in large ribosomal RNAs (Fig. 3a).
A region of the helix III-C-loop of 5S rRNA of higher vertebrata has an interesting structural feature. This region contains a palindromic duplex 5′-GAUCUC-3′/3′-CUCUAG-5′ which is shown by chemical cleavage and analysis of spatial structure of X. laevis 5S rRNA to be absolutely symmetrical with completely paired bases (including two unusual U:C pairs) [23]. The functional significance of this element is still unclear.Fig. 3. Structural features of loop C and helix III of 5S rRNA. a) C-Like motifs detected in ribosomal RNAs [22]. Pairing types are shown as follows: Hoogsteen pairs (squares); Watson-Crick pairs (circles); sugar-edge interactions (triangles). Open and closed symbols correspond, respectively, to cis- and trans-conformation of the nucleotide. Standard Watson-Crick pairs are shown by lines without additional symbols (G=C and A-U). b) A cross-eyed view of helix III of X. laevis 5S rRNA in solution [25] (Jena Library). Unpaired adenine residues are designated as A18 and A19. c) Helix III and loop C of H. marismortui 5S rRNA within the large ribosomal subunit [26] (Jena Library). The A-platform is indicated by an asterisk.
Helix III
Helix III seems to be one of the most important components of 5S rRNA in all living organisms. Both secondary and primary structures of this region are highly conservative. Thus, a recently detected mitochondrial 5S rRNA of Acanthamoeba castellanii possessing a very unusual nucleotide sequence and a weak resemblance with the secondary structure of 5S rRNA has been identified only due to the helix III and loop C which are strikingly similar to the corresponding elements of all known 5S rRNAs [24]. Such a strict conservation of this region of the molecule seems to be associated with the responsibility of just this element for 5S rRNA interactions with its most important partners, proteins of the eL5/L18-family, and, consequently, for 5S rRNA integration into the large ribosomal subunit. This interaction is especially important because only L18 (or its eukaryotic homolog eL5) of three ribosomal proteins capable of binding with 5S rRNA is present in all species.
What structural features make helix III such a good site for binding of ribosomal proteins? This may be caused by distortion of the A-conformation, which is typical for RNA helices by the presence of two (less frequently three) unpaired nucleotides producing a short loop directly in the middle of helix III (A50-A51 in Fig. 1b). This noticeable and very conservative feature suggests that this element should be important for maintenance of the specific structure of the helix and interaction with protein factors.
Native conformation of this bulge and helix III as a whole in solution was elucidated by NMR spectrometry [25]. Figure 3b shows asymmetric locations of unpaired adenines and the flanking guanines. Both adenine residues (A18 and A19 in the figure) are characterized by a predominant C2′-endo conformation of ribose. The majority of resulting structures display a specific stacking of two bulged bases with the guanine located right after them (G20). However, the bulged bases might possibly be arranged much less definitely, and then the stacking with the flanking pairs would be disturbed: their planes are nearly perpendicular to each other. Thus, this region might possess pronounced conformational mobility.
The C2′-endo form of adenine residues of ribose (instead of C3′-endo specific for RNA helices) leads to a local overwinding of the chain accompanied by translation of the whole pile of bases into the minor groove. This realignment opens the major groove and exposes A18 and the preceding guanine (G17). Thus, the whole element becomes quite favorable for binding with a protein factor.
It should be emphasized that all the above-described data refer to helix III structure in solution. Analysis of the structure of Haloarcula marismortui ribosomal 50S subunit [26] has shown a fundamentally different organization of the two-nucleotide bulge in the helix III region (Fig. 3c). Two adenines form the so-called A-platform with nearly coplanar bases. Obviously, integration of 5S rRNA into the ribosome is associated with a conformational rearrangement of this region. It has been shown on 5S rRNA of E. coli, X. laevis, and some archaea that binding with the eL5/L18-family proteins leads to reorganization of helix III. It seems also that in the case of H. marismortui production of A-platform can be induced by interaction with this protein factor.
Another unusual feature of helix III in various living organisms is the presence of post-transcriptional modifications. Such modifications have been found only in helix III and loop C of elements of the 5S rRNA secondary structure. Modifications of individual nucleotides are especially significant in the thermophilic archaeon Sulfolobus solfataricus and Pyrodictium occultum, which live at the temperature of 105°C. Stabilization of the 5S rRNA helices is achieved in two ways. First, helical structures virtually free of adenine and uracil residues are generated during adaptive evolution. In the second way, which is not characteristic of the majority of living organisms, adaptation is realized by a particular modification of nucleotide residues. Position 32 is occupied by 2′-O-methylcytidine instead of cytidine, and instead of cytidine-35 there is N4-acetyl-2′-O-methylcytidine. 2′-O-methylation stabilizes the C3′-endo conformation of ribose favoring maintenance of the A-shape specific for RNA helices. Being a good electron acceptor, the acetyl group increases acidity of the proton at N4, and this strengthens the hydrogen bond between the modified base and guanine. In the case of N4-acetyl-2′-O-methylcytidine, effects of the two modifications are additive [27]. Methylation of C-32 has also been found in the Achaean Sulfolobus acidocaldarius [28].
In addition to archaea, modification (pseudouridylation) in helix III is also specific for 5S rRNA of Saccharomyces cerevisiae (on residue 50) and Euglena gracilis [29]. Pseudouridine promotes more efficient stacking than uridine, and this ensures stabilization of the helical structure [30]. Obviously, in all cases modifications protect helix III and loop C against unfavorable structural rearrangements, and this seems to be associated with an especially important functional role of this region of the 5S rRNA molecule.
Helices IV and V and Loop D
The structure of the γ domain of 5S rRNA is slightly different in bacteria and archaea/eukaryotes, but in both cases it is virtually a single helix terminated by a loop. This domain plays a major role during interaction with the bacterial ribosomal protein L25 [31], eukaryotic TFIIIA, and 23S rRNA. It can be formally depicted as two helical regions (helices IV and V) separated by internal loop E and ended by terminal loop D. Such an unusual structure of loop E is considered in detail in the next section, and here we consider helices IV and V and loop D.
Helix IV of bacteria is a classical A-shaped duplex. The corresponding region of 5S rRNA has a more complicated structure. First, it always has an unpaired nucleotide on its 5′-side, but the type of this nucleotide is poorly conserved - it is often different even in systematically related species. Helix IV of higher eukaryotes is also specified by an unusual U:U juxtaposition with geometrical parameters similar to those of the wobble-pair G:U. In general, helix IV of eukaryotes is characterized by an increased content of non-canonical base pairs. The presence of many unusual structural elements in the same modulus has excited acute interest in both its structure and putative functional significance.
The helix IV and loop D conformations remained unclear for a long time. The majority of experimental data fit a traditional scheme where helix IV has a specific one nucleotide bulge and is terminated by a classical GNRA-type D-loop. However, the pattern of cleavage of some plant 5S rRNAs in this region suggests the existence of an enlarged loop D which can appear due to destabilization of helical structure caused by an unpaired base [32]. Approaches of molecular dynamics have elucidated this problem in the case of the helix IV-loop D region from 5S rRNA of lupine [33]. In this case, the D-loop has a classical GNRA structure, but it also can exist in two slightly different conformations. One of them is supported by water molecule, which mediates the bond between A and G, whereas in the other conformation the bases are in direct contact.
Helix IV has pronounced conformational mobility. The unpaired nucleotide can have different, energetically non-equivalent conformations. Its movement towards the major groove results in disruption of the adjacent base pairs and, therefore, is less advantageous than the movement towards the minor groove. In the last case a very favorable triple interaction with the foregoing nucleotide pair is generated, but it is kinetically difficult to obtain such a state [34].
The functional significance of helix IV and loop D is immense because just this region of the molecule is responsible for the interaction of 5S rRNA with 23S rRNA and is involved in the integration of the large subunit RNA component. Relatively recently, 5S rRNA was found to have a quite new feature: it displayed activity of a leadzyme, i.e. a ribozyme with Pb2+-dependent ribonuclease activity. These enzymatic properties are also determined by helix IV and loop D. The sequence of this region of the molecule is similar to that of the tRNAPhe-based leadzyme active site, but, as discriminated from the latter, 5S-leadzyme cuts RNA in trans, preferring dinucleotides CG. This specific feature of the mammalian 5S rRNA molecule is supposed to contribute to the in vivo toxicity of lead. Even low concentrations of Pb2+ can induce inactivation of various cellular RNAs at the cost of the apparently most abundant in the cell enzymatically active macromolecule [35].
By contrast, there are only few data on the helix. In the majority of cases, this helix has no loopings. Its size varies from 7-8 bp in eukaryotes/archaea to two pairs in bacteria. Only an absolutely conserved G:U pair on the boundary between the helix and loop A can be mentioned as a specific feature [14].
Loop E
The internal loop E which links helices IV and V seems to be the most interesting and unusual structural element of 5S rRNA. Its structure is fundamentally different in bacteria (and also in mitochondria and plastids) and in eukaryotes/archaea. Thus, according to terminology of Lescoute-Westhof [22], loop E exemplifies a corresponding (i.e. located equally) but not equivalent (i.e. equally organized spatially) motif. Taking into account that E loop-like structures also occur in many other RNAs, as will be further spoken about, this motif can also be assigned to recurrent ones. Differences in the loop E structure can be partially explained by different functional load of this element in representatives of different superkingdoms. Loop E in bacteria forms a site for binding with ribosomal protein L25, whereas in eukaryotes it interacts with quite another partner, TFIIIA (and with some of its protein homologs involved in generation of RNA-depositing 42S particles in oocytes of X. laevis). RNA-binding domains of these proteins are organized otherwise, and this somewhat determines the structure of their binding sites. In this connection, it is reasonable to consider separately the structure of E loops of both types.
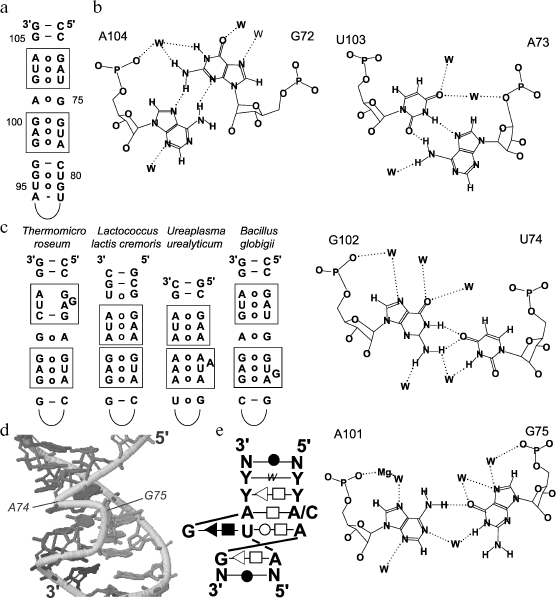
Bacterial type loop E. The consensus sequence of loop E and the context of its positioning in 5S rRNA of bacteria are shown in Fig. 4a. This region is a helix formed by continuous series of non-canonical pairs with two pronounced palindromic submotifs, which play a paramount role in the loop E interactions with protein L25 and 23S rRNA. Good order and a closed conformation of this element were revealed in studies on 5S rRNA cleavage by RNases specific for single-stranded regions. Loop E was shown to be fully insensitive to this treatment [27]. This conclusion was later confirmed by X-ray crystallographic analysis [36]. Such an unusual structure is maintained by numerous non-canonical interactions (Fig. 4b). These interactions are presented in detail in work [37] in which the authors compared biochemical and phylogenetic data with the crystal structure of loop E.
There is no distinct boundary between helix V and loop E. On the assumption that the helix V begins with the appearance of canonical nucleotide pairs, it is only two pairs in length. These two pairs are nearly always typical Watson-Crick G:C pairs responsible for the usual function of flanking elements: maintenance of terminal regions of a loop or of another non-A-helical structure. Pairs adjacent to helix IV play a similar role.Fig. 4. Structural features of the 5S rRNA γ-domain. a) Secondary structure of the γ-domain from E. coli 5S rRNA. Submotifs of loop E are in frames. Loop D is not shown. b) Nucleotide pairs that form the first submotif of loop E in E. coli 5S rRNA and the A101:G75 pair separating the submotifs [37]. W, water molecules. c) Variants of non-canonical organization of loop E in some bacteria [37]. d) Three-dimensional structure of loop E in 5S rRNA from X. laevis [58]. e) Consensus secondary structure of the motif of the eukaryotic type loop E [42]. Designations are the same as in Fig. 3a. W, water-mediated interaction.
Loop E can be subdivided onto three structurally independent regions. Two of them have very similar nucleotide sequences, which are parts of an imperfect palindrome. They are called submotifs, and in their limits the adjacent nucleotide pairs form a unified stacking system with virtually the same geometry of the pairs within both elements. These submotifs are characterized by the presence of so-called A-stacks in which stacking interactions link conservative adenines of the first two pairs from different chains. The third part of the loop is represented by only an A:G pair, which separates the submotifs. This unusual pair is supported by hydrogen bonds between AN6 and GO6 and between AN1 and GN1 through a water molecule that is rather often observed in such interactions (Fig. 4b).
In bacterial 5S rRNAs, individual nucleotides involved in formation of submotifs are substituted. The substitutions are strictly determined and in all cases satisfy the principle of isostericity: the element conformation and, correspondingly, the type of pairing are unified in every part of it. Only rarely loop E acquires some atypical features because of insertions or deletions distorting the structure of submotifs (Fig. 4c).
The conformation of loop E is very substantially supported by Mg2+ and Na+ [38]. Studies by molecular dynamics approaches have shown that on introducing Mg2+ (natural ligands) into a system that contains loop E from E. coli, its conformation becomes stabilized and more closed, and the major groove narrows slightly. On substitution of magnesium ions by sodium ions, the major groove widens slightly (to 4 Å), its width constantly changes, and a loose, “breathing” conformation arises.
The role of cations in the maintenance of the loop E structure is most clearly demonstrated by studies on its behavior in an ion-free medium. Even during the first nanosecond, the major groove extends up to 15 Å, and 2 nsec later its width becomes 20-22 Å (a fully denatured state) and remains unchanged. Base pairing continues up to the very end, and the pairs very slowly acquire more and more open conformation that suggests a pronounced plasticity of this structural element. Addition into the system of Na+ and especially Mg2+ leads to a rapid restoration of the loop geometry.
Structures similar to the bacterial type loop E and corresponding, at least, to one submotif, can also be detected in other RNAs. 16S (18S) rRNA is especially rich in them. Some analogs of both submotifs, including their exact copies, have been found in representatives of all systematic groups. In all cases the most widespread E-like motif found in 59.8% of known 16S rRNA sequences in the 581-583 and 758-760 positions is flanked from one side by a Watson-Crick pair and from the other side by a G:U pair, i.e. is very similar to the loop E from bacterial 5S rRNA. The bacterial E-loop-like structure is also present in domain IV of 7SL RNA from SRP-particle [39, 40].
Eukaryotic type loop E. The loop E of eukaryotic type is organized even more unusually. In addition to the location, it has in common with the bacterial analog a closed conformation roughly similar to helix A and exclusively non-canonical character of constituent nucleotide pairs. However, it is slightly shorter (4 bp), always has one unpaired guanine (i.e. it is an asymmetric loop), and has no repeated submotifs [41]. In fact, it seems to be similar to submotif 2 from loop E of 5S rRNA of Ureaplasma urealyticum and Bacillus globigii (Fig. 4c), but this similarity is rather relative.
The unusual conformation of loop E became obvious because this region could not be cleaved by agents specific for both single- and double-stranded RNA regions. Specific cleavage was obtained only with rhodium(III) bis(phenanthroline)(phenanthrenequinonediimine), which recognizes non-classic secondary structures and sites of tertiary interactions [23]. Considering specific features of interaction between this and RNA, loop E was supposed to have a markedly more open conformation than helix A, with its maintenance mainly conditioned by stacking between adjacent nucleotide pairs.
The geometry of chains constituting loop E is very different (Fig. 4d). One of the chains (the shorter one) corresponds as a whole to A-shaped RNA. The conformation of the other chain is rather peculiar due to presence of a “superfluous” G75. U73 and its neighbor A74 are linked by unusual bonds between ribose residues and between ribose and the base that results in A74 “reversion” and parallelism of both chains in this region. Such geometrical environment constrains the bulged G75 location in the major groove where it is involved in a triple interaction with the adjacent nucleotides.
Because of its most specific feature, the loop E motif was also called "bulged G-motif” or “S-motif”. In fact, this term comprises a whole set of structural elements with a highly conservative sequence in common and identical conformation [42]. Interactions that maintain the G-motif and its consensus sequence are shown in Fig. 4e. Both the bacterial loop E and this motif are characterized by abundance of various non-canonical pairs with nucleotide conformations typical not only for Watson-Crick pairing but also for Hoogsteen's1 linking and for sugar-edge or water-mediated interactions. Especially interesting is a uridine residue that pairs not only with adenosine of the opposite chain, but also with the bulged guanosine (corresponding to G75 of E the loop), which is its immediate predecessor. Such type of the binding combined with a continuous stacking leads to appearance of a specific S-turn with two changes of the chain direction.
1 Nitrogen base interacts by the Hoogsteen type if hydrogen bonds are produced with involvement of atoms at N6 (O6) and N7 of purines and N4 (O4) and C5 of pyrimidines. (Compare with Watson-Crick pairing where hydrogen bonds are generated by atoms at N1, N2 (in guanine), and N6 (O6) of purines and O2 (in cytosine), N3, and N4 (O4) of pyrimidines.) A pile of bases slightly “loose” because of the presence of three aromatic systems in the same plane is favorable for stacking with corresponding elements of other macromolecules. An unusual overall conformation with a significantly more open major groove than within the A-helix where non-canonical base pairs present a variety of chemical groups, suggests that loop E is an excellent site for interaction with very different partners. Among such partners, TFIIIA should be mentioned (see below). On this binding, the stacking with the loop E “triple platform” plays a significant role. Binding of 23S rRNA, which is also specific for loop E of eukaryotic/archaea type, is another important interaction.
In addition to loop E of eukaryotes, a sarcin/ricin (S/R) loop from the so-called “GTPase site” of 23S rRNA seems to exemplify the best-studied S-motif which is known as a site for attacks not only of sarcin and ricin which determine its name, but also of some other enzymes (e.g. some colicines). It is difficult to overestimate the functional importance of this element. The surface formed by the GAGA-tetraloop and the minor groove of the S/R loop interacts with translation factors EF-G and EF-Tu, which are absolutely necessary for functioning of the protein biosynthesis apparatus in all living organisms. Thus, this is another example of the bulged G-motif involvement in the structurally specific RNA-protein interaction [36].
The role of S-motif in RNAs of many plant viroids is rather peculiar. An element with the structure similar to that of the eukaryotic E-loop has been detected in RNA of potato spindle tuber viroid (PSTV) due to similar UV cross-linking patterns [43]. The cross-link location corresponded to the so-called “central conservative region” found in many plant viroids, and, thus, this structural element was supposed to play an important role in the life cycle of viroids. It was emphasized that this loop was located close to the site associated with autocatalytic processing of PSTV dimeric transcripts and supposed that it should be involved in formation of this site. The hypothesis was brilliantly confirmed in the work of Baumstark et al. [44]. On the basis of this study, a model of processing of pregenomic transcripts was proposed which included large-scale conformational rearrangements of the central conservative region. The original transcript with more than one genome in length contains three hairpins, two of which are terminated with fully conservative GNRA-tetraloops. The first site of cleavage appears in RNA in this conformation. Upon the first stage of processing, the 5′-fragment is released and a newly produced 5′-end is settled with formation of the E-loop of eukaryotic type (but with a bulged cytosine). This element is recognized as a signal for the second cleavage, which occurs in the 3′-terminal region of the molecule. This two-step processing results in ends that are exactly opposite to one another and immediately ligated with production of covalently closed genomic RNA of the viroid.
Thus, the unusual conformation of loop E promotes its activity as a specific element in very different processes: from binding of different macromolecules responsible for functioning of the apparatus of protein biosynthesis to processing of pregenomic RNA (the inevitable stage in the life cycle of the whole viroid family).
Conformational Mobility and Reorganization of 5S rRNA Structure
Studies of 5S rRNA structure under different conditions by physical, chemical, and enzymatic approaches led to the idea that this molecule should have alternative structures. Conformational changes in 5S rRNA were shown for various organisms on changes in temperature, polarity, and pH of the medium, and in some cases several alternative native tertiary structures could co-exist in solution. At present, it is difficult to predict such forms by modeling in silico because they are generated with involvement of non-canonical interactions. Therefore, physicochemical measurements and enzymatic or chemical hydrolysis are the only sources of our knowledge about 5S rRNA in solution, and they allow us to make conclusions concerning the structure of 5S rRNA and distribution of its conformers.
At first, it is reasonable to consider some reports resulting in a concept about conformational non-uniformity of 5S rRNA. Weidner and Crothers examined renaturation of E. coli 5S rRNA under different conditions and demonstrated several refolding pathways of this molecule. Thus, a rapid cooling from high temperature of this RNA solution produced approximately equal amounts of two conformers. But an initial slow cooling of the solution to 50-60°C followed by a rapid cooling resulted only in the “native” product. Note that stability of the structural nucleus of the refolding intermediates strongly depended on the Mg2+ concentration in the solution [45].
Kime and Moore analyzed NMR spectra of non-interchangeable protons of E. coli 5S rRNA and found that two native conformers of the molecule (A-form) existed that could be easily distinguished from the B-form [46]. Using chemical and enzymatic cleavage and modification of available nucleotides, Goringer et al. concluded that two stable conformers of E. coli 5S rRNA were different in secondary structure [47]. The A-form transition into the B-form was associated with disturbance of the helix II structure and, to a lesser degree, of structure of helices III and IV. Nevertheless, the data suggested that new structured regions could be produced upon transition into the B-form.
Kao and Crothers analyzed the pH dependence of early melting of E. coli 5S rRNA and also concluded that two native conformations should exist, with different compactness, diffusion constant, and some other physical parameters. The transition between the forms was induced by changes in ionic conditions and especially in pH (in particular, a basic catalyst presented by ribosomal protein could act as a switch, and this allowed the authors to suggest a possible functional significance of this conformational transition [48]).
5S rRNA from rat liver was also shown to have three conformations [49]. However, in this case, two additional structures resulted upon treatment with urea and EDTA, respectively. Native and urea-treated 5S rRNAs co-migrated in non-denaturing polyacrylamide gel, but were cleaved differently by nucleases. The differences were most pronounced in the helix III-loop C region and in helix V and loop D. As in the previous case, the biological significance of 5S rRNA conformational transitions is still unclear.
Another conformational transition was described for 5S rRNA of yeast. Similarly to E. coli 5S rRNA, its refolding depended on Mg2+. Binding of Mg2+ occurred sequentially and cooperatively, and every stage was accompanied by a relatively slow monomolecular conformational rearrangement that finally resulted in increase in the RNA ordering. Calculations of activation parameters of each stage of refolding indicated that these changes included disconnecting of some nucleotide pairs and certain transformations of the tertiary structure [50].
It should be noted that all above-described conformational rearrangements can be observed under conditions that are far from natural ones. In fact, pronounced overalls of ionic strength and pH in the cell are unlikely. They can occur only under conditions of a corresponding microenvironment, which, in particular, can be realized by protein interactions with 5S rRNA (the ribosome is an excellent example of such a system). Searching for alternative conformations performed with the paRNAss program (http://bibiserv.techfak.unibielefeld.de/parnass/) [51] has shown that, under conditions of constant temperature and ionic strength, 5S rRNAs of human and yeast S. cerevisiae are unable to undergo large-scale rearrangements. Nevertheless, these molecules are characterized by a considerable instability of individual modules: loops A and E, helices IV and V (in yeast 5S rRNA) and loop C (in human 5S rRNA). Transitions between different shapes are rather easy, and energy barriers are relatively low. Thus, they are not alternative conformations of 5S rRNA but only transient settings, which constantly transit into one another without a significant activation energy.
Thus, the structural organization of the 5S rRNA molecule has pronounced abilities for local rearrangements under stable ionic and temperature conditions, but a global reorganization of the tertiary structure of free 5S rRNA seems doubtful. Nevertheless, large-scale rearrangements in the spatial structure of individual elements and the whole molecule can be induced by binding with certain protein factors. In many cases these conformational transitions are very important functionally. Interactions of 5S rRNA with various proteins and also with 23S rRNA will be discussed in the next section.
INTERACTIONS WITH BIOLOGICAL MACROMOLECULES
Interaction with Transcription Factor IIIA
A great role of factor IIIA in the “fate” of 5S rRNA has been mentioned earlier. In addition to a direct involvement in transcription of 5S rRNA genes, TFIIIA acts as a factor providing for the export of 5S rRNA from the nucleus and its stabilization in the cytosol until its binding with ribosomal protein eL5. Thus, factor IIIA is of interest at least because it binds not only with the gene but also with its product. TFIIIA is found in all eukaryotic cells and seems to play a similar role everywhere. Note that it can specifically bind only eukaryotic 5S rRNA, and it can indicate the presence of common determinants of this RNA-protein interaction in all representatives of the superkingdom [52] and the absence of such determinants in bacterial 5S rRNAs.
Sites for 5S rRNA binding with TFIIIA were searched for by various research groups for the last two decades. In the earliest works, the general conformation of rRNA molecule was shown to play an important role in the interaction. Thus, native 5S rRNA rather rapidly produced complexes with TFIIIA, whereas denatured 5S rRNA bound only in the presence of a certain concentration of Mg2+, which were mainly responsible for the three-dimensional organization of 5S rRNA. Involvement of helices I, IV, and V in the interaction was shown [18], and, moreover, TFIIIA protected loops D and E and parts of helices I and II against cleavage by nucleases. Such were the first conclusions on locations of the site for binding with TFIIIA [53].
Later, these sites were described more in detail in work performed on 5S rRNA of X. laevis [54]. An unusual conformation of loop E plays a major role in the binding. Nucleotides G75, U76, and A100 involved in a triple interaction are the most important contributors to formation of the binding site. Point mutations in this module result in a considerable growth of the dissociation constant of the 5S rRNA complex with TFIIIA. Note that upon removal of U76 (a “missing nucleoside” approach) a decrease in the TFIIIA affinity for 5S rRNA is also the most pronounced [55].
Helix V is also involved in the interaction, but its role is less demonstrative. It seems that this structural element has only to preserve the A-helical conformation, whereas the sequence is less important. Thus, the G40C transversion resulting in C:C opposition increases threefold the dissociation constant of the complex. But the G70A transition associated with appearance of a non-canonical (although a Watson-Crick) pair A:C+ has no effect on the TFIIIA binding because parameters of this pair fit the limits permitted by the structure of A-form helix.
The above-mentioned important role of the common three-dimensional structure of 5S rRNA in the interaction with TFIIIA has inevitably attracted researchers' attention to loop A as a region directly responsible for mutual locations of the three domains of the molecule. The U109C substitution resulting in appearance of a Watson-Crick pair instead of the absolutely conservative G:U decreases mobility of this “hinge” and concurrently increases twofold the dissociation constant. A large series of mutations obtained by another group of researchers [56] has also revealed a correlation between loop A mobility and 5S rRNA affinity for TFIIIA. Mutation analysis [57] has shown that three of four nucleotides of the loop A are absolutely necessary for recognition by TFIIIA.
Data concerning the β-domain are fundamentally different. Point mutations in this region of the molecule have little influence on the binding with TFIIIA. The most significant role is played by helix II, in which maintenance of Watson-Crick interactions and unpaired cytosine has the greatest importance, although the authors estimate the contribution of this structural element with much attention to other works that have been mentioned above.
By contrast to the earlier studies, helix IV and loop D were shown to be insignificant for interaction with TFIIIA. The majority of mutations in this region of the molecule do not affect the stability of the complex. Only the C79G substitution resulting in appearance of an unfavorable opposition of G:G close to loop E increases KD threefold, which can be easily understood considering the role of loop E in the binding.
Relatively recently a three-dimensional structure has been obtained for the complex of the TFIIIA fragment which includes zinc fingers 4-6 (in total, TFIIIA contains nine zinc fingers of the CCHH-type) with the 5S rRNA fragment containing all elements required for the interaction (Fig. 5a). Obtaining of this structure completely confirmed all conclusions based on biochemical data. Three zinc fingers interact, respectively, with loop E, the major groove of helix V, and loop A [58]. Such a multisite binding provides for a specific function of TFIIIA, namely, the protection of 5S rRNA against degradation. However, it does not prevent the recognition of 5S rRNA by protein eL5, since the site for its binding is located in helix III and loop C that is required for its integration into the ribosome.
Interactions with Ribosomal Proteins of the eL5/L18-FamilyFig. 5. Interactions of 5S rRNA with protein factors. a) A cross-eyed view of the complex of 5S rRNA fragment with a X. laevis TFIIIA fragment which includes three zinc fingers [58] (Jena Library). The γ-domain is placed vertically, and the α-domain is above to the left. b) The 5S rRNA location in the large ribosomal subunit of H. marismortui [26] (Jena Library). The region occupied by protein L1 is indicated respectively. c) The 5S rRNA complex with ribosomal protein L18 from the large ribosomal subunit of H. marismortui [26] (Jena Library). d) A cross-eyed view of the complex of E. coli 5S rRNA fragment (which includes helix III and loop C) with protein L5 from Thermus thermophilus [76] (RCSB). e) A cross-eyed view of the complex of the γ-domain of E. coli 5S rRNA with protein L25 [80] (Jena Library). An additional helix α1 produced as a result of the interaction is indicated.
No doubt the 5S rRNA structure is interesting as it is. But it seems unreasonable to speak about ribosomal RNA turning off its natural environment presented by the ribosome, at least, from the viewpoint of its supposed functions. In the present and following sections, the structure of 5S rRNA within complex with ribosomal macromolecules will be discussed.
Figure 5b shows the location and spatial structure of 5S rRNA within the large ribosomal subunit, and just this component of RNA seems to play a main role in generation of the central protuberance. Ribosomal RNA is structurally a ribosomal “skeleton” for disposition of ribosomal proteins. 23S rRNA forms nearly all structures specific for the large subunit. Only the side protuberance produced by protein L1 and the central protuberance with 5S rRNA as a structural basis are exceptions. Similarly to large rRNAs, 5S rRNA is also “decorated” by proteins, which are different in number and type in representatives of different systematic groups. These proteins and also 23S rRNA are responsible for integration of the whole central protuberance into the large ribosomal subunit.
In this section, ribosomal proteins will be considered which are always found in complexes with 5S rRNA, independently of the systematic position of the living organism. In other words, the eL5/L18-family proteins are universal and utterly conservative ligands of 5S rRNA.
Although this family proteins in bacteria and eukaryotes (L18 and eL5, respectively) are significantly different in amino acid sequence and size, their pronounced homology allows the researchers to expect a similarity of tertiary structures and modes of interaction with 5S rRNA. 5S rRNA of archaea is bound with ribosomal protein L18 (Fig. 5c), which is more like eukaryotic protein eL5 than bacterial protein L18, and this is in agreement with similarity of 5S rRNA structure in representatives of just these two taxons [59].
The majority of studies concern the interactions of 5S rRNA with proteins of eukaryotes and archaea; therefore, binding of bacterial L18 will be considered only in brief. In the previous sections, we have already mentioned the main structural modules of 5S rRNA involved in this interaction. The majority of these modules are concentrated in the β-domain (helices II and III, loop C; see Fig. 5c) [19, 60, 61], although it is known that the protein prevents cleavage virtually of the whole RNA molecule [31]. Such an “enveloping” of 5S rRNA inevitably influences its general conformation. In fact, circular dichroism spectra and temperature dependence of UV absorption suggest the presence of considerable rearrangements in the secondary and, possibly, tertiary structure of 5S rRNA of E. coli on complexing with the ribosomal protein L18. Other ribosomal proteins that bind bacterial 5S rRNA (L5 and L25) not only fail to stabilize the higher levels of 5S rRNA organization but in the last case even decrease the ordering degree of the secondary structure. Thus, protein L18 seems to be mainly responsible for compactization and stabilization of 5S rRNA in the prokaryotic ribosome [62].
A 5S rRNA complex with ribosomal proteins isolated from a halophilic archaeon Halobacterium cutirubrum is very different in characteristics from a similar complex found in E. coli. Circular dichroism spectra suggest an increased stacking and slightly weakened interactions between base pairs within the bound RNA. The tertiary structure of 5S rRNA inside the complex is slightly more open. The secondary structure of the bound 5S rRNA was analyzed using different ribonucleases [63]. Based on the data, it was supposed that on binding with L18 the RNA should undergo considerable conformational changes resulting in an alternative packing with a completely rearranged γ-domain. The presence of a similar secondary structure was first proposed in the work [64] for free 5S rRNA, but only this case seems be its realization.
We think that such conclusions must be considered very carefully, in particular, having in mind that the crystal structure of 5S rRNA in the large ribosomal subunit of another archaeon H. marismortui completely corresponds to the classic Y-shaped organization (Fig. 1a). However, it has been established that at least the main sites of interaction with protein L18 undergo significant rearrangements (Fig. 3, b and c). The type of interaction between nucleotides changes in the region of loop C and the unpaired dinucleotide AA of the helix III is exposed outside producing A-platform, and another out-of-helix adenine together with adjacent nucleotides of helix II enters tertiary interactions of loop A constituents [26].
Interaction of 5S rRNA with the eukaryotic ribosomal protein eL5 is characterized in detail. In 1971, isolation of a 5S rRNA complex with a mammalian ribosomal protein (from rat liver and rabbit reticulocytes) was reported. 5S rRNA was shown to be released from the ribosome on removal of Mg2+ from the medium or on increase in the concentration of monovalent cations. Results of sedimentation and electrophoresis indicated that 5S rRNA was released not as a free substance, but as a stable complex with a ribosomal protein with molecular weight of 35 ± 2 kD [65].
Dissociation of 60S subunits from rat liver in solution of cesium sulfate produced three complexes, one of which with density of 1.40 g/ml corresponded to a ribonucleoprotein. This ribonucleoprotein contained 5S rRNA and protein eL5 in equimolar amounts [66]. These two works were a start for studies of 5S rRNA-protein interactions in ribosomes of eukaryotes.
By enzymatic analysis, protein eL5 was shown to protect virtually the whole 5S rRNA molecule against cleavage by nucleases, similarly to the case of TFIIIA [67]. The importance of this effect is obvious on taking into account the role of eL5 in the “life” of 5S rRNA in the cell.
The 5S rRNA complex with protein eL5 of X. laevis is characterized in detail [68]. This complex is stable under pronounced changes in ionic strength, and this suggests a significant role of non-electrostatic interactions. Under optimized conditions, the dissociation constant of the complex is 2 nM. Taking into consideration data on protection of 5S rRNA sites by the protein, the authors studied interaction of 5S rRNA mutants with point substitutions in the region between nucleotides 42 and 109 in order to detect a binding site for protein eL5. Helix III and loop C occurred to be the most important structural elements responsible for this interaction. Mutations in the region connecting these two elements led to more than an order of magnitude increase in the dissociation constant of the complex. Substitutions of conservative unpaired adenines in helix III were catastrophic for the binding. Other mutations affecting loop B, helix II (except the bulged cytosine), helices IV and V, and loop E did not cause a noticeable effect.
The yeast 5S rRNA complex with ribosomal protein eL5 manifests some features reminiscent of those of the above-described complex from H. cutirubrum. Conformations of free and bound 5S rRNA are different. Experiments with tritium exchange, incorporation of ethidium bromide, and thermal denaturation of 5S rRNA in both states have shown that dissociation of the complex is associated with changes in the general conformation of RNA. In the complex, the number of exposed double-stranded regions is decreased, and the denaturation kinetics suggests that the secondary structure of the bound RNA is reorganized. It was proposed that eL5 stabilizes a definite functional conformation of 5S rRNA that is unstable in solution [69]. These data complement the above presented reports about rearrangement of a main site for interaction with proteins of the eL5/L18-family--a dinucleotide bulge in helix III (see above) found in 5S rRNA of H. marismortui. These changes are certainly local. However, taking into account the large surface of the eL5 binding, it is reasonable to also suppose other conformational displacements, with a summarized effect similar to that observed by the authors of the work [69].
This behavior of 5S rRNA on binding with the protein eL5 was explained only recently. First, this complex is specified by its co-translational generation [70]. For a long time attempts to prepare in vitro such a complex from individual components were unsuccessful. 5S rRNA was shown to bind only with the growing chain of eL5 during translation of mRNA of the latter. The first 50 amino acids of the protein are absolutely necessary for the interaction and seem to produce an RNA-binding domain (but what part of eL5 really binds 5S rRNA is still unknown). Obviously, such an interaction between the complex components promotes a strict regulation of the 5S rRNA and eL5 ratio in the future ribosomal subunit; it also explains why the pathway of 5S rRNA movement from its gene to the nucleolus is so complex. Second, the interaction between 5S rRNA and protein eL5 is unusual because it is realized via the so-called mutual induced fit binding mechanism [71, 72]. In a free state, ribosomal protein eL5 is a rather poorly structured macromolecule. However, on binding with 5S rRNA its conformation is rearranged and stabilized. Influence of the partners is mutual, and 5S rRNA is also reorganized. Thus, both the protein and RNA act as partially unstructured chaperones responsible for correct refolding of each other with production of a functionally active conformation [73].
Unfortunately, no crystalline complex of 5S rRNA with protein eL5 has been obtained up to now. This makes difficult further studies of their interaction. Structures of whole ribosomal subunits are now known (but only of prokaryotic ones), which allow us to see not only relative locations of the components but also interacting residues of amino acids and nucleotides. However, one should remember that within the ribosome this complex is influenced by adjacent molecules and its microenvironment is fundamentally different. In particular, an approach using iodine-induced cleavage of phosphothioate derivatives of 5S rRNA has shown that integration of 5S rRNA-protein complex into the ribosome is associated with further rearrangements of RNA and changes in distribution of the protected regions [74].
Interaction with Ribosomal Protein L5 of Prokaryotes
L5 is another ribosomal protein that interacts with 5S rRNA, but its role is markedly less important that that of L18. L5 is characteristic only for bacteria and archaea (the only known exception among eukaryotes is the yeast S. cerevisiae) and is not reminiscent of eukaryotic eL5 in structure, features, and binding mode. Unfortunately, such confusion in nomenclature often creates complications, and it is necessary to define more accurately what protein factor is under special consideration.
For a long time a very low constant of L5 binding with 5S rRNA (2.3·106 M-1) prevented determination of sites of interaction with this protein. Note that L5 can bind 5S rRNA with high affinity only if the latter is already complexed with L18 (and the binding constant is about an order of magnitude increased) [75]. Thus, the site for binding with L5 is supposed to be not preformed but arise only upon the interaction with L18, or a protein-protein interaction with L18 is required for the L5 binding with 5S rRNA.
Using protection against ribonucleases, a cleavage-resistant fragment of E. coli 5S rRNA was detected [61] that specifically bound with Thermus thermophilus protein L5 (this protein binds similarly with 5S rRNAs from these two organisms, which are very alike in the primary structure). Note that limited cleavage by RNase A of isolated 5S rRNA and in the presence of L5 gave the same pictures. But if the ternary complex of 5S rRNA with L18 and L5 was under study, the protected region was different from that resulting on treatment by RNase of the 5S rRNA complex only with L18. This region was eight nucleotides larger and included loop C, which was believed by the authors to be the binding site for L5. Thus, L18 is directly involved in L5 binding with 5S rRNA. Interaction between these two proteins was also shown by the analysis of crystal structure of the large subunit of the H. marismortui ribosome [26].
In the next work [76], the authors carefully analyzed crystals of the L5 complex with a fragment of 5S rRNA, which included loop C and helix III (Fig. 5d). As compared to the free 5S rRNA, the C-loop structure in the bound fragment was markedly changed: many new bonds appeared, and not only in the loop itself but also between the loop and helix nucleotides. A pronounced role in building of a specific conformation of this region belongs to two loci of the three-way junction, one of which terminates helix III and the other is formed by nucleotides of the loop just above it. They are in the base of a “pyramid” with the apex formed by other residues of loop C. Loop C needs to be just of such geometry for binding with L5, and this structure seems to be specific for all bacteria and archaea: the primary structure of this region is extremely conservative.
The protein binding site also consists of very conservative residues of β2 and β3 strands and flanking loops, and, similarly to the loop C pyramid, it is supported by a complicated network of intramolecular interactions. Nonpolar groups of RNA and protein form a common hydrophobic nucleus of interaction, but, hydrogen bonds undoubtedly also play an important role in binding of partners. As differentiated from the case of L18, the RNA-binding domain of L5 is characterized in detail (in particular, in [77]). A concave surface of interaction, mobile loops involved in firmer binding, distribution of positively charged residues, and, finally, a specific sequence indicate that this domain belongs to the RRM-family (RNA Recognition Motif), although it is not its typical representative.
In conclusion, it should be noted that, in addition to 5S rRNA, L5 also interacts with 23S rRNA, as shown by analysis of crystal structure of the H. marismortui large subunit [26]. Thus, this protein is involved in 5S rRNA integration as a connecting link between two rRNAs that form the large subunit skeleton.
Interaction with Bacterial Ribosomal Protein L25
Protein L25 is the last one considered in the present review. This protein is specific only for bacteria and, thus, is the third factor that can bind 5S rRNA from bacterial ribosomes. Under different names, its homologs have been described for various species. They include TL5 from Th. thermophilus (again here is a nomenclature-associated confusion: it is individual for ribosomes of every species; at present, all homologous ribosomal proteins of bacteria are named in accordance with the nomenclature of E. coli, i.e. in this case it is L25 of Th. thermophilus), the common stress protein CTC of Bacillus subtilis and Aquifex aeolicus [78]. The community of binding sites, sequence homology, and conservation of RNA-binding domains (a 6-stranded β-barrel and two/three α-helices) indicate that these proteins are members of a separate family, and their tertiary structure is reminiscent of an anticodon-binding domain of glutaminyl-tRNA synthetases [79].
As in the case of the prokaryotic ribosomal protein L5, the binding site of L25 is quite local and limited by loop E and the proximal part of helix IV. Similarly to L5, protein L25 protects against cleavage by nucleases a rather limited part of the molecule (nucleotides 72-86 and 94-109) that is conditioned by compactness of this factor [60]. Crystals of the L25 complex with a 5S rRNA fragment, containing the site for L25 binding, were obtained, and this interaction was characterized in detail (Fig. 5e) [80]. Conformation of loop E within the complex is quite the same as in the free 5S rRNA (see above): there are a complete set of non-canonical interactions, obligatory pairing of all bases, and a double-helical structure. The loop E conformation within the complex with L25 and the binding efficiency are relatively independent of the presence of Mg2+. A crucial role of magnesium in maintenance of specific geometry of the loop has been mentioned above. Obviously, the protein is capable of compensating the loss of these cations and independently structuring the region to be bound.
Thus, the binding has no noticeable consequences for the RNA molecule. The same may be said about the other partner. Structures of the free and bound L25 are very similar: β-barrel and two α-helices (α2 and α3) nearly fully coincide on superposition of the isolated and interacting with 5S rRNA protein. However, there is a very important rearrangement directly associated with formation of the RNA-protein complex: a large unstructured loop between β1 and β2 strands typical for the free protein acquires conformation of the third α-helix (α1) and forms a part of the protein binding site. The interaction occurs at two sites. The first site is a side of the β-barrel. It is in contact with the widened minor groove of loop E in the region of A73:U103 and U77:A99 pairs and seems to be the main “RNA-recognizing” module of the protein, which reacts to a specified design of donor and acceptor groups in this region of the loop. As it has been said earlier, the second site is produced by the N-terminal part of helix α1, which interacts with the extended large groove of the loop and proximal part of helix IV. The binding surfaces include both regions with a high electrostatic potential and large uncharged regions and this suggests that RNP is produced due to different types of binding. Note that notwithstanding a quasi-symmetric structure of loop E, the binding is asymmetric. The absolute majority of amino acid residues involved in the interaction is bound with the 3′-side of the loop. Only eight of 20 nucleotides located in the binding site have contacts with amino acid residues, and only six of them are electrostatic, which indicates a definite specificity of the binding. Ribose residues form many bonds with protein groups. Some interactions seem to be mediated by water molecules.
Interactions of 5S rRNA with protein factors of its closest environment in the large ribosomal subunit were considered above. Figure 5 shows that three main groups of ribosomal proteins (such as eL5/L18, prokaryotic L5, and L25) demonstrate different types of binding with the same molecule. As a rule, proteins of the eL5/L18-family can protect nearly the whole 5S rRNA; the interaction with them occurs in many sites and is extremely specific with high affinity. The binding is associated with considerable rearrangements in structures of both partners. It is the main protein factor that is present in all ribosomes. The prokaryotic protein L5 is nearly in full contrast to L18/eL5. Its binding site and the protected region are extremely limited. The affinity for RNA increases only in the presence of bound L18. However, in this case both partners also undergo some conformational changes. If ribosomal protein L18 plays the leading role in the 5S rRNA integration into the large subunit via protein-protein interactions, L5 provides for uniting of rRNA components via two RNA-protein contacts. Finally, the exclusively bacterial protein L25 also interacts with a rather narrow region of RNA, although it protects nearly the whole γ-domain. As differentiated from other partners, this protein does not induce noticeable changes in the structure of bound RNA during the binding, and slight rearrangements in it are very significant functionally.
In all cases, RNA sites are produced with involvement of elements with non-canonical secondary structure, which expose a considerably greater number and variety of groups capable of interacting with protein. Because 5S rRNA has such elements in abundance, the multiplicity of protein factors binding with it and a great variety of strategies used for RNA-protein interaction are not surprising.
Interaction with 23S rRNA
Different integration modes of the RNA component, which forms a “skeleton” of the large ribosomal subunit, were already discussed. The first mode mainly mediated via the eL5/L18-family proteins is represented by protein-protein interactions between factors binding with each of two rRNAs. In a second mode, the prokaryotic protein L5 forms a “bridge” which concurrently binds the two RNA molecules. And the third and simplest mode is a direct interaction between rRNAs that will be described in the present section.
Study of RNA-RNA interactions is more difficult than investigation of binding between RNP components. This is caused not only by an extreme instability of RNA and dependence of the dissociation constant on multiple additional factors, but also by a limited number of approaches used in this case. In fact, there are two approaches: various cross-linking approaches between bases and analysis of spatial structures of natural complexes (i.e. ribosomal subunits). Both these strategies were used in studies of the interaction of 5S rRNA with 23S rRNA.
Russian researchers Dontsova et al. [81] and Dokudovskaya et al. [82] studied interaction between two ribosomal RNAs using intermolecular cross-linking. For this purpose, transcripts were prepared of E. coli 5S rRNA with randomly introduced substitutions of uridine by thiouridine. Upon photoinduction, the modified 5S rRNA produced cross-links with 23S rRNA in both 50S and 70S complexes. Thus, locations of the cross-links marked sites of interaction between the two rRNAs. This approach allowed the authors to reveal two regions of 23S rRNA specifically interacting with 5S rRNA. The first region (C2475) is on the apex of the helix-89 hairpin in domain V near the peptidyl transferase ring; the second region (A960) is on the extremity of helix-39 in domain II. In both cases, the interaction occurs with involvement of U89 of 5S rRNA loop D.
Obviously, these cross-links are mutually incompatible, and what domain of 23S rRNA will be covalently bound with U89 depends on many factors. When 50S subunits were reconstructed using 23S rRNA isolated with EDTA on sucrose gradient, the cross-linking with A960 was predominant. But this rRNA isolated in the presence of magnesium was cross-linked by the C2475 position. Thus, both regions of 23S rRNA seemed to be located near loop D of 5S rRNA, because thiouridine was a so-called “zero-length” reagent, i.e. a 1-2 Å deviation of the binding site completely prevented cross-linking. A possible functional significance of these contacts will be discussed in detail in the next section.
Unfortunately, this approach, in spite of its considerable advantages, does not allow all possible RNA-RNA interactions to be identified, at least because uridine residues are not always involved in them. Therefore, great expectations were associated with analysis of spatial structure of the large subunit of the H. marismortui ribosome [26]. In fact, 5S and 23S rRNAs were shown [83] to have at least one more specific contact to one another (Fig. 6a). This was a typical A-minor interaction of type 0. As at every binding of such type, adenine nucleotide enters into the minor groove of the partner's helix. The type 0 is specified by involvement of 2′-OH group of ribose in formation of a hydrogen bond, but the nitrogen base itself does not enter the minor groove. Therefore, in general the base type is not especially important (although in the case of adenine the contact is optimal) because ribose promotes the binding with any combination of the partner's groups. It is noteworthy that interaction between 5S and 23S rRNAs is symmetric. 5S rRNA gives for this the best of its “arsenal” presented by loop E in which the pair G:A together with the following inverse Hoogsteen pair U:A forms an inter-strand stacking of three adenines. 23S rRNA is utterly rich with such elements and for the interaction uses a similar structure from helix-38 of domain II. This results in helix-helix binding with involvement of six adenine residues.
Thus, the whole set of non-classical elements of 5S rRNA is functioning and promotes interactions with multiple protein and nucleic factors. This is especially surprising because they are parts of an extremely small molecule of RNA. It seems that in 5S rRNA Nature has reached its limit in the concentrating of functional sites and variety of interactions. And on this background the putative role of this molecule in the cell seems even more enigmatic. Functions of 5S rRNA in the ribosome are considered in the following section of this review.Fig. 6. Interaction of 5S rRNA with 23S rRNA and its functions. a) Secondary and three-dimensional structures of interaction between loop E of 5S rRNA and the A-site finger of H. marismortui 23S rRNA (Jena Library) [26, 83]. b) Scheme of the interaction network between the γ-domain of 5S rRNA and functional sites of 23S rRNA (after [12, 23]) (Jena Library). c) Secondary structures of the 23-nucleotide fragment from the human mitochondrial genome and a similar fragment of Bacillus subtilis 5S rRNA [114].
FUNCTIONS OF 5S rRNA
The first work concerning functions of 5S rRNA was published in 1971 [11]. It was reported that the E. coli ribosome assembled in the absence of 5S rRNA was unable to synthesize polypeptide. Thus, 5S rRNA is an essential structural and functional component of the ribosome [84]. But the exact role of 5S rRNA in different processes of the translational epicycle is still unclear. The 5S rRNA location within the large subunit is unique. Due to multiple direct (RNA-RNA) and mediated (RNA-protein and protein-protein) interactions, 5S rRNA has contacts with virtually all important functional regions of the ribosome: A- and P-sites, peptidyl transferase site, and GTPase site. Such a close location of 5S rRNA to these sites led to various hypotheses about functions of this molecule. The following functions were supposed to be the main ones: mediation in the transfer of a growing polypeptide through production of a covalent bond [85, 86], the GTPase activity of a 5S rRNA-protein complex [87, 88], positioning of tRNA at the cost of pairing of conservative regions of the two molecules [89, 90]. But none of these hypotheses have been confirmed. At present, it is established that 5S rRNA fails to directly interact with both tRNA and translation factors. Therefore, now researchers are inclined to believe that this molecule has mainly to regulate and coordinate interrelations and interdependence between functional sites of the ribosome [82, 91]. This standpoint is supported by both theoretical and experimental arguments.
A large-scale study of 5S rRNA of S. cerevisiae point mutants (the strain without a locus containing the cluster of 5S rRNA genes) has shown that mutations in three regions of the molecule result in consequences of different type and severity [12]. The first region represented by loop B-helix III-loop C (especially their 5′-sides) is the site for binding with protein eL5. Many mutations in this site are lethal and some of them strongly affect the translation accuracy (nonsense-suppression ability). This region plays a crucial role in formation of the contact between ribosomal subunits, and protein eL5 mediates the binding of 5S rRNA with peptidyl tRNA located in the P-site [92]. Loop E-helix IV presents the second functionally important region of 5S rRNA. Almost all mutations in this region are lethal (mainly because of an extremely high frequency of reading frame displacement). As mentioned, loop E acts as a binding site of 5S rRNA with helix-38 of 28S rRNA domain II (Fig. 6b). This helix is also called A-site finger (ASF). Thus, 5S rRNA also has a direct contact with the ribosome A-site. The third region is smaller in size and comprises the distal part of helix IV and loop D. Mutations in this region are associated with a decrease in the translation accuracy or even lethal. This region is also involved in interactions with large ribosomal RNA. These interactions are very diverse, which is exemplified by the large ribosomal subunit of H. marismortui. The helix-89 of 28S rRNA is in direct contact with helix-92, which in its turn interacts with aminoacyl-tRNA in the A-site. Thus, this is another link with this most important functional site. The same pathway via helix-92 and -91 leads to the sarcin-ricin loop and provides for the contact of 5S rRNA with the GTPase site. Helix-39, which also has a contact with loop D, promotes an additional access through helix-80 to peptidyl-tRNA in the P-site. As a result, we have a system of interrelated rRNA-protein structures which communicate all major functional sites of the ribosome. A branched network of interactions is responsible for conformational mobility of these sites, making them not independent from one another. It seems that each site “feels” the state of another and adequately reacts to it, and this results in a precise, accurate, and sequential functioning of the ribosome. And 5S rRNA is a central integrating link of this chain acting as a “manager”.
Not long ago functional roles of individual structural elements of yeast 5S rRNA were similarly analyzed in vivo by Kiparisov et al. [1], who came to the same conclusions. Moreover, this work markedly enriched the concept of the function of 5S rRNA as a “manager”. A careful analysis of artificial mutations and allelic variants of 5S rRNA allowed the authors to suppose that the multiple allelism of 5S rRNA could itself act as a mechanism for regulating the gene expression on the post-transcriptional level. Different alleles of 5S rRNA are responsible for different translation accuracy (i.e. different frequency of nonsense-suppression), and this can be used by the eukaryotic cell for production of minor variants of a protein during studies of development. At present, this is only a pretty hypothesis, but the finding of multiple transcriptionally active alleles of 5S rRNA in all eukaryotic cells seems to indicate their important functions.
The last work in this field [93] revealed a distinct interrelation between the conformational transitions of 5S rRNA and translational activity of the ribosome. Thus, ribosomes possessing 5S rRNAs with tertiary structure changed and fixed as a result of binding with N1-azidobenzamidinospermine demonstrated more efficient binding of tRNA and increased activity of peptidyl transferase and of translocation. Note that this agent induced the most serious changes in loop A, which is mainly responsible for general geometry of the molecule. Its structure becomes loose; thus the type of stacking between domains seems to be changed.
IMPORT OF 5S rRNA INTO MITOCHONDRIA OF MAMMALIAN CELLS
Pathways of 5S rRNA transport are rather complicated in eukaryotic cells (Fig. 2) and even more intricate in cells of higher vertebrates [94]. In 1994, a large amount of an RNA was detected by electrophoresis in denaturing polyacrylamide gel in preparation of nucleic acids isolated from mitochondria of bovine cells. This RNA corresponded in size to cytosolic 5S rRNA. Its amount in the matrix was higher than of any mitochondrial tRNA, and the sequence was identical to that of 5S rRNA encoded by the nuclear genome. Similar data were obtained for mitochondria of rat and chicken cells. Using various approaches, the authors found that the preparation was free of cytosolic RNAs. Thus, this RNA (the genes of which were located only in the nucleus) seemed to be imported into mitochondria from the cytosol.
At the time of this discovery and in the following years, import of different RNAs into mitochondria of various living organisms has been reported in many works. Thus, import of tRNA was found in Trypanosomatides (Leishmania, Trypanosoma) [95-99], Tetrahymena [100], some plants [101-105], marsupials [106], and also in the budding yeast S. cerevisiae [107]. At least one RNA, which is a component of MRP ribonuclease, can also penetrate into mitochondria of human cells [108]. RNA of the same size as 5S rRNA was found in human and mouse mitochondria [109, 110], but neither its nucleotide sequence nor exact location was established. Therefore, the work by Yoshionari et al. [94] is believed to be the first reliable report about this fact. Later, the discovery of the Japanese authors was confirmed in the work [111], which made it impossible to interpret the experimental data otherwise--5S rRNA encoded by nuclear DNA is really present in mitochondria of mammals.
To study the penetration of 5S rRNA into human mitochondria, an in vitro model of import of radiolabeled 5S rRNAs into isolated mitochondria has been elaborated in our laboratory [112]. Using this model, the 5S rRNA import was shown to require the presence of ATP, a system for its regeneration (i.e. phosphoenol pyruvate/pyruvate kinase), an intact apparatus for import of precursors of mitochondrial proteins, and, at least, of two yet unknown cytosolic protein factors involved in 5S rRNA binding and transfer towards mitochondria.
Some parameters of this in vitro model can be varied depending on the researcher's purpose. Using different protein fractions or individual proteins allows us to assess their ability to direct/suppress the import and establish the involvement of different protein factors in the import. Mutant 5S rRNAs (with changed structures of individual modules) as substrates are required to search for the import determinants, i.e. such elements of the RNA structure which are responsible for its interaction with the import factors and specific traffic into mitochondria. The validity of this in vitro model, which can adequately represent in vivo import, is especially evident if one remembers that 5S rRNA is encoded by multiple genes that virtually forbids using genetic approaches to search for the import determinants.
How can the 5S rRNA import be used? In fact, this RNA is a natural endogenous vector that moves into mitochondria. It was supposed that within this vector heterologous sequences could be transferred as insertions or terminal extensions, especially if they do not affect the import determinants and elements required for the effective expression, processing, and export of 5S rRNA. This hypothesis is now under examination.
A directed introduction of particular RNAs into the mitochondrial matrix is promising for studies of expression and replication of the mitochondrial genome. The same principle may serve as a basis for application of the import. For instance, some hereditary diseases are caused by mutations in mitochondrial DNA, and mitochondria with both normal and mutant genomes can be present in the cell in different ratios (so-called heteroplasmy). Injection of RNA complementary to the mutation region into “bad” mitochondria can markedly lower replication efficiency of the mutant DNA, because the produced DNA-RNA duplex (more stable than DNA-DNA) prevents movement of the replicative complex. Thus, mitochondria with wild type DNA obtain a selective advantage as compared to their mutant “fellows”, which results in elimination of the mutants and “sanitation” of the mitochondrial population.
Possible Role of 5S rRNA in Mammalian Mitochondria
For a long time, mitochondrial ribosomes (mitoribosomes) of animals were known to possess a rather specific set of proteins and RNAs. Only some of these proteins are homologous to prokaryotic ribosomal proteins, whereas for other proteins no homologs have been found in all known living organisms. The RNAs are about twofold shorter than their bacterial homologs (sedimentation coefficients of mitochondrial rRNAs are 16S and 12S, respectively, for mammals), but the most striking is a complete absence of 5S rRNA. How mitoribosomes can function without this most important component is still unclear. It is known that the major part of the “lost” rRNA in mitoribosomes is replaced by ribosomal proteins (that is why the RNA/protein ratio in mitoribosomes is inverse and approximately equal to 1 : 3 [112]), but neither of them is likely to compensate the “loss” of 5S rRNA. A quantitative evaluation of the 5S rRNA content in human mitochondria performed in our laboratory [113] has shown that the number of copies of this RNA approximately corresponds to the number of mitoribosomes, and this does not contradict a possible involvement of cytosolic 5S rRNA in production of functional mitochondrial ribosomal subunits. But up to now no detection of 5S rRNA in preparation of mammalian mitoribosomes has been reported. This association might be very unstable, but now this remains only a hypothesis.
However, this hypothesis is indirectly confirmed by some findings. Thus, a 23-nucleotide rudimentary sequence homologous to a region of bacterial 5S rRNA was found in its typical context (within the rRNA gene cluster) in the genome of human mitochondria [114]. Note that this “rudiment” is virtually identical to helix III and loop C of gram-positive bacteria (Fig. 6c). Possibly, some ancestral forms of mitochondria contained a complete set of rRNAs, but it underwent a considerable reduction during evolution. And 5S rRNA either totally disappeared (with retention up to the last moment of its most important structural element, the site for interaction with protein L18), or was substituted by the imported cytosolic 5S rRNA.
Moreover, all mitochondria of vertebrata contain the protein MRP-L18, which is a pronounced homolog of the ribosomal protein L18 [115, 116]. Considering that it is the major and absolutely ubiquitous 5S rRNA-binding protein and that sites of interaction with proteins of the eL5/L18-family are similar for 5S rRNAs of different origin, a serious question can be raised: what is the role of this factor in mitoribosomes in the absence of its traditional partner? There is a hope that further studies will allow us to answer this question.
Dozens of RNA types are now known that are different in structure, location, interaction with other biological molecules, and, finally, in functions, the number of which seems to be well beyond the limits of the “classic triad” mRNA-tRNA-rRNA. Despite a limited set of monomers, structures of RNA display a striking variety and intricacy, which sometimes are no less than those of protein molecules. This determines an extremely broad circle of processes with RNA molecules as major players, and these processes are associated with the basis of metabolism of every living system--reproduction and realization of genetic information.
In the light of recent discoveries of small non-coding RNAs it seems very surprising that, despite nearly forty years of studies, the 5S ribosomal RNA molecule familiar from school textbooks seems to be the most enigmatic representative of the RNA world. In fact, a careful study of this relatively small molecule shows that it is characterized by an unusually sophisticated organization, which includes a number of non-canonical elements. This molecule includes unusual structures and interactions and presents combinations of quite unexpected variants of packing, from the classic double helix of the A-form to loop E which seems to contradict not only the principle of complementary pairing but even more universal “law” of anti-parallelism of chains. Many of these elements also found in other RNAs are crucial in very different processes. But some elements seem to be unique for 5S rRNA.
Despite multiple careful studies of 5S rRNA interactions with proteins and RNAs, the major function of this molecule still remains unclear. An inevitable component of virtually every ribosome, utterly conservative in structure, 5S rRNA seems to play an important role during translation. However, all attempts to associate 5S rRNA with any specific stage of the elongation cycle have been unsuccessful. Recently, the hypothesis about a strictly definite specific function of 5S rRNA is replaced by the idea of multiplicity of its functions, and this idea puts this molecule into cross-roads of the main translational processes. This small RNA can coordinate activities of different functional sites of the ribosome and act as a regulatory link required for any complicated system.
But the list of unique properties 5S rRNA is still not exhausted. A complicated scheme of intracellular transfers shows its difference from other RNAs. The triple localization (the nucleus, cytosol, and mitochondria) of 5S rRNA is provided for by interactions with various carrier proteins, which are not sufficiently identified. While our knowledge about the functions of 5S rRNA in the cytosol becomes more definite, its role in mitochondria remains enigmatic and even intriguing.
Mitochondrial ribosomes of animals seem to contain no 5S rRNA-like molecule characterized by a stable association with the large ribosomal subunit. The mitochondrial genome also lacks the gene of 5S rRNA. However, the major and absolutely universal 5S rRNA-binding ribosomal protein L18 is present in the central protuberance of mammalian mitoribosomes. Yet it is not clear if 5S rRNA encoded by the nuclear genome is involved in mitochondrial translation or performs some other function. We can only hope that all these questions will be answered very soon.
The authors are very grateful to O. A. Kolesnikova for her help in preparing the material of the review and for comprehensive support.
This work was supported by the Russian Foundation for Basic Research (project No. 03-05-22001-NTsNI a).
REFERENCES
1.Kiparisov, S., Petrov, A., Meskauskas, A., Sergiev,
P. V., Dontsova, O. A., and Dinman, J. D. (2005) Mol. Gen.
Genom., 274, 235-247.
2.Studnicka, G. M., Eiserling, F. A., and Lake, J. A.
(1981) Nucleic Acids Res., 9, 1885-1904.
3.Toots, I., Metspalu, A., Villems, R., and Saarma,
M. (1981) Nucleic Acids Res., 9, 5331-5343.
4.Nishikawa, K., and Takemura, S. (1977) J.
Biochem., 81, 995-1003.
5.Delihas, N., and Andersen, J. (1982) Nucleic
Acids Res., 10, 7323-7344.
6.Pelham, H. R. B., and Brown, D. D. (1980) Proc.
Natl. Acad. Sci. USA, 77, 4170-4174.
7.Steitz, J. A., Berg, C., Hendrick, J. P., la
Branche-Chabot, H., Metspalu, A., Rinke, J., and Yario, T. (1988) J.
Cell Biol., 106, 545-556.
8.Rudt, F., and Pieler, T. (1996) EMBO J.,
15, 1383-1391.
9.Picard, B., and Wegnez, M. (1979) Proc. Natl.
Acad. Sci. USA, 76, 241-245.
10.Szymanski, M., Barciszewska, M. Z., Erdmann, V.
A., and Barciszewski, J. (2003) Biochem. J., 371,
641-651.
11.Erdmann, V. A., Fahnestock, S., Higo, K., and
Nomura, M. (1971) Proc. Natl. Acad. Sci. USA, 68,
2932-2936.
12.Smith, M. W., Meskauskas, A., Wang, P., Sergiev,
P. V., and Dinman, J. D. (2001) Mol. Cell. Biol., 21,
8264-8275.
13.Wagner, R., and Garrett, R. A. (1978) Nucleic
Acids Res., 5, 4065-4075.
14.Szymanski, M., Barciszewska, M. Z., Erdmann, V.
A., and Barciszewski, J. (2000) Mol. Biol. Evol., 17,
1194-1198.
15.White, S. A., Nilges, M., Huang, A., Brunger, A.
T., and Moore, P. B. (1992) Biochemistry, 31,
1610.
16.Raue, H. A., Lorenz, S., Erdmann, V. A., and
Planta, R. J. (1981) Nucleic Acids Res., 9,
1263-1269.
17.Lee, Y., and Nazar, R. N. (2003) J. Biol.
Chem., 278, 6635-6641.
18.Andersen, J., and Delihas, N. (1986) J. Biol.
Chem., 261, 2912-2917.
19.Peattie, D. A., Douthwaite, S., Garrett, R. A.,
and Noller, H. F. (1981) Proc. Natl. Acad. Sci. USA, 78,
7331-7335.
20.Xiong, Y., and Sundaralingam, M. (2000)
RNA, 6, 1316.
21.Nishikawa, K., and Takemura, S. (1978) J.
Biochem., 84, 259-266.
22.Lescoute, A., Leontis, N. B., Massire, C., and
Westhof, E. (2005) Nucleic Acids Res., 33, 2395-2409.
23.Chow, C. S., Hartmann, K. M., Rawling, S. L.,
Huber, P. W., and Barton, J. K. (1992) Biochemistry, 31,
3534-3542.
24.Bullerwell, C. E., Schnare, M. N., and Gray, M.
W. (2003) RNA, 9, 287.
25.Huber, P. W., Rife, J. P., and Moore, P. B.
(2001) J. Mol. Biol., 312, 823.
26.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) Science, 289,
905-920.
27.Bruenger, E., Kowalak, J. A., Kuchino, Y., Mc
Closkey, J. A., Mizushima, H., Stetter, K. O., Brunel, C., Romby, P.,
Westhof, E., Ehresmann, C., and Ehresmann, B. (1991) J. Mol.
Biol., 221, 293-308.
28.Kiprekar, F., Douthwaite, S., and Roepstorff, P.
(2000) RNA, 6, 296-306.
29.Miyazaki, M. (1974) J. Biochem.,
75, 1407-1410.
30.Helm, M. (2006) Nucleic Acids Res.,
34, 721-733.
31.Douthwaite, S., Garrett, R. A., Wagner, R., and
Feunteun, J. (1979) Nucleic Acids Res., 6, 2453-2470.
32.Ciesiolka, J., and Krzyzosiak, W. J. (1996)
Biochem. Mol. Biol. Int., 39, 319-328.
33.Sarzynska, J., Kulinski, T., and Nilsson, L.
(2000) Biophys. J., 79, 1213-1227.
34.Barthel, A., and Zacharias, M. (2006) Biophys.
J., 90, 2450-2462.
35.Barciszewska, M. Z., Wyszko, E., Bald, R.,
Erdmann, V. A., and Barciszewski, J. (2003) J. Biochem.,
133, 309-315.
36.Correll, C. C., Freeborn, B., Moore, P. B., and
Steitz, T. A. (1997) Cell, 91, 705-712.
37.Leontis, N. B., and Westhof, E. (1998)
RNA, 4, 1134-1153.
38.Reblova, K., Spakova, N., Stefl, R., Csaszar, K.,
Koa, J., Leontis, N. B., and Sponer, J. (2003) Biophys. J.,
84, 3564-3582.
39.Lukavsky, P., Billeci, T. M., James, T. L., and
Schmitz, U. (1997) in Molecular Modelling of Nucleic Acids
(Leontis, N. B., and Santa Lucia, J., eds.) American Chemical
Society, Washington, DC, pp. 122-149 (cited after Leontis and Westhof,
1998).
40.Lentzen, G., Moine, H., Ehresmann, B., and
Wintermeyer, W. (1996) RNA, 2, 244-253.
41.Wimberly, B., Varani, G., and Tinoco, I., Jr.
(1993) Biochemistry, 32, 1078-1087.
42.Correll, C. C., Beneken, J., Plantinga, M. J.,
Lubbers, M., and Chan, Y.-L. (2003) Nucleic Acids Res.,
31, 6806-6818.
43.Branch, A. D., Benenfeld, B. J., and Robertson,
H. D. (1985) Proc. Natl. Acad. Sci. USA, 82,
6590-6594.
44.Baumstark, T., Schroder, A. R. W., and Riesner,
D. (1997) EMBO J., 16, 599-610.
45.Weidner, H., and Crothers, D. M. (1977)
Nucleic Acids Res., 4, 3401-3414.
46.Kime, M. J., and Moore, P. B. (1982) Nucleic
Acids Res., 10, 4973-4983.
47.Goringer, H. U., Szymkowiak, C., and Wagner, R.
(1984) FEBS J. (EJB), 144, 25-34.
48.Kao, T. H., and Crothers, D. M. (1980) Proc.
Natl. Acad. Sci. USA, 77, 3360-3364.
49.Toots, I., Misselwitz, R., Bohm, S., Welfle, R.,
Villems, R., and Saarma, M. (1982) Nucleic Acids Res.,
10, 3381-3389.
50.Maruyama, S., and Sugai, S. (1980) J.
Biochem., 88, 151-158.
51.Giegerich, R., Haase, D., and Rehmsmeier, M.
(1999) Pacific Symposium on Biocomputing, pp. 126-137.
52.Hanas, J. S., Bogenhagen, D. F., and Wu, C. W.
(1984) Nucleic Acids Res., 12, 2745-2758.
53.Huber, P. W., and Wool, I. G. (1986) Proc.
Natl. Acad. Sci. USA, 83, 1593-1597.
54.Rawling, S. L., Matt, G. D., and Huber, P. W.
(1996) J. Biol. Chem., 271, 869-877.
55.Darsillo, P., and Huber, P. W. (1991) J. Biol.
Chem., 266, 21075-21082.
56.Baudin, F., Romaniuk, P. J., Romby, P., Brunel,
C., Westhof, E., Ehresmann, B., and Ehresmann, C. (1991) J. Mol.
Biol., 218, 69-81.
57.Theunissen, O., Rudt, F., and Pieler, T. (1998)
Eur. J. Biochem., 258, 758-767.
58.Lu, D., Searles, M. A., and Klug, A. (2003)
Nature, 426, 96-100.
59.McDougall, J., and Wittmann-Liebold, B. (1994)
Eur. J. Biochem., 221, 779-785.
60.Huber, P. W., and Wool, I. G. (1984) Proc.
Natl. Acad. Sci. USA, 81, 322-326.
61.Gongadze, G. M., Perederina, A. A.,
Meshcheryakov, V. A., Fedorov, R. V., Moskalenko, S. E., Rak, A. V.,
Serganov, A. A., Shcherbakov, D. V., Nikonov, S. V., and Garber, M. B.
(2001) Mol. Biol. (Moscow), 35, 521-526.
62.Bear, D. G., Schleich, T., Noller, H. F., and
Garrett, R. A. (1977) Nucleic Acids Res., 4,
2511-2526.
63.Nazar, R. N., Willick, G. E., and Matheson, A. T.
(1979) J. Biol. Chem., 254, 1506-1512.
64.Nishikawa, K., and Takemura, S. (1974) FEBS
Lett., 40, 106-109.
65.Blobel, G. (1971) Proc. Natl. Acad. Sci.
USA, 68, 1881-1885.
66.Isoda, N., Tanaka, T., and Ishikawa, K. (1981)
J. Biochem., 90, 551-554.
67.Aoyama, K., Tanaka, T., Hidaka, S., and Ishikawa,
K. (1984) J. Biochem., 95, 1179-1186.
68.Scripture, J. B., and Huber, P. W. (1995) J.
Biol. Chem., 270, 27358-27365.
69.Yeh, L.-C. C., Horowitz, P. M., and Lee, J. C.
(1988) J. Biol. Chem., 263, 17412-17417.
70.Lin, E., Lin, S.-W., and Lin, A. (2001)
Nucleic Acids Res., 29, 2510-2516.
71.DiNitto, J. P., and Huber, P. W. (2003) J.
Mol. Biol., 330, 979-992.
72.Williamson, J. R. (2000) Nature Struct.
Biol., 7, 834-837.
73.Tompa, P., and Csermely, P. (2004) FASEB
J., 18, 1169-1175.
74.Shpanchenko, O. V., Dontsova, O. A., Bogdanov, A.
A., and Nierhaus, K. H. (1998) RNA, 4, 1154-1164.
75.Spierer, P., Bogdanov, A. A., and Zimmermann, R.
A. (1978) Biochemistry, 17, 5394-5398.
76.Perederina, A., Nevskaya, N., Nikonov, O.,
Nikulin, A., Dumas, P., Yao, M., Tanaka, I., Garber, M., Gongadze, G.,
and Nikonov, S. (2002) RNA, 8, 1548-1557.
77.Nakashima, T., Yao, M., Kawamura, S., Iwasaki,
K., Kimura, M., and Tanaka, I. (2001) RNA, 7,
692-701.
78.Korepanov, A. P., Gongadze, G. M., and Garber, M.
B. (2004) Biochemistry, 69, 607-611.
79.Stoldt, M., Wohnert, J., Gorlach, M., and Brown,
L. R. (1998) EMBO J., 17, 6377-6384.
80.Stoldt, M., Wohnert, J., Ohlenschlager, O.,
Gorlach, M., and Brown, L. R. (1999) EMBO J., 18,
6508-6521.
81.Dontsova, O., Tishkov, V., Dokudovskaya, S.,
Bogdanov, A., Doring, T., Rinke-Appel, J., Thamm, S., Greuer, B., and
Brimacombe, R. (1994) Proc. Natl. Acad. Sci. USA,
91, 4125-4129.
82.Dokudovskaya, S., Dontsova, O., Shpanchenko, O.,
Bogdanov, A., and Brimacombe, R. (1996) RNA, 2,
146-152.
83.Nissen, P., Ippolito, J. A., Ban, N., Moore, P.
B., and Steitz, T. A. (2001) Proc. Natl. Acad. Sci. USA,
98, 4899-4903.
84.Dohme, F., and Nierhaus, K. H. (1976) Proc.
Natl. Acad. Sci. USA, 73, 2221-2225.
85.Raacke, I. D. (1971) Proc. Natl. Acad. Sci.
USA, 68, 2357-2360.
86.Fahnestock, S. R., and Nomura, M. (1972) Proc.
Natl. Acad. Sci. USA, 69, 363-365.
87.Horne, J. R., and Erdmann, V. A. (1973) Proc.
Natl. Acad. Sci. USA, 70, 2870-2873.
88.Ogata, K., Terao, K., and Uchiumi, T. (1980)
J. Biochem., 87, 517-524.
89.Forget, B. G., and Weissman, S. M. (1967)
Science, 158, 1695-1699.
90.Pace, B., Matthews, E. A., Johnson, K. D.,
Cantor, C. R., and Pace, N. R. (1982) Proc. Natl. Acad. Sci.
USA, 79, 36-40.
91.Bogdanov, A. A., Dontsova, O. A., Dokudovskaya,
S. S., and Lavrik, I. N. (1995) Biochem. Cell. Biol., 73,
869-876.
92.Meskauskas, A., and Dinman, J. D. (2001)
RNA, 7, 1084-1096.
93.Kouvela, E. C., Gerbanas, G. V., Xaplanteri, M.
A., Petropoulos, A. D., Dinos, G. P., and Kalpaxis, D. L. (2007)
Nucleic Acids Res., 1-12.
94.Yoshionari, S., Koike, T., Yokogawa, T.,
Nishikawa, K., Ueda, T., Miura, K., and Watanabe, K. (1994) FEBS
Lett., 338, 137-142.
95.Simpson, A. M., Suyama, Y., Dewes, H., Campbell,
D. A., and Simpson, L. (1989) Nucleic Acids Res., 17,
5427-5444.
96.Lye, L.-F., Tom Chen, D.-H., and Suyama, Y.
(1993) Mol. Biochem. Parasitol., 58, 233-246.
97.Mottram, J. C., Bell, S. D., Nelson, R. G., and
Barry, J. D. (1991) J. Biol. Chem., 266, 18313-18317.
98.Hancock, K., LeBlanc, A. J., Donze, D., and
Hajduk, S. L. (1992) J. Biol. Chem., 267,
23963-23971.
99.Schneider, A., Martin, J., and Agabian, N. (1994)
Mol. Cell. Biol., 14, 2317-2322.
100.Rusconi, C. P., and Cech, T. R. (1996)
EMBO, 15, 3286-3295.
101.Akashi, K., Sakurai, K., Hirayama, J.,
Fukuzava, H., and Ohyama, K. (1996) Curr. Genet., 30,
181-185.
102.Brubacher-Kauffmann, S., Marechal-Drouard, L.,
Cosset, A., Dietrich, A., and Duchkne, A.-M. (1999) Nucleic Acids
Res., 27, 2037-2042.
103.Glover, K. E., Spencer, D. F., and Gray, M. W.
(2001) J. Biol. Chem., 276, 639-648.
104.Chen, H.-C., Viry-Moussaïd, M., Dietrich, A.,
and Wintz, H. (1997) Biochem. Biophys. Res. Commun.,
237, 432-437.
105.Marechal-Drouard, L., Weil, J.-H., and
Guillemaut, P. (1988) Nucleic Acids Res., 16,
4777-4788.
106.Dorner, M., Altmann, M., Poobo, S., and Morl,
M. (2001) Mol. Biol. Cell, 12, 2688-2698.
107.Martin, R. P., Schneller, J. M., Stahl, A. J.,
and Dirheimer, G. (1979) Biochemistry, 18,
4600-4605.
108.Puranam, R. S., and Attardi, G. (2001) Mol.
Cell. Biol., 21, 548-561.
109.Wong, T. W., and Clayton, D. A. (1986)
Cell, 45, 817-825.
110.King, M. P., and Attardi, G. (1993) J. Biol.
Chem., 268, 10228-10237.
111.Magalhaes, P. J., Andreu, A. L., and Schon, E.
A. (1998) Mol. Biol. Cell, 9, 2375-2382.
112.Suzuki, T., Terasaki, M., Takemoto-Hori, C.,
Hanada, T., Ueda, T., Wada, A., and Watanabe, K. (2001) J. Biol.
Chem., 276, 21724-21736.
113.Entelis, N. S., Kolesnikova, O. A., Dogan, S.,
Martin, R. P., and Tarassov, I. A. (2001) J. Biol. Chem.,
276, 45642-45653.
114.Nierlich, D. P. (1982) Mol. Cell. Biol.,
2, 207-209.
115.O'Brien, T. W., Liu, J., Sylvester, J. E.,
Mougey, E. B., Fischel-Ghodsian, N., Thiede, B., Wittmann-Liebold, B.,
and Graack, H.-R. (2000) J. Biol. Chem., 275,
18153-18159.
116.Koc, E. C., Burkhart, W., Blackburn, K., Moyer,
M. B., Schlatzer, D. M., Moseley, A., and Spremulli, L. L. (2001) J.
Biol. Chem., 276, 43958-43969.
117.Lescoute, A., and Westhof, E. (2006)
RNA, 23, 83-93.