REVIEW: Generation of Recombinant Antibodies and Means for Increasing Their Affinity
E. P. Altshuler, D. V. Serebryanaya*, and A. G. Katrukha
Department of Biochemistry, Faculty of Biology, Lomonosov Moscow State University, 119999 Moscow, Russia; E-mail: DariaSerebryanaya@gmail.com* To whom correspondence should be addressed.
Received April 21, 2010; Revision received June 24, 20010
Highly specific interaction with foreign molecules is a unique feature of antibodies. Since 1975, when Keller and Milstein proposed the method of hybridoma technology and prepared mouse monoclonal antibodies, many antibodies specific to various antigens have been obtained. Recent development of methods for preparation of recombinant DNA libraries and in silico bioinformatics approaches for protein structure analysis makes possible antibody preparation using gene engineering approaches. The development of gene engineering methods allowed creating recombinant antibodies and improving characteristics of existing antibodies; this significantly extends the applicability of antibodies. By modifying biochemical and immunochemical properties of antibodies by changing their amino acid sequences it is possible to create antibodies with properties optimal for certain tasks. For example, application of recombinant technologies resulted in antibody preparation of high affinity significantly exceeding the initial affinity of natural antibodies. In this review we summarize information about the structure, modes of preparation, and application of recombinant antibodies and their fragments and also consider the main approaches used to increase antibody affinity.
KEY WORDS: recombinant antibody fragment, affinity, maturation, modeling, in vitro evolution, displayDOI: 10.1134/S0006297910130067
Abbreviations: CH, constant domain of heavy chain; CL, constant domain of light chain; CDR, complementarity determining regions; Fab, antigen-binding fragment; Fc, crystallizable fragment; FR, framework regions; Fv, variable fragment; HC, immunoglobulin heavy chain; Ig, immunoglobulins; IgG, immunoglobulins G; LC, immunoglobulin light chain; SDR, specificity determining regions; VH, variable domain of heavy chain; VL, variable domain of light chain.
The development of the method of monoclonal antibody preparation by
means of the hybridoma technology proposed by Keller and Milstein in
1975 significantly influenced both fundamental science and clinical
practice. Owing to a unique feature of specific and high affinity
interaction with antigens, monoclonal antibodies are actively used in
scientific studies and in therapy of various diseases. Antibodies now
represent one third of the total number of proteins used for therapy of
various diseases in developed countries [1].
During the last 15 years the development of recombinant DNA technologies and creation of combinatorial gene libraries and also appearance of various bioinformatics resources and methods of computer-aided modeling of three-dimensional structures of protein molecules have made it possible to prepare and improve recombinant antibodies. Using recombinant technologies it is possible to insert various modifications into antibody sequences, influence their biochemical and immunochemical properties [2], and increase antibody affinity. A clear advantage of recombinant technologies also is the possibility of preparation of recombinant analogs of existing antibodies and also de novo creation of antibodies specific to various antigens.
Many reviews highlighting various methods of recombinant antibody preparation have been published [2-9]. However, from our viewpoint little attention has been given to the problem and methodology of artificially increasing the affinity of recombinant antibodies. This review includes two main parts. In the first one we consider in detail the structure, preparation, and practical application of recombinant antibodies and their fragments. In the second part we systematize the main approaches used for preparation of high affinity recombinant antibodies: from the methods of random or site-directed mutagenesis to recombinant DNA technologies for antibody production. Special attention is given to computer-aided modeling that predicts three-dimensional structure of antibodies and identifies amino acid residues that make the major contribution to antibody–antigen interaction and which can be replaced by other residues to create antibodies with increased affinity.
ANTIBODIES: STRUCTURE, FUNCTIONS, AFFINITY MATURATION in
vivo, AND PRACTICAL APPLICATION
Antibodies (immunoglobulins) are a family of glycoproteins existing in blood plasma and tissue fluids of vertebrates; they can recognize and bind foreign substances (antigens). The antigen site that specifically interacts with an antibody is known as an epitope, and the antibody site interacting with an antigen is known as a paratope [10]. In this chapter, using immunoglobulins G, we consider structure of antibodies, their main functions, the process of their affinity maturation in vivo, and also some aspects of application of antibodies in scientific studies, diagnostics, and therapy of various diseases.
Structure of Immunoglobulins
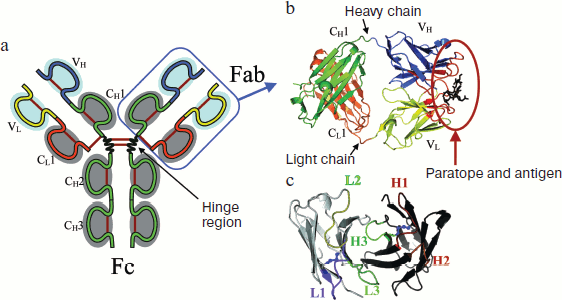
In most mammals five different classes of immunoglobulins (Ig) have been found: IgG, IgA, IgM, IgD, and IgE. Antibodies of various classes differ in size, charge, amino acid composition, and content of the carbohydrate component. Normally, IgG predominate in human serum and they usually represent about 70-75% of the total amount of serum antibodies [10-12]. The IgG antibody molecule is composed of four polypeptide chains: two identical heavy chains (HC) with molecular mass of 50-77 kDa and two identical light chains (LC) with molecular mass of 25 kDa (Fig. 1; see color insert). Each LC contains two domains: one variable domain (VL, variable domain of the light chain) and one constant domain (CL1, constant domain of the light chain); each HC contains one variable domain (VH, variable domain of the heavy chain) and three constant domains (CH1-3, constant domains of the heavy chain). Each domain is mainly a β-sheet structure formed by seven antiparallel β-strands forming two β-sheets. The structure of each Ig domain is stabilized by an intradomain disulfide bond. In addition, disulfide bonds are also formed between light and heavy chains as well as between heavy chains. Each pair of variable domains VH and VL located within neighboring heavy and light chains form one variable fragment (Fv). Amino acid variability differs inside variable domains. The highest level of variability is in the so-called hypervariable regions known as complementarity determining regions (CDR), which determine complementarity of antigen–antibody interaction. Variable domains of heavy and light chains contain three CDR (VLCDR1-3, VHCDR1-3). In the amino acid sequence of the variable domain these regions alternate with four relatively invariant regions known as the framework regions (FR1-FR4), which represent about 80% of the amino acid sequence of V-domains. The role of these regions is the maintenance of similar three-dimensional structure of the V-domain, which is required for specific interaction of the hypervariable regions with antigen [10, 12].
The variable domains of light and heavy chains (VH and VL) together with the nearest constant domains (CH1 and CL1) form antibody Fab-fragments (fragment, antigen binding). The remaining part represented by the C-terminal heavy chain constant domains is defined as the Fc-fragment (fragment, crystallizable). Fab and Fc fragments are linked by the hinge region, which provides relative flexibility of fragments of the antibody molecules and contains disulfide bonds linking heavy chains to each other [10, 12].Fig. 1. Structure of the IgG molecule. a) General scheme. The IgG molecule consists of two light chains (LC) and two heavy chains (HC). Each LC consists of a variable domain (VL) and a constant domain (CL1), and each HC consists of one variable domain (VH) and three constant domains (CH1-3). Two fragments are recognized in the IgG structure: the antigen-binding Fab-fragment and the Fc-fragment performing effector functions. The Ig structure is stabilized by disulfide bonds (marked with red lines) connecting LC and HC and also linking HC to each other. There are disulfide bonds inside each domain. b) Interaction between an antigen and the antibody Fab-fragment (three-dimensional structure of Fab-fragment from PDB, code 1CFN). The Fab-fragment includes LC and the HC N-terminal region including VH and CH1 domains. Each Fab-fragment contains the antigen binding site (paratope) located in the region of heavy and light chain variable domains. CDR loops are marked with red color. c) The antigen-binding site structure (three-dimensional structure of Fab-fragment from PDB, code 1CFN). The antigen-binding site of each Fab-fragment consists of amino acid residues of six hypervariable loops (CDR-loops) located on the surface of the LC and HC variable domains. The hypervariable loops are marked with color (L1-3 are the LC CDR loops shown in the order of their location in the amino acid sequence; H1-3 are the HC CDR loops).
As mentioned above, CDR play a key role in the antibody–antigen interaction. These hypervariable regions are loops located between β-sheets, which significantly vary in amino acid sequence and length in various antibodies. CDR provide complementarity of paratope and epitope surfaces, and this determines antigen recognizing capacity of apical regions of the variable domains. Epitope recognition on the complementary antigen surface by the three-dimensional structure formed by six loops of CDR is a basis for antibody–antigen interaction. In the case of antibodies specific to small antigens (such as low molecular weight compounds) only some CDR are involved into interaction with the antigen [13]. Usually VHCDR (especially VHCDR3) rather than VLCDR make major contribution into contact formation with the antigen and the geometrical center of the antibody–antigen contact is usually located near VHCDR3 [13]. CDR residues directly involved in contact with the antigen are called specificity determining regions (SDR). Analyzing three-dimensional structure of the antibody–antigen complex, it is possible to identify amino acid residues directly involved in contact with the antigen. Certain evidence exists that FR may also be involved into antigen recognition, but their role is rather modest: the proportion of such sites may reach 15% of surface contact sites participating in antibody–antigen contacts [13-15].
Antibody–antigen complexes are characterized by high complementarity in their interacting surfaces. The interaction surface of protein antigen and antibody varies from 1400 to 2300 Å2 with approximately equal contribution of antibody (650-900 Å2) and antigen [16]. Usually from 5 to 15 residues of protein antigens and approximately the same number of antibody residues are involved in the antibody–antigen interaction [16, 17]. However, smaller antigens (e.g. low molecular weight antigens) have smaller contact area during antibody binding. The shape of the antibody–antigen contacting surface significantly depends on the antigen size. The antibody contact surface becomes more concave as the antigen becomes smaller. Usually the antibody contact surface involved in binding of protein molecules is planar, whereas the surface binding smaller antigens represents a groove (in the case of peptides) or a single cavity (in the case of low molecular weight nonpeptide substances) [13, 18]. The spatial structure of the antigen binding site is determined by the length and amino acid composition of CDR loops. The CDR3 loops are characterized by significant flexibility, which makes possible mutual epitope–paratope adaptation. In the case of small antigens (including peptides) the conformation flexibility may be more pronounced compared with protein antigens. The main types of conformation flexibility include concerted translocations of several CDR loops and changes in side group location in some CDR residues. Antigens are also characterized by conformational changes during their interactions with antibodies.
The interaction between epitope and paratope amino acid residues mainly occurs due to hydrogen bonds, electrostatic interactions, and to a lesser extent van der Waals interactions [13, 19]. Some studies revealed that the structures of many complexes formed by protein antigens and antibodies contain water molecules in the paratope regions within common structure. It is suggested that in such complexes water molecules are required for filling complementarity gaps between the protein antigen molecule and the antibody [18, 20, 21].
The antibody antigen binding energy is mainly determined by a small number of interactions between epitope and paratope amino acid residues. Substitution of such amino acid residues in the natural antigen or antibody variants for alanine is often accompanied by decreased binding of antibody and antigen (see below). Selection of alanine as the candidate for possible substitution is determined by the fact that this is the mildest mode of elimination of interactions determined by the amino acid side group without appearance of additional interactions. Experiments of epitope and paratope mapping revealed so-called hotspots: the amino acids that make the major contribution to the antibody–antigen interaction. Such residues are usually located in the central part of the epitope–paratope interaction site. Peripheral residues contacting with the aqueous medium make significantly smaller contribution to the antibody–antigen binding energy [16]. In addition to amino acid residues directly involved into interaction with the antigen, there are other residues that play an important role in antigen recognition; they play a structural role and determine correct spatial positioning of the contact surfaces. These residues maintain certain conformation of CDR loops required for interaction with antigen [22].
Immunoglobulin Functions
B-Lymphocytes synthesize antibodies in response to the appearance of antigens in humans and other vertebrate organisms. Antibodies are involved in several processes occurring in the immune system: 1) prevention of penetration of foreign (including pathogenic) molecules into cells and their degradation due to complex formation with antibodies; 2) stimulation of elimination of foreign objects by macrophages and other cells by means of antibody interaction with their surface antigens (opsonization); 3) triggering of mechanisms of foreign particle degradation by stimulating the immune response (activation of the complement system).
Antibodies of all isotypes have two main functions: 1) recognition and binding of antigens; 2) effector functions that include binding to cell membrane receptors (e.g. phagocytes) and interaction with the first component of the complement system for initiation of the classical pathway of the complement cascade. Effective recognition and binding of antigens by antibodies requires high antibody specificity needed for recognition of the particular antigen and high antibody affinity for formation of stable antibody–antigen complex. Variable domains of antibodies are involved in the first function. Constant domains maintain structural organization of variable domains; they also play effector functions promoting neutralization and elimination of potential pathogens. Neutralization of some viruses and toxins requires only interaction with the antibody, whereas elimination of pathogenic microorganisms or endogenous cells not controlled by the organism needs triggering of other effector mechanisms such as phagocytosis and cytotoxic reactions. In this case antibody Fab-fragments bind to surface cell antigens, whereas Fc-fragments of such antibodies may interact with complement system proteins, which kill cells by membrane perforation or with Fc-cell receptors localized on the surface of immune system cells capable of phagocytosis [23]. Thus, the presence of the Fc-fragment and Fab-fragments in the antibody molecule accounts for its capacity to recognize particular antigen and to activate the system of elimination of foreign substances and some endogenous cells.
To recognize and bind each particular antigen, antibodies should pass through the affinity maturation process.
Affinity Maturation of Antibodies in vivo
Antibodies recognize millions of different antigens due to huge diversity of their binding sites. Information about different variants and antibody sequence is not originally coded in the genome; it is created in B-lymphocytes by recombination of a limited number of gene segments. In human and mouse genomes there are families of IgG genes grouped into three independent genetic loci: two of them code IgG light chains and one locus codes IgG heavy chain. Each of these families includes many genes coding variable domains and single genes coding constant domains. These groups of genes are located at significant distance from each other. In the first stage LC and HC are formed due to recombination of genes coding for one variable and one constant IgG region. The nucleotide sequences D and J located between V- and C-genes code small fragments of the polypeptide chain that are included into the variable region. B-Lymphocyte maturation is accompanied by recombination of V, D, and J genes; this leads to approaching of nucleotide sequences coding variable and constant domains and also promoters preceding each gene and enhancers located in the intron between the J- and C-segments and in the distal region after the C-genes. Thus, recombination results in formation of a transcription unit containing introns and exons. Splicing of primary RNA transcript leads to intron removal, and the exons form an mRNA molecule that will be then translated into the corresponding protein [24]. Such scenario creates molecular diversity of the recognition site located on the continuous surface formed by CDR loops. The resulting antibodies known as germline antibodies demonstrate rather low affinity and specificity to their antigens [13, 25].
During the next stage of antibody maturation the affinity and specificity of germline antibodies increase due to somatic hypermutation of the antibody variable regions. The somatic hypermutation is the machinery generating numerous antibody variants due to appearance of random point mutations in gene segments accompanied by selection of the most suitable clones. During somatic hypermutation clone selection in the immune system occurs as follows. The membrane form of immunoglobulins immobilized on the B-cell surface interacts with its antigen. The resulting antibody–antigen complex undergoes internalization followed by subsequent antigen processing, and resulting antigen-derived peptides interact with major histocompatibility complex II and are presented by T-cells. Subsequent antigen presentation by T-cells is accompanied by activation of B-cells. If affinity of antibodies exposed on the B-cell surface is not high enough the antigen dissociates from the antibody within the time interval required for internalization, and activation of this particular B-cell does not occur. If the lifetime of the antibody–antigen complex significantly exceeds the time interval required for internalization, B-lymphocyte activation occurs. In this case the immune system is unable to perform subsequent selection among several variants of high affinity antibodies. Mutations occur unevenly in both CDR-loops and FR with an average frequency of 1 per 103 base pairs during one cycle. This increases immunoglobulin diversity originated from recombination of gene segments. The resulting antibodies are known as mature antibodies [25, 26].
Studies of mechanisms of somatic hypermutation have shown that during antibody maturation variable domain mutations mainly appear in certain CDR and FR sites. These sites are called hotspots, whereas the sites rarely involved into the mutagenesis process are called as coldspots. Usually, coldspot amino acid residues coincide with the residues crucial for formation of variable domain conformation. It is known that in 50-60% of cases hotspots are located inside the consensus nucleotide sequence RGYW (R = A or G, Y = C or T, W = A or T) and serine codons AGC and AGT or neighboring nucleotides [27, 28]. Among other trinucleotides and also dinucleotides there is a hierarchy of mutations, and mutations in V-genes occur at a frequency that is a million times greater than spontaneous mutation in other genes. In CDR and FR there are differences in codon composition, and CDR contain codons in which mutations occur most frequently [25]. In the primary antibody population differences in antibody sequence are mainly located in the center of the antigen binding site, and the differences also cover the peripheral part of the antigen binding site due to somatic hypermutation [29].
During secondary immune response IgG antibodies specific to protein antigens are characterized by dissociation constant (KD) values ranging from 10–7-10–10 M–1. The maximally possible rate of antibody antigen complex formation is determined by the rate of diffusion of protein molecules. In this case the association rate constant (ka) is about 106 M–1·sec–1. The dissociation rate constant (kd) may reach up to 10–4 sec–1, and its value is determined by in vivo selection of primary antibody variants [8, 30]. According to the hypothesis of affinity ceiling [30-32] selection of antibodies with higher affinity occurs less effectively and subsequent increase in affinity does not increase selection efficiency. This may be attributed to the fact that for antibodies with KD values of 10–10 M–1 the half-lifetime of the antigen complex with the B-cell receptor is 30 min. This value is 2-3-fold higher than the time required for endocytosis of the antibody complex with the B-cell receptor (about 8.5 min). Thus, subsequent increase in affinity (half-lifetime for the complex) does not promote adaptive increase in B-cell proliferation [31, 33]. Consequently, in theory artificial mutagenesis similar to somatic hypermutation and artificial selection of antibody variants of the highest affinity can result in production of antibodies with higher affinity compared to natural antibodies.
In the case of primary antibodies their association with antigens is accompanied by an increase in enthalpy (due to numerous weak and strong antibody–antigen interactions) with simultaneous decrease in entropy (loss of mobility in the structure of the free antibody). In the case of dissociation the opposite situation occurs (enthalpy of the system decreases and entropy increases). Relative increase in affinity of the final antibody variants occurs due to removal of entropy problems by decreasing mobility of a free antibody [13]. Primary antibodies are usually characterized by higher flexibility and mobility of their antigen-binding loops. This helps them to recognize a set of similar antigens by forming different variants of complementary surfaces due to the flexible loop structure. Interaction of primary antibodies with antigens can be described by the induced fit model, when the epitope structure influences formation of a complementary paratope during the binding process. However, maturation of antibodies accompanied by increase in their affinity and specificity results in decrease in flexibility of the antigen-binding loops, and the process of antibody antigen interaction becomes increasingly similar to the lock and key model. After affinity maturation, the antigen binding site looses its mobility and remains fixed in the conformation, the best complementarity to a certain epitope. Thus, increase in antibody affinity and specificity occurs due to reduction of loop mobility in the antigen-binding site [34]. On the other hand, for some antibodies increase in conformational mobility allows increase in spatial complementarity of paratopes to the epitopes by the induced fit interaction. This effect accounts for the increase in affinity of some antibodies after introducing glycine into the CDR sequence; glycine increases conformational mobility without any significant change in the antigen interaction surface [35-37].
In most proteins conservative mutations, i.e. substitutions for amino acids with similar physicochemical properties, represent the major part of preserved amino acid substitutions; this leaves fewer chances to change the protein conformation. However, this is not always true for variable domains of antibodies because substitutions for amino acids with opposite properties sometimes result in an increase in antibody affinity. Such mutations usually occur in the CDR but sometimes in FR as well; since FR residues play an important role in formation and maintenance of stability of protein structure, their mutations may also influence antibody affinity. In general, appearance of conservative mutations is more typical for FR whereas appearance of non-conservative mutations accompanied by significant changes in physicochemical properties is more typical for CDR. In the case of amino acid residues located inside the protein molecule conservative mutations are more typical than for residues susceptible to the solvent. During antibody maturation the CDR residues susceptible to the solvent are usually substituted for hydrophobic, polar, and polarized residues, which increase interaction with the antigen. Although theoretically for each amino acid position 19 substitution variants exist, in fact the number of observed mutations is significantly lower due to energetic barriers, genetic code degeneration, and also most amino acid substitutions occur due to single nucleotide substitution. In addition, transitions (i.e. substitutions of purine base for purine or pyrimidine base) dominate over transversions (i.e. substitutions of purine base for pyrimidine or vice versa). Transitions usually occur two times more frequently than transversions, and so the number of possible variants for amino acid substitution at each position decreases to 13-15 [5, 25, 38].
Affinity of mature antibodies is determined by mutations introduced into the primary antibody sequence. Antibodies containing in the paratope many residues capable of forming hydrogen bonds and ionic interactions demonstrate high affinity to the antigen [25]. Mutations promoting antigen–antibody bond formation involving water bridges also increase antibody affinity [13]. This is also accompanied by an increase in peripheral hydrophobic and van der Waals interactions due to the increase in hydrophobic surface area. Such increase in hydrophobic interactions is a consequence of small coordinated rearrangements of amino acid residues on the periphery of the contacting surface, whereas the most energetically important interaction site remains unaltered. Somatic hypermutation not only results in an increase in affinity by amino acid substitutions at the antigen binding site, but it also increases interaction between VL-VH domains, thus increasing stability and decreasing plasticity of the antibody molecule. At the same time these changes improve antibody binding with antigen [37, 39-41]. The number of somatic mutations that appear during antibody maturation positively correlates with the increase in affinity of antibodies [13, 32, 42]. Thus, it appears that amino acid substitutions that appear during somatic hypermutation not only increase chemical reactivity of CDR residues but also maintain their structure.
Preparation and Practical Use of Antibodies
Antibodies are widely used in both scientific studies and also in practical medicine due to their ability to recognize any antigen with high affinity and specificity.
The easiest method for preparation of antibodies specific to a particular antigen is immunization, i.e. in vivo administration of the antigen in a mixture with certain additives, adjuvants that increase the immune response of the animal (mouse, rabbit, goat, sheep, etc.). Blood of the immunized animal can be used for isolation of polyclonal antibodies, a heterogeneous (by structure, epitope specificity, and affinity) population of antibodies specific to the administered antigen. Preparation of antibodies characterized by identical structure and specificity requires isolation of antibody-producing lymphocytes and their subsequent immortalization (i.e. formation of cells with countless replicative capacity). The immortalization is performed by fusion of the lymphocytes with tumor cells. The cell line representing a product of lymphocyte fusion with the tumor cells is known as a hybridoma. Hybridomas can secrete antibodies that are absolutely the same in their properties (structure, affinity, specificity). Antibodies produced by the hybridoma cell line are known as monoclonal antibodies [43].
Antibodies are one of the most widely used instruments in modern biochemistry, cytology, and clinical chemistry. They are used for qualitative and quantitative determination of various substances by the methods of enzyme-linked immunosorbent assay [44, 45] and immunoblotting [46]; antibodies can be used to study intracellular and extracellular localization of proteins and distribution and expression level of proteins in various cells of organisms by the methods of immunohistochemistry and immunocytochemistry. Antibodies can also be used for cell sorting by flow cytometry and for purification of various proteins.
In medicine antibodies are widely used in diagnostics and in therapy of various diseases. In diagnostics various immunochemical methods are used for detection of the presence of proteins or determination of protein concentration. The list of such proteins is very long and is constantly increasing. It includes protein markers of cardiovascular diseases (troponins I and T [47-51], creatine kinase [52], myoglobin [53], B-type natriuretic peptide and its precursor [54-56], etc.), diabetes mellitus (glycated hemoglobin [57], microalbumin, insulin, proinsulin, C-peptide [58], etc.), cancer diseases [59], pathologies associated with pregnancy, renal disease (cystatin C [60]), autoimmune diseases (antibodies to endogenous cell components of the body [61]), allergies (total antibody content, content of antibodies to particular antigens), endocrine system disorders (hormones, their precursors, products of their metabolism and proteins interacting with them [62, 63]), various infectious diseases [64-67], etc.
Antibodies are also used for determination of blood groups [68], diagnostics of pregnancy [69], and detection of drug or toxin content in human blood and also in foodstuffs.
Therapeutic use of full-length antibodies obtained from serum of immunized animals or using hybridoma technologies is significantly complicated by the fact that the constant domains of animal antibodies are strongly immunogenic for humans, and this causes production of endogenous antibodies specific to animal antibodies [6]. Thus, for therapeutic purposes recombinant antibodies or their fragments are used; they are obtained from monoclonal antibodies with significantly reduced immunogenicity (by means of gene engineering manipulations) or prepared de novo using various display methods (considered below in the section “Practical Application of Recombinant Antibodies in Clinical Practice and Scientific Studies”).
RECOMBINANT ANTIBODIES: DERIVATIVES AND FRAGMENTS, METHODS FOR
THEIR PREPARATION, AND PRACTICAL IMPORTANCE
Recombinant antibodies are antibodies obtained by means of gene engineering. Using gene engineering methods it is possible to express light and heavy chains of immunoglobulins as individual proteins, to create the whole set of various antibody fragments, and also to change such antibody properties as affinity, number and specificity of paratopes, domain composition, molecule mobility, spatial orientation of antigen binding sites, molecular weight, isoelectric point, and potential immunogenicity.
Types of Recombinant Antibody Fragments
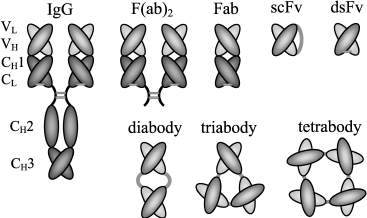
Preparation of several different types of recombinant antibody fragments has been described in the literature (Fig. 2 shows some of them). The following types of recombinant antibodies are now recognized: Fv, variable fragment; scFv, single chain Fv in which light and heavy chain fragments are connected by a polypeptide linker; (scFv)2, fragment that consists of two scFv molecules connected by a disulfide bond; dsFv, variable fragment stabilized by additional intramolecular disulfide bond; Fab- and (Fab)2, fragments identical in their structure with fragments formed during proteolysis of full-length IgG antibodies by papain and pepsin, respectively [70]; VH, heavy chain variable domain. There are antibody derivatives known as “diabodies”, “triabodies”, and “tetrabodies”; their molecules consist of two, three, or four identical or different fragments of antibodies of the same or different specificity linked together. The Fv fragment is the smallest antibody fragment that retains antigen binding capacity. It consists of variable fragments of light (VL) and heavy (VH) chains, each of which is stabilized by an intramolecular disulfide bond. However, contacts between VH and VL involve only ionic interactions and therefore the structure of this fragment is very unstable. Structural stability of Fv fragments can be achieved by either insertion of interchain disulfide bonds (dsFv) or a linker amino acid sequence between variable domains of the HC and LC chains (scFv), and the resulting antigen-binding protein molecule is expressed as a single polypeptide chain [71-73].
The scFv fragments are the “simplest” object for cloning and expression; however, they frequently exhibit lower stability and, sometimes, lower affinity; during therapeutic use they are characterized by shorter half-lifetime in the blood circulation compared with larger fragments [37, 74-78]. The complexes consisting of two (“diabodies” or (scFv)2), three (“triabodies”), and four scFv-monomers (“tetrabodies”) often demonstrate even higher affinity to the antigen due to increased number of antigen-binding sites. This occurs when scFv-monomers are derived from identical antibodies and the antigen contains many identical epitopes, or in the case when the scFv-monomers are derived from different antibodies to the same antigen and all antigen epitopes are susceptible to them. The (scFv)2 fragments obtained by disulfide bond linking between C-terminal cysteine residues demonstrate better in vivo transport into tissues compared with the scFv-fragments during their use as therapeutic agents. This occurs due to longer half-lifetime in the bloodstream and higher structural stability compared with scFv. Recombinant antibody derivatives containing the Fc-fragment are characterized by even longer half-lifetime; the long-term presence of such antibody derivatives in tissues is determined by interaction with FcRB receptors (Fc Brambell receptor) located on the surface of hemopoietic and endothelial cells [79, 80].Fig. 2. Main types of fragments and derivatives of recombinant antibodies. Designations: VL, LC variable region; VH, heavy chain variable region; CL, constant region of light chain; CH1, CH2, and CH3 are constant regions of heavy chain; IgG, full length immunoglobulin G. Light gray contour lines show disulfide bonds between various chains, dark gray lines show linker sequences inserted for stabilization of the recombinant antibody structure (prepared using [74, 75]).
Preparation of Recombinant Antibodies and Their Fragments
There are two principally different approaches for preparation of recombinant antibodies. The first consists in generation of recombinant analogs of monoclonal antibodies or their fragments using genetic materials from hybridoma cells producing monoclonal antibodies. The second approach is based on the development of libraries containing numerous variants of recombinant antibody sequences followed by subsequent selection of antibodies of required specificity and affinity, i.e. de novo formation of antibodies of necessary specificity.
Creation of recombinant analogs of antibodies. The method of preparation of recombinant antibodies or their fragments identical to antibodies obtained by the standard hybridoma method includes the following main stages [81-84]: 1) isolation of total RNA from hybridoma cells secreting antibodies; 2) first strand cDNA synthesis (reverse transcription); 3) amplification of heavy and light chain cDNAs by polymerase chain reaction [80]; 4) preparation of molecular-genetic constructs containing resulting cDNA; 5) expression (or coexpression) in a suitable expression system [85]; 6) isolation and purification of the expressed protein [85]. Using this method it is possible to obtain both full-length antibodies and their fragments and also antibody light and heavy chains as individual proteins. Use of gene engineering methods for antibody preparation allows manipulations with nucleotide and consequently amino acid sequence of light and heavy chains.
Creation of recombinant antibodies and their fragments de novo. The method for preparation of recombinant antibodies or their fragments in vitro using selection in antibody fragment libraries for antigen binding capacity is the alternative approach for preparation of recombinant antibodies [86]. Methods used for solution of this problem are known as display methods. The advantage of these methods is the possibility of simultaneous work with nucleotide and amino acid sequence of each particular antibody variant. Preparation of recombinant antibody fragments using this approach includes three steps: creation of recombinant antibody DNA libraries, expression and presentation of antibody fragments on a cell surface or phage particles (depending on the display method used), and selection of suitable antibody fragments by their interaction with the antigen. If a full-length antibody specific to a corresponding antigen is needed, initially an antibody fragment is prepared and then used as a basis for reconstruction of full-length nucleotide and amino acid sequences of this antibody.
The display principle consists of the use of nucleotide sequence of the target protein and product of its expression in a single molecular complex. Using this approach it is possible to select necessary genetic material (DNA or RNA) by ability of the product of its expression (immunoglobulin or its fragment) to interact with this antigen. Using several repeated selection cycles it is possible to obtain antibodies demonstrating high affinity interaction with the antigen. Several display methods for preparation of antibodies are now known: phage, ribosomal, mRNA, and cell display [9, 87-95]. All these approaches can be used not only for creation of antibodies de novo, but also for improvement of parameters of exiting antibodies (see section “Increasing Antibody Affinity”).
Antibody fragments rather than full-length antibodies are usually the basis for creation and testing of antibody libraries. Since scFv fragments are the smallest stable antibody units that can bind antigens, it is much easier to use them to increase affinity in each type of display than full-length antibodies or Fab-fragments. There are several principles for creation of libraries of recombinant antibody fragments [77]: 1) on the basis of antibody genes obtained from immunized donors (library enriched with antibodies to certain antigens); 2) on the basis of primary antibody genes (see section “Affinity Maturation of Antibodies”); 3) on the basis of nucleotide sequence of antibodies obtained from B-lymphocytes [96]; 4) on the basis of sequences of the most stable variable antibody domains with randomized residues in artificially synthesized CDR-loops [97].
The display technology is used for selection of antibodies with needed properties from the huge number of variants (108-109) available in the library. The common feature of all these methods is the joining of nucleotide and amino acid sequences of the selected antibody fragment in a single object, and their difference consists of the type of such object: from the mRNA complex with the antibody fragment synthesized on this template to the cell carrying the antibody fragment on its surface. Although such recently developed methods as cell display and ribosomal and mRNA displays are becoming increasingly popular, phage display still remains the most widely used and the simplest method.
Cell display. The cell display method uses various prokaryotic or eukaryotic cell systems for expression of various antibody variants and also for selection of the expressed antibodies by certain characteristics. In this case the created library of hybrid DNA simultaneously contains nucleotide sequences of antibody fragments and a transmembrane domain of one of the membrane proteins, and the antibodies synthesized after cell transformation (or transfection) by such library represent hybrid proteins consisting of a native antibody fragment exposed to the extracellular space and the transmembrane domain responsible for anchoring the antibody fragment in the membrane. Thus, the cell functions as a link between the expressing antibody variant and the genes that code it. Clones with the highest affinity are selected by a variant of flow cytometry known as fluorescence activated cell sorting (FACS) [90, 95]. This method is based on the sorting of cells containing antibody fragments of their surface together with a fluorescently labeled antigen. Differences in fluorescence intensity emitted by cells distinguish variants possessing affinity to antigen from variants not capable of antigen binding, and they serve as the criterion for sorting of the material.
There are prokaryotic and eukaryotic displays. The prokaryotic display is characterized by high transformation efficiency and simple use, but it has limited applicability because some antibody fragments containing potential glycosylation sites are functionally inactive when synthesized in the bacterial system. Several transmembrane proteins have been proposed for anchorage of synthesized antibody molecules on the surface of bacterial cells. These transmembrane proteins are used for creation of chimeric proteins containing antibody fragments. For example, the outer membrane protein OmpA is often used for anchorage of the antibody scFv fragments [90, 91].
Using the eukaryotic display it is possible to express both various antibody fragments and full-length antibodies because a protein molecule synthesized in such system has all eukaryotic-specific posttranslational modifications needed for functioning. The yeast display is one of the most widely used versions of the eukaryotic display because yeast cells are the simplest ones for gene engineering manipulations and cultivation. Recombinant antibody fragments are exposed on the yeast cell surface as a part of hybrid proteins that consist of the antibody or its fragment and the Aga2p agglutinin subunit, which is covalently linked by means of two disulfide bonds with Aga1 agglutinin anchored in the yeast cell wall [92-95]. A similar strategy has also been developed for antibody expression in mammalian cells [94].
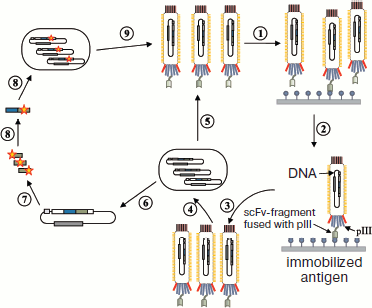
Phage display. In the phage display method [87] a gene of the hybrid protein joining an antibody fragment and a phage surface protein is inserted into the bacteriophage genome. Ig fragments are exposed on the surface of viral particles (Fig. 3; see color insert). Phage particles are selected by affinity of antibody fragments to the antigen. Usually the phage M13 is used for preparation of the phage display, and antibody fragments are expressed as hybrid proteins with the phage envelope proteins pIII, pIV, and pVIII [80]. Selection of phage particles is then accompanied by phage cDNA isolation and determination of nucleotide sequence of antigen fragments. This sequence is used for subsequent generation of full-length recombinant antibodies [8, 9, 87, 98, 99].
Ribosome display. Ribosome display is another display method [80, 88]; it uses a cell-free system for synthesis of a polypeptide chain on the mRNA template. Protein synthesis in this system is accompanied by formation of the ternary protein–ribosome–mRNA complex (Fig. 4). This complex is then isolated from solution using capacity of the synthesized antibody fragment to bind the target antigen. Using this method it is possible to select simultaneously the highest affinity antibody fragments together with their mRNA. In this case a ribosome functions as a stabilizer of the complex. The mRNA is then subjected to reverse transcription; resulting cDNA is amplified by PCR and the resulting PCR products are used for plasmid construction for recombinant antibody fragments. Prokaryotic and eukaryotic displays have been described in the literature [80].Fig. 3. Increasing antibody affinity using the phage display method. A library of 108-109 variants of antibody fragments (usually antibody scFv-fragments are used) exposed on the surface of M13 phage particles is incubated with an immobilized antigen (1). Several washing steps remove low affinity phage particles (2), and then remaining bound phage particles are eluted (3). Particles selected after the first cycle are amplified by infecting E. coli cells (4) for generation of a library enriched with variants of antibody fragments demonstrating high affinity interaction with the antigen. This cycle is repeated from two to four times (5) until domination of the fraction of high affinity antibodies. Affinity of selected antibodies can be further increased by isolation of their genes from phage particles (6) and creation of new variants using the mutagenesis method (7). The resulting genes are cloned into plasmids (8), which are used for transformation of E. coli cells (8); this yields a new library of antibody fragments exposed on the surface of phage particles (9); this library can be used for the next selection (1) and subsequent increase in affinity. The use of phage display results in selection of high affinity antibodies, and their genes can be isolated from phage particles (6) for subsequent cloning and expression (adapted from [117]).
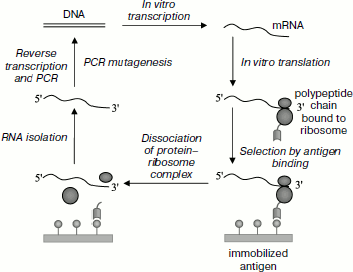
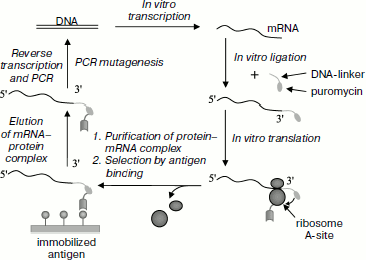
mRNA display. The mRNA display method (also known as “in vitro virus” or “RNA–peptide fusion”) is also used for preparation of recombinant antibodies [89, 100]. The strategy of this method is based on the linkage of in vitro synthesized polypeptide chain with its mRNA template by means of the puromycin linker, which ligates two of these molecules. Thus, corresponding mRNA can be selected by immunochemical properties of the synthesized protein product (Fig. 5). Subsequent steps are basically the same as described in the “Ribosome Display” section.Fig. 4. Ribosome display. A library of antibody genes (e.g. scFv fragments) is transcribed and translated in vitro. The resulting mRNA lacks a stop codon and this preserves the ribosome–protein complexes, which are subjected to subsequent selection using the immobilized antigen similarly to selection of the phage display. The selected complexes are used for preparation of mRNA used as a template for reverse transcription and PCR. Using point substitutions inserted into the nucleotide sequence, it is possible to create new variants of antibody fragments that can be used in the subsequent cycle. A DNA molecules coding high affinity variants of antibodies selected during several display cycles can be obtained by the method of reverse transcription of mRNA molecules and PCR and used for subsequent cloning and expression (summarized from [102, 213]).
Thus, one can see that there are many methods for selection and preparation of antibody variants of the highest affinity using gene libraries. The phage display method is now most widely used. However, later approaches employing in vitro systems of protein translation, which do not require transformation (mRNA and ribosome display), can be used for screening of larger libraries than in the case of phage display [75, 101, 102]. One limitation of display methods is the irregular expression of various forms of antibodies. Sometimes even the presence of a single amino acid substitution blocks expression of the mutant form of the antibody [103]. In addition, in the case of libraries containing a large number of variants (e.g. 1010_1013) there are problems associated with impossibility of analyzing each variant. This should be taken into consideration during creation and use of such libraries.Fig. 5. Method of mRNA display. A library of antibody genes (e.g. scFv fragments) is transcribed in vitro. Using transcribed mRNA ligation with puromycin, it is possible to create mRNA–synthesized protein complexes using a DNA-linker. Terminating in vitro translation the ribosome stops at the site of DNA–RNA linkage, puromycin binds to the ribosomal A-site, and then the synthesized polypeptide chain is translocated to puromycin. The resulting covalent mRNA–protein complex can be used for affinity selection similar to the phage display selection and then (after elution) for reverse transcription and cDNA amplification by PCR. Diversity of sequence variants can be achieved in cDNA by mutagenesis (adapted from [102]).
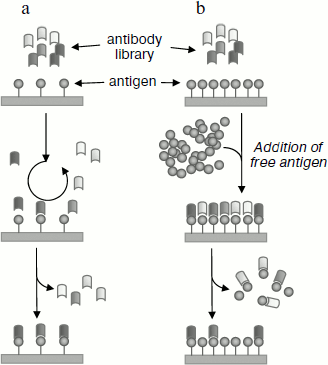
Methodological approaches for selection of antibody variants of highest affinity using antigens. Preparation of high affinity antibodies requires discrimination between high and low affinity variants. This can be achieved using methods of selection based on kinetic or thermodynamic parameters of antibodies (Fig. 6) [102, 104]. For selection by thermodynamic parameters (under equilibrium state) a library of antibodies is incubated with an antigen used at lower concentrations than a designed KD value for the antigen–antibody complex. After reaching equilibrium in this system antibodies of higher affinity remain preferentially bound to the antigen, whereas antibodies of lower afinity are unbound. Subsequent sorting of the resulting immune complexes yields antibodies of the highest affinity. However, this method requires prolonged incubation (up to one week!) to achieve equilibrium, and so only small amounts of antibodies with improved properties can be found. Selection by kinetic parameters [94, 105, 106] (“off-rate selection”) consists of incubation of an antibody library with a small amount of labeled (or adsorbed) antigen whose concentration is close to the KD value of the analyzed antigen–antibody complex. During the next step this mixture is incubated with excess of the free antigen, and duration of incubation increases at each subsequent step of selection. Addition of free antigen excess makes dissociation of the tested antibodies from labeled (or adsorbed) antigen almost irreversible. Weakly bound antibody molecules characterized by the highest rate of dissociation form a complex with the free antigen and then are removed during sorting, and the antibody molecules characterized by the lowest dissociation rate remain bound to the (labeled or adsorbed) antigen and are then selected for subsequent manipulations.
Expression of recombinant antibodies. Depending on experimental design, recombinant antibodies and their fragments can be expressed in various systems: bacterial (E. coli), yeast (Pichia pastoris), baculovirus (sf9 cell line), and in mammalian cells (e.g. CHO and HEK293 cell lines) [107]. The bacterial and yeast expression systems are used for expression of small nonglycosylated antibody fragments (VL, VH, scFv, dsFv) and also Fab-fragments. In the case of full-length glycosylated antibodies, mammalian cells and baculovirus expression systems are used most frequently. High yield of recombinant fragments (20-1000 mg/liter culture medium) is often achieved using bacterial and yeast expression systems. However, expression may be accompanied by appearance of various problems complicating preparation of functional antibody fragments: 1) formation of incorrect disulfide bonds followed by accumulation of functionally inactive target protein and oligomers of synthesized fragments; 2) loss of functional activity caused by mutations; 3) proteolysis of the synthesized protein by intracellular peptidases. Although in the case of eukaryotic and baculovirus systems the yield of the target product is somewhat lower (1-10 and 10-100 mg/liter, respectively), almost all the synthesized target protein is functionally active.Fig. 6. Two strategies for selection of antibodies with the highest affinity (see explanations in text). a) Equilibrium selection. Antibodies are incubated with a limited amount of an antigen. After reaching equilibrium, antibodies of higher affinity remain preferentially bound to the antigen. b) Selection by kinetic parameters. Antibodies are incubated with sufficient amount of the immobilized antigen and then the free antigen is added. Only antibodies possessing the lowest dissociation rate (and therefore higher affinity) remain bound to the antigen.
Thus, using the above-considered methods it is possible to prepare recombinant antibodies or their fragments suitable for all scientific and applied tasks.
Practical Use of Recombinant Antibodies
Advantages of recombinant antibodies and their fragments over monoclonal antibodies significantly extend field of their application, first of all due to the possibility of their use as therapeutic agents for treatment of various diseases.
As mentioned above, the possibility of modification of amino acid sequences and introduction of various changes needed for particular tasks are important advantages of antibodies obtained using gene engineering technologies. Using gene engineering methods it is possible to overcome such unwanted features of monoclonal antibodies complicating their use in clinical practice as increased immunogenicity. To decrease immunogenicity of monoclonal antibodies sites inducing immune response (these are usually constant domains) are replaced by corresponding sequences of human antibodies. Such antibodies are known as “chimeric”, and they preserve antigen specificity due to their unique variable domains. Subsequent decrease in immunogenicity is achieved by preparing humanized (almost totally replaced) antibodies, which contain only sites of mouse antibody variable domains directly involved in the interaction with the antigen, and the remaining parts of the mouse antibody are substituted by sequences of human immunoglobulins [14].
Thus, the antibody modifications described above have significantly extended their applicability in clinical practice. Therapeutic use of antibodies includes treatment of cancer, autoimmune, respiratory, cardiovascular, infectious, and many other diseases. Therapeutic antibodies are also used to decrease probability of transplant rejection. Use of recombinant antibody fragments is now successfully developed [108, 109]. Certain difficulty complicating the use of therapeutic antibodies is changeability of molecular targets, such as viral and bacterial proteins, and also the possibility of immune reaction, which can develop in a patient even in response to almost totally humanized antibodies.
Improvement of recombinant antibodies for scientific tasks can be achieved by changing affinity and specificity of antibodies; it is possible to insert additional amino acid residues into an antibody sequence for subsequent conjugation with other molecules (including labels) and also add short amino acid fragments (tags) to the antibody sequence, which significantly simplify antibody purification [80, 110]. In addition, gene engineering technologies create antibodies to antigens that are not immunogenic or are toxic for animals and therefore difficult to produce using the hybridoma technology.
One of the most promising directions in the development of technology of recombinant antibody preparation is the preparation of antibodies of higher affinity than in initial natural antibodies. In the next chapter we consider in detail modern methods developed for artificially increasing antibody affinity.
INCREASING ANTIBODY AFFINITY
Increase in antibody affinity is a natural process associated with maturation of antibodies in vertebrate organisms. Studies of mechanisms of this process revealed the main principles applicable for artificial changes in antibody sequences required for preparation of variants of higher affinity.
Natural approaches for increasing antibody affinity (recombination of gene segments and somatic hypermutation) have some limitations. Recombination of gene segments is limited by a set of existing gene segments used for construction of the particular antibody gene, somatic hypermutation mainly involves hotspots of mutagenesis, and single nucleotide substitutions dominate in this process. Thus, parts of antibody sequences remain basically unchanged, and some types of mutations that need more than one nucleotide substitution in the codon become significantly less probable [33]. Artificial mutagenesis increases the number of antibody variants compared with natural variants, for example, by substituting those amino acid residues that are rarely mutated in vivo [111]. Using artificial mutagenesis it is possible to perform rare (under natural conditions) amino acid substitutions determined by changes of more than one nucleotide in the codon. Using an effective method of selection, it is possible to obtain recombinant antibodies with affinity exceeding the “affinity ceiling” values for natural antibodies and dissociation constant values reaching 10–10 M and below.
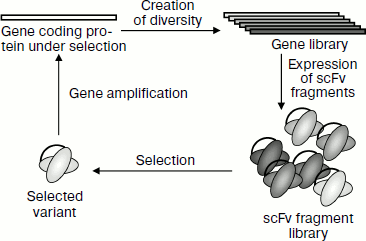
Although many different software products exist for computer-aided modeling of antibody–antigen interaction, it is very difficult (basically impossible) to predict what mutation or several mutations will result in an increase in antibody affinity. Thus, it is often necessary to obtain and test many various mutants. This can be carried out using the method of “in vitro evolution” [7, 101, 102], which is also based (as natural increase in affinity) on the principle of creation of diversity followed by subsequent selection of antibody variants of the highest affinity. According to this principle, antibodies of the highest affinity are selected and their genes are amplified, and then they are subjected to mutagenesis and selection and this whole cycle repeats (Fig. 7). Thus, for increase in antibody affinity its phenotype should be physically imposed to the genotype. Mutant antibody forms of the highest affinity are selected using the same display methods as in the case of preparation of recombinant antibodies de novo.
Systems for increasing antibody affinity are subdivided into “all in vitro” approaches, which are based on all steps of “molecular evolution” in vitro (creation of genetic diversity, expression of all possible variants, selection of the best variants), and “partially in vitro” approaches in which some of the above-mentioned steps occurs inside cells and some in vitro [101, 102]. The “all in vitro” approaches include ribosome display [80, 88] and mRNA-display [89, 100]. The “partially in vitro” approaches include phage [87] and cell [90-95] displays, which require the step of transformation of antibody gene library (inserted into a plasmid or a phage replicon) in bacterial, yeast, or mammalian cells for expression. In the case of the cell display the expressed antibodies are transported to the cell surface, and in the case of the phage display bacterial cells are used for production of bacteriophage particles [103].Fig. 7. Scheme illustrating increasing antibody affinity by the method of “in vitro evolution” using the example of creation of the scFv fragment (adapted from [101]).
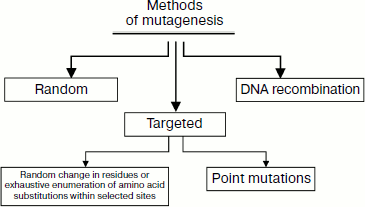
During the last 20 years many different methods have been developed to create genetic diversity in vitro; they can be subdivided into the methods of random and targeted mutagenesis. The first group includes various mutagenesis methods that are used in the case when clear information about amino acid residues that should be substituted is absent, and also the methods of DNA recombination. The second group includes site-directed mutagenesis and also methods that can predict antigen structure and its complex with the antibody; these are computational methods of protein structure analysis (in silico) and crystallography analysis (Fig. 8).
Methods of Random MutagenesisFig. 8. Classification of mutagenesis methods used to increase antibody affinity. All mutagenesis methods can be subdivided into three groups: site-directed mutagenesis, random mutagenesis, and recombination. Random mutagenesis consists of random change of any amino acid residues in the antibody sequence. The characteristic feature of site-directed mutagenesis is that it is limited to a number of amino acid positions. Site-directed randomization generates random substitutions within selected amino acid positions. The other variant of targeted mutagenesis is exhaustive enumeration of amino acid substitutions within selected sites. Point mutagenesis is used for insertion of substitutions of selected amino acids. Using DNA recombination, it is possible to increase diversity of libraries obtained by means of site-directed and random mutagenesis and also to pool several “useful” mutations within the same amino acid sequence.
The major advantage of random mutagenesis is the following: this method does not need any information about antibody structure and interacting amino acid residues. The main disadvantage is the appearance of a significant proportion of functionally inactive mutant forms; this results in creation of large libraries and complicates selection of functionally active mutants. Experimental libraries obtained by random mutagenesis do not represent all possible variants of amino acid substitutions because most of them require changes in more than one nucleotide in a codon; for example, substitution of 10 of 18 amino acids lacking negative charge for negatively charged amino acid requires substitution of two nucleotides in a codon [112].
Diversity of variants of nucleotide sequences of antibodies obtained by random mutagenesis can be additionally extended by the combination of several sequence variants obtained using in vitro DNA recombination (DNA-shuffling) (see below) [99, 113-115].
Modification of antibody amino acid sequence to increase affinity may also result in such unwanted side effects as change in antibody specificity (cross-reactivity), stability, effectiveness of expression and folding, toxicity to producing cells, and tendency to oligomerization. In the case of therapeutic antibodies an additional side effect may also include reduced half-lifetime, increase in immunogenicity, altered tissue distribution of antibodies, and tendency to amyloid structure formation. All these facts can be a consequence not only of “favorable” mutations specially introduced by a researcher, but also of unwanted mutations occurring due to errors of PCR reaction or induced by primers. Thus it is necessary to test selected antibodies with increased affinity for maintenance of their other important biochemical properties [116, 117].
Random mutagenesis. Random mutagenesis is a random change of any amino acid residues over the whole antibody sequence. Its major advantage is the possibility of its use when primary data on the structure of the complex of interacting molecules is absent. However, success of this approach requires mutagenesis frequency control, because most amino acid substitutions are not favorable, and at a high rate of mutagenesis they may result in appearance of a large number of functionally inactive antibody variants. Introduction of two mutations per 1000 base pairs is considered as optimal for the increase in scFv affinity; in this case the highest number of point (33%) and double (22%) mutations per gene and the minimal number of genes (22%) lacking mutations were observed [118].
Several approaches are used to perform random mutagenesis. One of them known as the error-prone PCR consists of introduction of amino acid substitutions caused by inaccurate functioning of DNA-polymerases under nonstandard conditions of PCR [92, 119, 120]. An important advantage of this approach is the possibility of mutagenesis frequency control and limitation of a mutation region by the size of the amplified fragment. Changes in several parameters of PCR can increase the number of incorrectly inserted nucleotides. These include a certain concentration of bivalent cation and deoxyribonucleotide triphosphate ratio in the reaction mixture and also the number of PCR cycles. The increase in Mn2+ and Mg2+ in the reaction mixture for PCR increases the number of mutations during amplification. Addition of 0.5 mM Mn2+ (final concentration) causes a 5-fold increase in the number of mutations without any influence on PCR effectiveness, whereas addition of Mg2+ not only increases the number of errors but also decreases effectiveness of amplification during PCR. The ratio of deoxyribonucleotide triphosphates in the reaction mixture is the other factor increasing the number of incorrectly inserted nucleotides. Increased content of one of them and inclusion of nucleotide analogs (e.g. 8-oxo-dGTP and dPTP) can also result in a number of incorrectly inserted nucleotides [121]. Probability of appearance of wrong nucleotides at a low mutagenesis rate is also proportional to the number of PCR cycles [99, 118]. The other approach used for appearance of random mutations is the use of so-called “mutator strains” of E. coli cells providing accumulation of mutations due to switched off mechanisms of correction of synthesized DNA [122, 123]. The mutD5 strain is one such strain; it has a deficient DNA polymerase III ε-subunit, which normally possesses 3′-5′-exonuclease and removes incorrectly inserted nucleotides. The rate of mutagenesis in the bacterial “mutator strains” depends on conditions of bacterial growth: in an ordinary medium containing minimum nutrients the mutagenesis rate is about 50-times higher than in the wild type strain, and in enriched media this rate may be increased by 103-105 times. Among appearing mutations transitions dominate, but transversions and frame shifts are also possible. The rate of mutagenesis of certain nucleotides strongly depends on the nucleotide environment. Mutagenesis of antibody sequences shares similarity with somatic hypermutation. First, mutagenesis frequency is similar to that seen in somatic hypermutation (3-5 mutations per 104 base pairs); the ratio of various mutation types is similar to the ratio observed in somatic hypermutation; third, mutations are irregularly distributed over the sequence, and many mutations are located near certain consensus sequences (“hotspots”) such as the AGY-motif [122]. The advantage of this approach over PCR-based approaches is the possibility of more convenient control of the mutagenesis rate and also lack of necessity of many gene engineering manipulations. Disadvantages of this method include the need for several rounds of mutagenesis and also constant control of antibody expression, because appearance of mutations may occur at any site of the vector sequence and stop antibody expression [124].
Increasing antibody affinity by the recombination method. Recombination of DNA sites carrying antibody genes is often used to increase the number of variants obtained by random mutagenesis and for preparation of combinations of several mutations. Depending on types of combining sites, there are recombination of heavy and light chain variable domains (chain shuffling), CDR loops (CDR shuffling), and also combinations of random homologous fragments of sequences of several mutant antibody variants (DNA shuffling) [125].
There are several methods for antibody recombination. One is based on fragmentation and partial degradation of a pool of DNA molecules coding amino acid sequences of several antibody variants by DNase I endonuclease followed by subsequent PCR to obtain hybrid DNA molecules (“sexual PCR”, DNA shuffling). This procedure consists of three steps. The first step includes preparation of a set of random fragments of two or more parent sequences, which are then subjected to PCR in the absence of primers. During this stage recombination occurs due to annealing of partially complementary fragments acting as primers. The final step consists of PCR using primers limiting the initial gene to obtain a reasonable number of full-length molecules from the pool of DNA fragments [111, 114, 126]. Another method is overlap extension PCR [127], which can be used for shuffling of both various combinations of light and heavy chains and various CDR sites [125]. The other PCR-based approach consists of so-called “StEP” amplification (staged extension process): in this case complementary DNA synthesis (elongation) is periodically interrupted by denaturation of double-stranded DNA followed by formation of DNA duplexes (annealing) between an under-synthesized DNA fragment and a new template molecule, and then chain elongation continues. This results in formation of DNA strands composed of sites complementary to various DNA templates [128]. There is a method used for recombination that includes treatment of a pool of DNA molecules carrying antibody genes with the same set of restriction endonucleases followed by subsequent ligation of the resulting DNA fragments. In this case the order of DNA fragments in the resulting molecular genetic constructs is preserved due to unique features of the restriction sites, but newly formed constructs represent a product of pooled fragments formed from several DNA molecules. It should be noted that DNA recombination can also be used for combining several mutant variants obtained by the methods of targeted mutagenesis (see below).
Although the random approaches for creation of genetic diversity are still used, now the technology for increasing affinity is intensively developed towards creation of genetic diversity by means of targeted mutagenesis methods.
Targeted Mutagenesis
Approaches for selection of potential targets for mutagenesis. Application of point mutagenesis requires information about amino acid residues that need to be mutated. Several approaches are used for selection of mutagenesis sites. For example, this may be information about a position of mutagenesis hotspots in the antibody sequence. Data on three-dimensional structure of the antibody–antigen complex is especially valuable for selection of sites for targeted mutagenesis.
Data on three-dimensional structure of antibodies and their complexes with antigens can be obtained using experimental approaches (X-ray analysis or nuclear magnetic resonance) or by means of in silico modeling (if X-ray data are not available) [129]. Analysis of three-dimensional structure has some advantages compared with antibody sequence analysis because it can predict which amino acid modification would increase antibody affinity (taking into consideration contribution of individual amino acids into maintenance of stability of the antibody structure and interaction with antigen). Use of antibody sequence analysis and analysis of three-dimensional structures and antibody–antigen complexes followed by subsequent targeted mutagenesis (rational design) has some advantages compared with random mutagenesis. First, this approach significantly reduces the number of amino acid residues subjected to mutagenesis to several amino acid positions and simultaneously increases probability of detection of favorable mutations. In addition, the process of targeted mutagenesis can be subdivided into several parallel testings of small libraries. A small size of these libraries (compared with the size of libraries obtained during random mutagenesis) significantly reduces the number of tested antibody variants and significantly decreases risk of false negative result, i.e. risk of missing of an improved variants due to insignificant completeness of the library or due to impossibility of effective screening of all existing variants by the display technology [130].
There are two factors limiting the rational design methods. The first factor is impossibility of absolutely accurate prediction of the effects of even single amino acid substitutions on mutant affinity by means of currently existing calculation algorithms. However, in some cases accuracy of predictions of consequences of possible rearrangements by the method of in silico modeling is reasonably high, and good examples of design of higher affinity antibodies based on prediction of changes in existing antibodies have already been reported [131, 132].
The second limiting factor is the necessity for validation of an astronomically high number of possible mutations [133]. For consideration of all possible amino acids at each position (saturating mutagenesis) of the sequence of LC and HC variable domains (about 230 residues) testing of 10300 antibody variants is needed, and this is technically impossible.
In silico approaches. Although results obtained using experimental approaches are more reliable, harder work is needed compared with in silico analysis. Pilot simplification by simulating mutation consequences predicted by in silico analysis may significantly facilitate subsequent experimental manipulations. It should be noted that computer-aided approaches cannot fully substitute for experimental work, but they can significantly decrease the number of variants for direct experimental search for favorable mutations [134]. Thus, the major task of in silico analysis in terms of increasing antibody affinity is the study of the effects of amino acid substitution on the antibody structure and stability of the immune complexes for detection of substitutions that are suitable for increasing affinity during antibody–antigen interaction.
Sequence prediction of unfavorable mutations. In the absence of information about three-dimensional structure of the antibody of interest and its amino acid residues involved in interaction with a particular antigen, it is possible to reduce significantly the number of residues that have to be mutated to increase antibody affinity: in silico analysis of antibody sequence can exclude high probability mutations of residues that will impair antibody structure. These include FR residues, conservative CDR residues, and also residues with side chains located inside domains. Computer modeling methods employ various approaches for prediction of mutation effects in the antibody sequence; they can use various input data for subsequent analysis from amino acid sequence to atomic coordinates in a three-dimensional structure. They can combine available information to increase prediction accuracy (methods used in programs SNPs3d [135], SIFT [136], PolyPhen [137-140], and SNAP [141]). Some programs can evaluate the energetic effect of amino acid substitution on stability of antibodies and their complexes with antigens (programs FOLD-X [142], CHARMM [143-145], SNPeffect [146], LS-SNP [147], I-Mutant2.0 [148], and PoPMuSiC [149]). Thus use of in silico approaches for antibodies with unknown spatial structure significantly decreases the size of screening libraries due to exclusion of potentially unfavorable mutations from the list of generated mutations.
Identification of hotspots for mutagenesis. The method of hotspot mutagenesis is also used to increase antibody affinity. It is based on the in vitro use of the same principle that is used for natural increase in antibody affinity during somatic hypermutation. As in the case of the search for unfavorable mutations, the search for hotspots for mutagenesis involves antibody sequence analysis.
The localization of amino acid residues corresponding to hotspots for mutagenesis (i.e. nucleotide sequences that are changed during somatic mutation most frequently) consists of three steps: 1) search of V gene regions corresponding to variable domains of the analyzed antibody; 2) search for consensus sequences typical for hotspot mutagenesis in the V gene sequences; 3) identification of residues corresponding to hotspots for mutagenesis in the particular antibody. For this purpose the search of V gene sequences closely related to variable domain sequences of this antibody is performed using the IGMT database [150]. In the found V gene sequences all sites corresponding to the consensus sequences RGWY (A/G G C/T A/T) and also AGC and AGT should then be localized [27, 28]. During mutagenesis of CDR-H3 residues one should avoid hotspots containing nucleotides added during recombination of gene segments (V(D)J recombination) in the process of antibody gene generation.
Two types of hotspots are recognized in nucleotide sequence: germline hotspots (primary) contained in primary antibodies and gene segments from which these antibodies have been obtained, and non-germline hotspots (secondary) that are absent in primary antibodies and obviously represent a result of somatic hypermutation [27, 28, 33, 151-153]. Results of mutagenesis studies using these two types of hotspots have shown that changes in primary hotspots are much more effective for increasing antibody affinity [33]. During mutagenesis it is necessary to randomize all codons involved (partially or totally) in the selected hotpots [27].
In silico modeling of three-dimensional structure of antibody–antigen complex and in silico mutagenesis. Generation of three-dimensional structure of an antibody and its complex with the antigen represents a much more difficult task compared with sequence analysis. Increasing antibody affinity by in silico methods is performed by predicting mutations that would increase: a) the number of antibody residues interacting with the antigen; b) strength of antibody–antigen interaction. Such changes usually result in an increase in antibody affinity due to the decrease in dissociation (koff). Increase in association rate may be achieved using an approach based on changes in the surface charge of the antibody molecule due to insertion of charged residues near interacting surfaces. It is believed that appearance of such residues in the antibody structure facilitates correct orientation of the antigen molecule versus the antibody without formation of new bonds between the antibody and the antigen [154, 155].
Existence of three-dimensional structure of the antibody–antigen complex obtained at high resolution is an important precondition for increasing antibody affinity. However, for most antibodies their crystal structures remain unknown, and therefore models of their three-dimensional structures are used. The modeling of the antigen–antibody complex consists of three steps: 1) modeling of three-dimensional structures of the antigen and antibody separately and optimization of resulting models; 2) docking of antibody and antigen models followed by formation of a virtual complex and, finally; 3) analysis of the energy of the antibody–antigen complex for all possible mutant forms with subsequent selection of mutant forms with minimum energy.
Two major approaches are used for modeling of three-dimensional structure of proteins: homology modeling and de novo modeling. Homology modeling uses three-dimensional structures available in the PDB (Protein Data Bank) as the template for modeling of proteins with unknown structure. The initial search yields proteins demonstrating the highest sequence similarity with the protein under study, and then the model is generated by homology with the template protein: it consists in overlapping of the sequence of the investigated protein onto the template composed on the basis of homologous structures [156, 157]. According to studies performed within CASP (Critical Assessment of Techniques for Protein Structure Prediction), modeling of protein structure by homology can be correctly performed when structures of homologous proteins share at least 30% identity [157]. In the case of proteins lacking homologs with known structure and reasonable similarity over the whole sequence, the method of protein threading (similar to the homology method) using homology of some structural elements with known tertiary structure is employed [158, 159]. There are many automated tools for homology modeling of three-dimensional structures of various proteins. These include MODELLER [160], SWISS-MODEL [161], I-TASSER [162], 3D-JIGSAW [163], ESyPred3D [164], Phyre [165], CPHmodels [166], etc. They can be rapidly and easily used for modeling of various proteins, but they do not take into consideration characteristic features of antibodies, and therefore a low probability exists that the use of these programs will result in generation of models of good quality.
The idea of the second approach is the modeling of protein folding (Rosetta [167] and TASSER [168] programs). This approach involves generation and subsequent evaluation of multiple folding variants depending of potential energy values. The completed search yields a conformation having significantly lower energy compared with all other possible conformations. To reproduce a natural mechanism of protein structure formation, researchers try to identify structural elements in the analyzed sequence that are initially folded and remain unchanged during subsequent folding. The procedure of de novo modeling is very sophisticated; it requires huge computer resources, and these algorithms are frequently used together with homology modeling for optimization of resulting models (for example, for modeling of loop structures).
Antibody modeling has some specific features. The sequence of the framework regions is so conservative that the task is usually limited to modeling of six CDR loops [169]. However, generation of the model of the antibody–antigen complex by the docking method requires a very accurate paratope model, and this significantly complicates the task [129].
Detection of correspondence between amino acid residues from different antibody sequences and correct homology modeling of CDR loops requires a system for designation of amino acid positions in the sequences of variable domains. It is also important to determine principles of identification of antibody regions involved in interaction with an antigen. For these purposes the following criteria are used: profile of amino acid variability [170], topology of amino acid residues [171-173], an optimized model of three-dimensional structure of the AbM loops interacting with the antigen [174], sequences of V genes coding κ and λ LC, and also sequences of HC and T-cell receptor domains [150, 175] and the role of amino acid residues in the interaction with the antigen [176].
Thus, one can see that no universal numeration of amino acid residues has been yet developed and “universal borders” of the antigen binding sites still need to be determined. Researchers do not try to reassess CDR borders – they just identify alternative sets of amino acid residues overlapping with the CDR that can be used for solution of various tasks [169].
For generation of three-dimensional structures of CDR loops by the method of homology modeling, the structures of antibody loops related to various structural groups (canonic classes) determined by Chotia [171-173] are used as templates. For this purpose antibody loops related to the same class as the loops of the modeled antibody and coinciding with its loop sequences by maximal number of amino acid positions are selected from the structural database. The VHCDR3 loops are rather variable, and therefore they cannot be clearly subdivided into canonic classes; nevertheless, they can also be subdivided into groups. For modeling of CDR loops that cannot be referred to certain groups, the method of ab initio loop modeling and the method of conformational search are used [174, 177]. Sometimes a structure of the same length that is characterized by similar distance between points of attachment to the FR is used as a template [169]. In the case of modeling of the variable domain FR, it is necessary to check homologous structures found in databases and used as templates for the presence of sites of low resolution and to replace such sites by consensus conformations of other known structures [178].
Several special programs are available for modeling of variable fragments of antibodies: WAM (Web Antibody Modeling) [179, 180], PIGS (Prediction of Immunoglobulin Structure) [181], and RosettaAntibody [182]. These programs use combined methods of analysis for generation of antibody models and so their application is more effective than automated modeling by means of universal programs for protein modeling. The method used in programs specialized on antibody modeling is based on generation of a model using variable domain FR as a template by transplanting template loops or by modeling of loop structures de novo. The PIGS and WAM servers do not optimize the resulting models with high resolution and do not consider thermodynamic parameters for modeling [182], whereas the primary model generated by the RosettaAntibody program undergoes subsequent optimization initially at low and then at high resolution.
The RosettaAntibody program for antibody modeling is the most developed tool for antibody modeling. It combines various principles of modeling of CDR loops such as use of canonic classes, homologous antibody loops, available structural databases, conformational search [174, 177], and de novo modeling of the H3 loop in the absence of its structure in the structural database. In addition, the RosettaAntibody program performs multistage ptimization of the resulting structure and mutual orientation of VL and VH domains taking into consideration mutual location of the H3 loop FR modeled de novo and also other loops. For antibodies sharing high similarity with the found template, the prediction accuracy of structures obtained by the RosettaAntibody program allows use of the resulting structures for generation of models of the antigen–antibody complex [129].
Modeling of mutant forms of antibodies based on three-dimensional structure of an initial antibody (in silico mutagenesis) is a simpler task compared with modeling of antibodies with unknown structure because amino acid substitutions in CDR loops influence only the local environment and leave the general domain structure unchanged [178].
Despite diversity of program products developed for protein modeling, researchers often face the problem when the most energetically favorable structure of a protein molecule determined in silico does not necessarily coincide with its original structure [129, 178]. This is associated with complexity of correct evaluation of all intramolecular interactions and also with complexity of evaluation of interaction of the molecule with the solvent [129]. In this connection the most promising approach is the generation of several most reasonable models (ten in the case of RosettaAntibody) and subsequent validation of correctness of these models by virtual docking of the antibody model molecules to the antigen. Taking into consideration the conformational mobility of antibody and antigen during modeling of a structure of their complex, it is possible to compensate some inaccuracies in the modeling of conformation of loops interacting with the antigen.
Protein–protein docking can be defined as prediction of a structure of the complex of two proteins (e.g. antibody with antigen) provided that their structures are known. Various programs for docking use either “rigid body” approximation (proteins are considered as “rigid bodies” with fixed conformation) or they are considered as mobile structures [183]. The programs that perform “rigid” docking, include Zdock/Rdock [183], ClusPro [184], 3D-dock [185], and RosettaDock [129, 186]. These programs do not take into consideration conformational changes in the protein backbone during protein interactions, but they consider mobility of amino acid side chains. Two models exist for flexible docking that take into consideration intramolecular mobility of proteins [186]: 1) the model of selection of conformation; according to this model a free protein exists as a set of conformers with low energy and one of them can bind to another protein; 2) the model of induced fit; according to this model, interaction of two proteins influence structures of both interacting partners. In addition, there are programs that use a hybrid model; according to this model, initial selection of a conformation is then accompanied by primary complex formation followed by subsequent induced fit of interacting proteins. The programs performing flexible docking include Autodock [187], Dock [188], FlexX [189], Glide [190], Gold [191], HADDOCK [186, 192], ICM [193], and Situs [194]. The complete list of existing programs for docking is rather long and constantly changing. Autodock is considered the most popular program for docking [195]. Although there are certain achievements in in silico prediction, modeling of protein molecule flexibility and those changes that occur during complex formation still represents one of the major problems limiting in silico approaches for modeling of protein complexes [115].
Various conformations obtained by the docking method and affinity of various mutant variants are evaluated and ranged by scoring functions that discriminate antibody variants capable of complex formation from variants that cannot form complexes. The same functions should predict the most probable structure of the complex among all structures generated by a computer program [195].
Rational design based on modeling results. After analysis of an antibody sequence and/or three-dimensional structure of the antigen–antibody complex (or its model), it is important to select amino acid residues that will be mutated in direct experiments. Several criteria now exist for rational amino acid selection for subsequent targeted mutagenesis.
Amino acid residues of CDR loops and, possibly, some FR residues that may be involved into direct antigen binding or may influence antigen interaction with contacting residues should be preferentially used for targeted mutagenesis [13, 196]. Usually mutagenesis of FR residues involved in maintenance of the immunoglobulin domain can cause dramatic consequences [38], and it seems unlikely that the effect of alteration of amino acids distantly located from the antigen binding site on the antibody affinity can be correctly predicted. During mutagenesis accompanying affinity maturation FR mutations increasing affinity due to loss of stability are not preserved [25]. Although examples of mutations that occur distantly from the binding site and increase antibody affinity are known in the literature [92], their consequences cannot be predicted in silico.
CDR mutations should not involve residues that can play structural functions (form parts of the domain “internal core”, internal salt bridges, hydrogen bonds, etc.). Usually these are conservative residues, and any substitution of these residues causes decrease in affinity (deleterious sequence features) [76, 143]. CDR residues susceptible to a solvent and involved in solvent interaction are considered as the best candidates for mutagenesis [25, 197]. Such mutations have minor effect on the general domain structure, but they may be involved in interactions with the antigen and the increase in affinity caused by mutations of these residues can be predicted most accurately.
Selection of hotspots (amino acid positions that are modified during somatic hypermutation most frequently) for subsequent mutagenesis increases probability of de novo preparation of antibodies with higher affinity because it simulates a natural process of antibody maturation. However, this approach may be less effective for the increase in affinity of recombinant antibodies obtained from monoclonal antibodies (which already passed through somatic hypermutation and demonstrate high affinity) because their hypervariable positions have been already optimized during maturation.
Decreasing the number of enumerated types of amino acids at each position is one of the approaches limiting the total number of antibody gene variants subjected to further selection. This can be achieved by exclusion of rare amino acids either found in CDR or rarely appearing during natural antibody maturation in B-cells [25, 198-200]. On the other hand, introduction of substitutions that have not been found in natural antibodies may increase affinity that has not been achieved due to limitations of somatic hypermutation. In addition, the number of amino acid variant at each position can be limited by the set of amino acids representing the main physicochemical groups of amino acids [200]. However, frequently increase in affinity is induced by “conservative” substitutions that only slightly modify existing interactions rather than mutations dramatically influencing amino acid features at this particular position. The other possible variant of limitation of enumerated amino acids at each position is the fact that most substitutions of amino acids significantly varying in their properties often yield inactive protein. Thus, mutagenesis should be limited to substitutions for similar amino acids (see the section “Parsimonious Mutagenesis”) [17, 201]. However, this approach does not take into consideration cases when nonconservative substitutions increase affinity.
To increase the number of screened positions, one of the following approaches is used: the “sequential” approach consists in optimization of one CDR and then of another CDR using for each new step of the “in vitro evolution” antibodies optimized in the previous step; the parallel approach uses independent optimization of various antibody sites followed by subsequent pooling of all favorable mutations.
For the increase in affinity of monoclonal antibodies prepared in vivo, mutagenesis of amino acid residues located on the periphery of the antigen binding site may have advantages over mutagenesis of central residues because the main energetic hotspots located in the center of the antigen-binding site have usually already been optimized during maturation and, therefore, their substitution may only decrease binding energy. Peripheral residues are more promising for mutagenesis [13, 39, 121, 202] because their changes may involve other residues (which have not participated in antigen binding) in antigen binding with minimal risk of impairments of existing bonds. Increase in antibody affinity by peripheral residues may occur due to increase in the antigen interacting hydrophobic surface inaccessible to the solvent; approaching and the increase in complementarity of the interacting surfaces of the antigen and the antibody; the increase in electrostatic interactions and also altered mobility of antigen binding site residues [99, 121, 201, 203, 204]. Increase in affinity may occur due to increased mobility of the antibody molecule (owing to increase in complementarity of epitope and paratope surfaces) and also due to its decrease by fixing the antibody conformation demonstrating optimal antigen binding.
When designing antibody with higher affinity for potential use in a diagnostic system, it is reasonable to limit its mobility to minimize the possibility of nonspecific interactions of the resulting antibody with other structurally similar molecules. In silico mutagenesis, i.e. modeling of mutant antibodies followed by subsequent evaluation of antigen binding energy allows prediction of which point substitutions may result in increase in antibody affinity. This is the most rational approach that needs the use of three-dimensional structure of high resolution. Although the number of known crystal structures of antibody–antigen complexes is rather modest, it is constantly increasing. Development of methods for prediction of three-dimensional structure together with the increase in accuracy of the methods used for modeling of mutated forms will improve successful applicability of in silico mutagenesis [134, 142, 143, 145, 205].
Thus, after generation of the three-dimensional structure of the antibody–antigen complex and identification of amino acid residues that contribute to formation of this complex, subsequent targeted mutagenesis of these residues yields several variants of antibody mutants to be subjected to subsequent selection.
Methods of targeted mutagenesis. Targeted mutagenesis is based on the idea that increase in affinity can be achieved by mutagenesis of only those amino acid residues that are located within the antigen binding site and can be involved in antigen interaction. Several subtypes of targeted mutagenesis have been developed depending on substitutable residues and modes of their substitutions (Fig. 8). In some ways targeted mutagenesis is similar to in vivo somatic hypermutation: mutagenesis covers a limited number of CDR amino acid residues or a small number of residues selected by means of in silico approaches [101, 102, 152, 206]. Targeted mutagenesis can include substitution of single residues for other (earlier determined) residues (point mutagenesis), randomization within selected residues, or complete enumeration of all amino acid positions within selected sites. Point mutagenesis is applicable for antibodies with known three-dimensional structure when it is already known which amino acid substitution has to be performed in order to get the desired effect.
Point mutagenesis. Point mutagenesis (also known as site-specific mutagenesis) is used for substitution of one amino acid at a certain position for another earlier determined amino acid. Amino acid substitutions are carried out by PCR using primers containing the desired mutations. Enumeration of various types of amino acids in the same position is achieved by using primers degenerate in this particular position (i.e. using the set of primers differing by just one amino acid inserted into this position by means of mutation). Such primers have identical nucleotide sequences except for the randomized nucleotide triplet corresponding to the amino acid selected for mutagenesis.
Targeted randomization and mutagenesis with complete enumeration of variants. This group contains some methodical approaches that include both random changes of selected residues (randomization) and also enumeration of all possible variants of amino acid substitutions within selected positions of residues (saturation mutagenesis).
Optimization of CDR residues. Since simultaneous optimization of all CDR residues is impossible due to limited sizes of tested libraries, three approaches are used to solve this problem: sequential modification of CDR residues (“CDR walking”) [106], parallel modification of CDR residues, and randomization of some CDR loops. The first approach implies sequential optimization of amino acid residues at various CDR sites. A fragment sequence of antibody demonstrating increased affinity and obtained due to optimization of previous CDR variants serves as a starting material for residue optimization in each new cycle. The strategy of parallel optimization is an alternative for the former approach; in this case various CDR fragments undergo independent optimization, and then the best variants are pooled in a single gene. This strategy is based on experimental data demonstrating an additive effect during combination of non-interacting mutations [207], and this frequently gives positive results [105, 208, 209]. The advantage of this strategy is the rapid preparation of the final product. However, lack of the additive effect during combination of several mutations has also been reported [106]. Sometimes saturating mutagenesis of only one or two principal CDR loops (usually VL-CDR3 and VH-CDR3 and sometimes VH-CDR2) results in an increase in affinity, possibly due to major energetic contribution of these loops into the interaction with the antigen [210-212].
Look-through mutagenesis. Look-through mutagenesis has been developed for total optimization of all six CDR loops to improve antigen binding. Among 20 proteinogenic amino acids, this approach uses just nine (alanine, serine, histidine, leucine, proline, tyrosine, aspartate, glutamine, and lysine) representing the main amino acid groups classified on the basis of physicochemical properties of their side chains. For each CDR amino acid position a series of mutant forms in which a wild type residue is substituted for one of nine selected amino acids is created. Then the CDR sites coding single amino acid substitutions are linked to each other by DNA recombination followed by formation of scFv fragments containing combinations of single amino acid substitutions in various CDR. After selection of resulting clones by their antigen-binding capacity, genes of clones demonstrating improved characteristics are sequenced for identification of favorable mutations, which are then combined for detection of synergistic mutations [200].
“Parsimonious mutagenesis”. Parsimonious mutagenesis is the method of preparation of antibody variants with higher affinity by means of simultaneous and total mutagenesis of CDR residues in small libraries. Decrease in the library size can be achieved by decreasing the number of degenerated coding sequences (one codon for each amino acid is selected) and limitation of the number of enumerated amino acids at each position. Although, theoretically, for each amino acid position 19 substitution variants exist, the method of parsimonious mutagenesis uses only those variants that share chemical and/or steric similarity with wild type residues. This approach is also based on the principle that only a small proportion of amino acids make the most substantial contribution to antigen binding, whereas other amino acid residues of the antigen binding site may be mutated for formation of additional contacts with the antigen [17, 201].
Using the above described methods (random mutagenesis, in silico and rational design methods, targeted mutagenesis) it is possible to prepare libraries of genetic variants of investigated antibodies. High affinity variants are subsequently selected by means of various display methods that have been discussed in the chapter “Preparation of Recombinant Antibodies de novo”. It should be noted that in some cases it is nearly impossible to prepare an antibody with affinity exceeding the affinity of its natural analogs. This may be attributed to the fact that during natural maturation of antibodies in vivo an optimal combination of antibody residues demonstrating the maximal affinity towards the corresponding antigen has already been chosen during antibody selection. In this case subsequent attempts to increase affinity of such antibody by using combinatorial methods will not result in a derivative with higher affinity.
CONCLUSIONS
Antibodies are widely used tools for scientific studies, diagnostics, and therapy of various diseases. In many cases practical use of recombinant antibodies and their fragments has certain advantages over full-length antibodies obtained by the hybridoma method. Changing amino acid sequence of antibodies by gene engineering approaches, it is possible to improve such antibody characteristics as specificity, affinity, stability, pharmacokinetic parameters, effector parameters, tissue penetrating capacity of antibodies, and their immunogenicity. By modifying these antibody features it is possible to develop a new generation of useful antibodies with improved properties for solution of scientific problems and problems of clinical medicine. Subsequent promise for improvement of antibody affinity includes preparation of antibodies to therapeutically relevant antigens and also creation of antibodies with ultrahigh affinity of 10–12 and even 10–15 M [92, 96, 106, 121, 210, 213]. In the future creation of antibodies with such high affinity will significantly extend field of antibody application in scientific studies, diagnostics, and also as highly specific and effective therapeutics.
The authors are grateful to Dr. A. G. Laman (Branch of Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Pushchino, Moscow Region), Professor N. B. Gusev, and Dr. A. V. Kharitonov (Department of Biochemistry, Faculty of Biology, Moscow State University) for reading this review and their valuable comments.
REFERENCES
1.Hagemeyer, C. E., von Zur Muhlen, C., von
Elverfeldt, D., and Peter, K. (2009) Thromb. Haemost.,
101, 1012-1019.
2.Hudson, P. J., and Souriau, C. (2001) Expert
Opin. Biol. Ther., 1, 845-855.
3.Arbabi-Ghahroudi, M., Tanha, J., and MacKenzie,
R. (2005) Cancer Metastasis Rev., 24, 501-519.
4.Buchner, J., and Rudolph, R. (1991)
Biotechnology (N. Y), 9, 157-162.
5.Cowell, L. G., and Kepler, T. B. (2000)
J. Immunol., 164, 1971-1976.
6.Donzeau, M., and Knappik, A. (2007) Meth. Mol.
Biol., 378, 14-31.
7.Honegger, A. Trobleshooting Antibody
Fragments, URL: http://www.bioc.uzh.ch/antibody/Introduction/VirtualSeminars/AE2003/source/slide01.htm.
8.Maynard, J., and Georgiou, G. (2000)
Annu. Rev. Biomed. Eng., 2, 339-376.
9.Winter, G., Griffiths, A. D., Hawkins, R. E., and
Hoogenboom, H. R. (1994) Annu. Rev. Immunol., 12,
433-455.
10.Schroeder, H. W., Jr., and Cavacini, L. (2010)
J. Allergy Clin. Immunol., 125, S41-52.
11.Volkov, V., Kayushina, R., Lapuk, V., Shtykova,
E., Varlamova, E., Malfois, M., and Svergun, D. (2003) Crystallogr.
Rep., 48, 98-105.
12.Padlan, E. A. (1994) Mol. Immunol.,
31, 169-217.
13.Sundberg, E. J. (2009) Meth. Mol. Biol.,
524, 23-36.
14.Kashmiri, S. V., de Pascalis, R., Gonzales, N.
R., and Schlom, J. (2005) Methods, 36, 25-34.
15.Padlan, E. A., Abergel, C., and Tipper, J. P.
(1995) FASEB J., 9, 133-139.
16.Duquesnoy, R. J. Structural and Functional
Definitions of Epitopes Reacting with Mouse Monoclonal Antibodies,
URL: HLAMatchmaker Website: http://www.hlamatchmaker.net.
17.Balint, R. F., and Larrick, J. W. (1993)
Gene, 137, 109-118.
18.Muller, Y. A., Chen, Y., Christinger, H. W., Li,
B., Cunningham, B. C., Lowman, H. B., and de Vos, A. M. (1998)
Structure, 6, 1153-1167.
19.Saul, F. A., and Alzari, P. M. (1996) Meth.
Mol. Biol., 66, 11-23.
20.Nakanishi, T., Hori, M., Tsumoto, K., Yokota, A.,
Kondo, H., and Kumagai, I. (2007) Photon Factory Activity
Report, 25B, 217.
21.Yokota, A., Tsumoto, K., Shiroishi, M., Kondo,
H., and Kumagai, I. (2003) J. Biol. Chem., 278,
5410-5418.
22.Chi, S. W., Maeng, C. Y., Kim, S. J., Oh, M. S.,
Ryu, C. J., Han, K. H., Hong, H. J., and Ryu, S. E. (2007)
Proc. Natl. Acad. Sci. USA, 104, 9230-9235.
23.Raghavan, M., and Bjorkman, P. J. (1996) Annu.
Rev. Cell Dev. Biol., 12, 181-220.
24.Jung, D., Giallourakis, C., Mostoslavsky, R., and
Alt, F. W. (2006) Annu. Rev. Immunol., 24, 541-570.
25.David, M. P., Asprer, J. J., Ibana, J. S.,
Concepcion, G. P., and Padlan, E. A. (2007) Mol. Immunol.,
44, 1342-1351.
26.Berek, C., and Milstein, C. (1987) Immunol.
Rev., 96, 23-41.
27.Chowdhury, P. S. (2003) Meth. Mol. Biol.,
207, 179-196.
28.Ho, M., and Pastan, I. (2009) Meth. Mol.
Biol., 525, 293-308.
29.Tomlinson, I. M., Walter, G., Jones, P. T., Dear,
P. H., Sonnhammer, E. L., and Winter, G. (1996) J. Mol. Biol.,
256, 813-817.
30.Foote, J., and Eisen, H. N. (1995)
Proc. Natl. Acad. Sci. USA, 92, 1254-1256.
31.Batista, F. D., and Neuberger, M. S.
(1998) Immunity, 8, 751-759.
32.Cauerhff, A., Goldbaum, F. A., and Braden, B.
C. (2004) Proc. Natl. Acad. Sci. USA, 101,
3539-3544.
33.Ho, M., Kreitman, R. J., Onda, M., and Pastan, I.
(2005) J. Biol. Chem., 280, 607-617.
34.Thorpe, I. F., and Brooks, C. L., 3rd (2007)
Proc. Natl. Acad. Sci. USA, 104, 8821-8826.
35.Davies, D. R., and Padlan, E. A. (1992) Curr.
Biol., 2, 254-256.
36.Braden, B. C., and Poljak, R. J. (1995) FASEB
J., 9, 9-16.
37.Acierno, J. P., Braden, B. C., Klinke, S.,
Goldbaum, F. A., and Cauerhff, A. (2007) J. Mol. Biol.,
374, 130-146.
38.Jung, S., Spinelli, S., Schimmele, B., Honegger,
A., Pugliese, L., Cambillau, C., and Pluckthun, A. (2001) J.
Mol. Biol., 309, 701-716.
39.Li, Y., Li, H., Yang, F., Smith-Gill, S. J., and
Mariuzza, R. A. (2003) Nat. Struct. Biol., 10,
482-488.
40.Sundberg, E. J., Urrutia, M., Braden, B. C.,
Isern, J., Tsuchiya, D., Fields, B. A., Malchiodi, E. L., Tormo, J.,
Schwarz, F. P., and Mariuzza, R. A. (2000) Biochemistry,
39, 15375-15387.
41.Masuda, K., Sakamoto, K., Kojima, M., Aburatani,
T., Ueda, T., and Ueda, H. (2006) FEBS J., 273,
2184-2194.
42.Wedemayer, G. J., Patten, P. A., Wang, L. H.,
Schultz, P. G., and Stevens, R. C. (1997) Science, 276,
1665-1669.
43.Bartal, A. H., and Hirshaut, Y. (1987) Methods
of Hybridoma Formation, Humana Press.
44.Engvall, E., and Perlmann, P. (1971)
Immunochemistry, 8, 871-874.
45.Van Weeman, B., and Schuurs, A. (1971)
FEBS Lett., 15, 232-235.
46.Towbin, H., Staehelin, T., and Gordon, J. (1979)
Proc. Natl. Acad. Sci. USA, 76, 4350-4354.
47.Katrukha, A. G., Bereznikova, A. V., Esakova, T.
V., Pettersson, K., Lovgren, T., Severina, M. E., Pulkki, K.,
Vuopio-Pulkki, L. M., and Gusev, N. B. (1997) Clin. Chem.,
43, 1379-1385.
48.Katrukha, A., Bereznikova, A., Filatov, V., and
Esakova, T. (1999) Clin. Chem. Lab. Med., 37,
1091-1095.
49.Katrukha, A., Bereznikova, A., and Pettersson, K.
(1999) Scand. J. Clin. Lab. Invest. (Suppl.), 230,
124-127.
50.Koshkina, E. V., Krasnosel’skii, M.,
Fedorovskii, N. M., Goriacheva, E. V., Polupan, A. A., Aref’ev,
A. A., and Katrukha, A. G. (2009) Anesteziol. Reanimatol.,
42-46.
51.Adams, J. E., 3rd, Bodor, G. S., Davila-Roman, V.
G., Delmez, J. A., Apple, F. S., Ladenson, J. H., and Jaffe, A. S.
(1993) Circulation, 88, 101-106.
52.Feng, Y. J., Chen, C., Fallon, J. T., Lai, T.,
Chen, L., Knibbs, D. R., Waters, D. D., and Wu, A. H. (1998)
Am. J. Clin. Pathol., 110, 70-77.
53.Vaidya, H. C. (1994) J. Clin. Immunoassay,
17, 35-39.
54.Katrukha, A. G., Bereznikova, A. V., Filatov, V.
L., Esakova, T. V., Kolosova, O. V., Pettersson, K., Lovgren, T.,
Bulargina, T. V., Trifonov, I. R., Gratsiansky, N. A., Pulkki, K.,
Voipio-Pulkki, L. M., and Gusev, N. B. (1998) Clin. Chem.,
44, 2433-2440.
55.Filatov, V. L., Katrukha, A. G., Bereznikova, A.
V., Esakova, T. V., Bulargina, T. V., Kolosova, O. V., Severin, E. S.,
and Gusev, N. B. (1998) Biochem. Mol. Biol. Int., 45,
1179-1187.
56.Semenov, A. G., Postnikov, A. B., Tamm, N. N.,
Seferian, K. R., Karpova, N. S., Bloshchitsyna, M. N., Koshkina, E. V.,
Krasnoselsky, M. I., Serebryanaya, D. V., and Katrukha, A. G.
(2009) Clin. Chem., 55, 489-498.
57.Krishnamurti, U., and Steffes, M. W.
(2001) Clin. Chem., 47, 1157-1165.
58.Clark, P. M. (1999) Ann. Clin. Biochem.,
36 (Pt. 5), 541-564.
59.Ludwig, J. A., and Weinstein, J. N. (2005)
Nat. Rev. Cancer, 5, 845-856.
60.Arthur, J. M., Janech, M. G., Varghese, S. A.,
Almeida, J. S., and Powell, T. B. (2008) Contrib. Nephrol.,
160, 53-64.
61.Nakamura, R. M., and Binder, W. L. (1988)
Arch. Pathol. Lab. Med., 112, 869-877.
62.Ekins, R. (1992) Clin. Chem., 38,
1289-1293.
63.Shivaraj, G., Prakash, B. D., Sonal, V., Shruthi,
K., Vinayak, H., and Avinash, M. (2009) Eur. Rev. Med. Pharmacol.
Sci., 13, 341-349.
64.Cummings, M. C., Lukehart, S. A., Marra, C.,
Smith, B. L., Shaffer, J., Demeo, L. R., Castro, C., and McCormack, W.
M. (1996) Sex. Transm. Dis., 23, 366-369.
65.Stamm, W. E., Cole, B., Fennell, C., Bonin, P.,
Armstrong, A. S., Herrmann, J. E., and Holmes, K. K. (1984) J. Clin.
Microbiol., 19, 399-403.
66.Black, C. M. (1997) Clin. Microbiol. Rev.,
10, 160-184.
67.Safford, J. W., Abbott, G. G., Craine, M. C., and
MacDonald, R. G. (1991) J. Clin. Pathol., 44,
238-242.
68.Malomgre, W., and Neumeister, B. (2009)
Anal. Bioanal. Chem., 393, 1443-1451.
69.Cole, L. A. (2009) Expert Rev. Mol.
Diagn., 9, 721-747.
70.Andrew, S. M., and Titus, J. A. (2001) Curr.
Protoc. Immunol., Unit 2.8, DOI: 10.1002/0471142735.im0208s21,
Online Posting Date: 2001.
71.Hust, M., Jostock, T., Menzel, C., Voedisch, B.,
Mohr, A., Brenneis, M., Kirsch, M. I., Meier, D., and Dubel, S.
(2007) BMC Biotechnol., 7, 14.
72.Nieba, L., Honegger, A., Krebber, C., and
Pluckthun, A. (1997) Protein Eng., 10,
435-444.
73.Whitlow, M., Bell, B. A., Feng, S. L., Filpula,
D., Hardman, K. D., Hubert, S. L., Rollence, M. L., Wood, J. F.,
Schott, M. E., Milenic, D. E., et al. (1993) Protein Eng.,
6, 989-995.
74.Jain, M., Kamal, N., and Batra, S. K. (2007)
Trends Biotechnol., 25, 307-316.
75.Wark, K. L., and Hudson, P. J. (2006) Adv.
Drug Deliv. Rev., 58, 657-670.
76.Chowdhury, P. S., and Vasmatzis, G. (2003)
Meth. Mol. Biol., 207, 237-254.
77.Conrad, U., and Scheller, J. (2005) Comb.
Chem. High Throughput Screen., 8, 117-126.
78.Borrebaeck, C. A., Malmborg, A. C., Furebring,
C., Michaelsson, A., Ward, S., Danielsson, L., and Ohlin, M. (1992)
Biotechnology (N. Y), 10, 697-698.
79.Telleman, P., and Junghans, R. P. (2000)
Immunology, 100, 245-251.
80.Lo, B. K. C. (ed.) (2004) Antibody
Engineering. Methods and Protocols, Humana Press.
81.Benhar, I., and Pastan, I. (1994) Protein
Eng., 7, 1509-1515.
82.Tachibana, H., Cheng, X. J., Watanabe, K.,
Takekoshi, M., Maeda, F., Aotsuka, S., Kaneda, Y., Takeuchi, T., and
Ihara, S. (1999) Clin. Diagn. Lab. Immunol., 6,
383-387.
83.Tachibana, H., Takekoshi, M., Cheng, X. J.,
Maeda, F., Aotsuka, S., and Ihara, S. (1999) Am. J. Trop. Med.
Hyg., 60, 35-40.
84.Wlad, H., Ballagi, A., Bouakaz, L., Gu, Z., and
Janson, J. C. (2001) Protein Expr. Purif., 22,
325-329.
85.Saviranta, P., Haavisto, T., Rappu, P., Karp, M.,
and Lovgren, T. (1998) Bioconj. Chem., 9, 725-735.
86.Presta, L. G. (2005) J. Allergy Clin.
Immunol., 116, 731-736.
87.Smith, G. P. (1985) Science, 228,
1315-1317.
88.Hanes, J., and Pluckthun, A. (1997) Proc.
Natl. Acad. Sci. USA, 94, 4937-4942.
89.Liu, R., Barrick, J., Szostak, J. W., and
Roberts, R. W. (2000) Meth. Enzymol., 1, 268-293.
90.Daugherty, P. S., Chen, G., Olsen, M. J.,
Iverson, B. L., and Georgiou, G. (1998) Protein Eng., 11,
825-832.
91.Georgiou, G., Stathopoulos, C., Daugherty, P. S.,
Nayak, A. R., Iverson, B. L., and Curtiss, R., 3rd (1997) Nat.
Biotechnol., 15, 29-34.
92.Boder, E. T., Midelfort, K. S., and Wittrup, K.
D. (2000) Proc. Natl. Acad. Sci. USA, 97,
10701-10705.
93.Boder, E. T., and Wittrup, K. D. (1998)
Biotechnol. Prog., 14, 55-62.
94.Boder, E. T., and Wittrup, K. D. (1997)
Nat. Biotechnol., 15, 553-557.
95.Siegel, R. W. (2009) Meth. Mol. Biol.,
504, 351-383.
96.Vaughan, T. J., Williams, A. J., Pritchard, K.,
Osbourn, J. K., Pope, A. R., Earnshaw, J. C., McCafferty, J., Hodits,
R. A., Wilton, J., and Johnson, K. S. (1996) Nat. Biotechnol.,
14, 309-314.
97.Knappik, A., Ge, L., Honegger, A., Pack, P.,
Fischer, M., Wellnhofer, G., Hoess, A., Wolle, J., Pluckthun, A., and
Virnekas, B. (2000) J. Mol. Biol., 296, 57-86.
98.Luginbuhl, B., Kanyo, Z., Jones, R. M.,
Fletterick, R. J., Prusiner, S. B., Cohen, F. E., Williamson, R. A.,
Burton, D. R., and Pluckthun, A. (2006) J. Mol. Biol.,
363, 75-97.
99.Krebber, A., Bornhauser, S., Burmester, J.,
Honegger, A., Willuda, J., Bosshard, H. R., and Pluckthun, A. (1997)
J. Immunol. Meth., 201, 35-55.
100.Tabuchi, I., Soramoto, S., Nemoto, N., and
Husimi, Y. (2001) FEBS Lett., 508, 309-312.
101.Matsuura, T., and Yomo, T. (2006) J.
Biosci. Bioeng., 101, 449-456.
102.Pluckthun, A., Schaffitzel, C., Hanes, J., and
Jermutus, L. (2000) Adv. Protein Chem., 55, 367-403.
103.Ulrich, H. D., Patten, P. A., Yang, P. L.,
Romesberg, F. E., and Schultz, P. G. (1995) Proc. Natl. Acad. Sci.
USA, 92, 11907-11911.
104.Levin, A. M., and Weiss, G. A. (2006)
Mol. Biosyst., 2, 49-57.
105.Hawkins, R. E., Russell, S. J., and Winter,
G. (1992) J. Mol. Biol., 226, 889-896.
106.Yang, W. P., Green, K., Pinz-Sweeney, S.,
Briones, A. T., Burton, D. R., and Barbas, C. F., 3rd (1995) J. Mol.
Biol., 254, 392-403.
107.Verma, R., Boleti, E., and George, A. J. (1998)
J. Immunol. Meth., 216, 165-181.
108.Dimitrov, D. S., and Marks, J. D. (2009)
Meth. Mol. Biol., 525, 1-27.
109.Dubel, S. (2007) Appl. Microbiol.
Biotechnol., 74, 723-729.
110.Groner, B., Hartmann, C., and Wels, W.
(2004) Curr. Mol. Med., 4, 539-547.
111.Stemmer, W. P. C. (1995) Nat.
Biotechnol., 13, 549-553.
112.Lippow, S. M., Wittrup, K. D., and Tidor, B.
(2007) Nat. Biotechnol., 25, 1171-1176.
113.Crameri, A., Raillard, S. A., Bermudez, E., and
Stemmer, W. P. (1998) Nature, 391, 288-291.
114.Stemmer, W. P. (1994) Nature,
370, 389-391.
115.Chodorge, M., Fourage, L., Ravot, G., Jermutus,
L., and Minter, R. (2008) Protein Eng. Des. Sel., 21,
343-351.
116.Cline, J., Braman, J. C., and Hogrefe, H. H.
(1996) Nucleic Acids Res., 24, 3546-3551.
117.Hoogenboom, H. R., and Chames, P. (2000)
Immunol. Today, 21, 371-378.
118.Martineau, P. (2002) Meth. Mol. Biol.,
178, 287-294.
119.Schlapschy, M., Gruber, H., Gresch, O.,
Schafer, C., Renner, C., Pfreundschuh, M., and Skerra, A. (2004)
Protein Eng. Des. Sel., 17, 847-860.
120.Fujii, I. (2004) Meth. Mol. Biol.,
248, 345-359.
121.Zahnd, C., Spinelli, S., Luginbuhl, B.,
Amstutz, P., Cambillau, C., and Pluckthun, A. (2004) J. Biol.
Chem., 279, 18870-18877.
122.Low, N. M., Holliger, P. H., and Winter, G.
(1996) J. Mol. Biol., 260, 359-368.
123.Greener, A., Callahan, M., and Jerpseth, B.
(1996) Meth. Mol. Biol., 57, 375-385.
124.Irving, R. A., Kortt, A. A., and Hudson, P. J.
(1996) Immunotechnology, 2, 127-143.
125.Lantto, J., Jirholt, P., Barrios, Y., and
Ohlin, M. (2002) Meth. Mol. Biol., 178, 303-316.
126.Bikbulatova, S. M., Mingazetdinova, S. R.,
Chemeris, A. V., and Vakhitov, V. A. (2009) Usp. Sovr. Biol.,
129, 323-335.
127.Pogulis, R. J., Vallejo, A. N., and Pease, L.
R. (1996) Meth. Mol. Biol., 57, 167-176.
128.Zhao, H., Giver, L., Shao, Z., Affholter, J.
A., and Arnold, F. H. (1998) Nat. Biotechnol., 16,
258-261.
129.Sivasubramanian, A., Sircar, A., Chaudhury, S.,
and Gray, J. J. (2009) Proteins, 74, 497-514.
130.Barderas, R., Desmet, J., Timmerman, P.,
Meloen, R., and Casal, J. I. (2008) Proc. Natl. Acad. Sci. USA,
105, 9029-9034.
131.Essen, L. O., and Skerra, A. (1994) J. Mol.
Biol., 238, 226-244.
132.Schiweck, W., and Skerra, A. (1995)
Proteins, 23, 561-565.
133.Marvin, J. S., and Zhu, Z. (2005) Drug
Design Reviews-Online, 2, 419-425.
134.Teng, S., Michonova-Alexova, E., and Alexov, E.
(2008) Curr. Pharm. Biotechnol., 9, 123-133.
135.Yue, P., Li, Z., and Moult, J. (2005) J.
Mol. Biol., 353, 459-473.
136.Ng, P. C., and Henikoff, S. (2003) Nucleic
Acids Res., 31, 3812-3814.
137.Ramensky, V., and Sunyaev, S. (2009) Mol.
Biol. (Moscow), 43, 286-294.
138.Sunyaev, S., Ramensky, V., and Bork, P. (2000)
Trends Genet., 16, 198-200.
139.Sunyaev, S., Ramensky, V., Koch, I., Lathe, W.,
3rd, Kondrashov, A. S., and Bork, P. (2001) Hum. Mol. Genet.,
10, 591-597.
140.Ramensky, V., Bork, P., and Sunyaev, S. (2002)
Nucleic Acids Res., 30, 3894-3900.
141.Bromberg, Y., and Rost, B. (2007) Nucleic
Acids Res., 35, 3823-3835.
142.Schymkowitz, J. W., Rousseau, F., Martins, I.
C., Ferkinghoff-Borg, J., Stricher, F., and Serrano, L. (2005)
Proc. Natl. Acad. Sci. USA, 102, 10147-10152.
143.Teng, S., Madej, T., Panchenko, A., and Alexov,
E. (2009) Biophys. J., 96, 2178-2188.
144.Brooks, B. R., Bruccoleri, R. E., Olafson, B.
D., States, D. J., Swaminathan, S., and Karplus, M. (1983) J.
Comput. Chem., 4, 187-217.
145.Boas, F. E., and Harbury, P. B. (2008) J.
Mol. Biol., 380, 415-424.
146.Reumers, J., Schymkowitz, J., Ferkinghoff-Borg,
J., Stricher, F., Serrano, L., and Rousseau, F. (2005) Nucleic Acids
Res., 33, D527-532.
147.Karchin, R., Diekhans, M., Kelly, L., Thomas,
D. J., Pieper, U., Eswar, N., Haussler, D., and Sali, A. (2005)
Bioinformatics, 21, 2814-2820.
148.Capriotti, E., Fariselli, P., and Casadio, R.
(2005) Nucleic Acids Res., 33, W306-310.
149.Dehouck, Y., Grosfils, A., Folch, B., Gilis,
D., Bogaerts, P., and Rooman, M. (2009) Bioinformatics,
25, 2537-2543.
150.Lefranc, M. P., Pommie, C., Ruiz, M.,
Giudicelli, V., Foulquier, E., Truong, L., Thouvenin-Contet, V., and
Lefranc, G. (2003) Dev. Comp. Immunol., 27, 55-77.
151.Chowdhury, P. S. (2002) Meth. Mol.
Biol., 178, 269-285.
152.Yau, K. Y., Dubuc, G., Li, S., Hirama, T.,
Mackenzie, C. R., Jermutus, L., Hall, J. C., and Tanha, J. (2005) J.
Immunol. Meth., 297, 213-224.
153.Chowdhury, P. S., and Pastan, I. (1999) Nat.
Biotechnol., 17, 568-572.
154.Marvin, J. S., and Lowman, H. B. (2003)
Biochemistry, 42, 7077-7083.
155.Selzer, T., Albeck, S., and Schreiber, G.
(2000) Nat. Struct. Biol., 7, 537-541.
156.Chugunov, A. (2007) URL: http://biomolecula.ru/content/189.
157.Chugunov, A. (2008) URL: http://biomolecula.ru/content/264.
158.Guo, J. T., Ellrott, K., and Xu, Y. (2008)
Meth. Mol. Biol., 413, 3-42.
159.Xu, J., Jiao, F., and Yu, L. (2008) Meth.
Mol. Biol., 413, 91-121.
160.Eswar, N., Webb, B., Marti-Renom, M. A.,
Madhusudhan, M. S., Eramian, D., Shen, M. Y., Pieper, U., and Sali, A.
(2007) Curr. Protoc. Protein Sci., Published Online 2007 in
Wiley Interscience, Unit 2.9.1-2.9.31, DOI:
10.1002/0471140864.ps0209s50.
161.Schwede, T., Kopp, J., Guex, N., and Peitsch,
M. C. (2003) Nucleic Acids Res., 31, 3381-3385.
162.Zhang, Y. (2008) BMC Bioinformatics,
9, 40.
163.Bates, P. A., Kelley, L. A., MacCallum, R. M.,
and Sternberg, M. J. (2001) Proteins, Suppl. 5, 39-46.
164.Lambert, C., Leonard, N., de Bolle, X., and
Depiereux, E. (2002) Bioinformatics, 18, 1250-1256.
165.Kelley, L. A., and Sternberg, M. J. (2009)
Nat. Protoc., 4, 363-371.
166.Lund, O., Frimand, K., Gorodkin, J., Bohr, H.,
Bohr, J., Hansen, J., and Brunak, S. (1997) Protein Eng.,
10, 1241-1248.
167.Qian, B., Raman, S., Das, R., Bradley, P.,
McCoy, A. J., Read, R. J., and Baker, D. (2007) Nature,
450, 259-264.
168.Zhang, Y., and Skolnick, J. (2004) Proc.
Natl. Acad. Sci. USA, 101, 7594-7599.
169.Martin, A. C. R., and Allen, J. (2007)
Bioinformatics Tools for Antibody Engineering, in Handbook of
Therapeutic Antibodies.
170.Wu, T. T., and Kabat, E. A. (1970) J. Exp.
Med., 132, 211-250.
171.Al-Lazikani, B., Lesk, A. M., and Chothia, C.
(1997) J. Mol. Biol., 273, 927-948.
172.Chothia, C., and Lesk, A. M. (1987) J. Mol.
Biol., 196, 901-917.
173.Chothia, C., Lesk, A. M., Tramontano, A.,
Levitt, M., Smith-Gill, S. J., Air, G., Sheriff, S., Padlan, E. A.,
Davies, D., and Tulip, W. R. (1989) Nature, 342,
877-883.
174.Martin, A. C., Cheetham, J. C., and Rees, A. R.
(1989) Proc. Natl. Acad. Sci. USA, 86, 9268-9272.
175.Honegger, A., and Pluckthun, A. (2001) J.
Mol. Biol., 309, 657-670.
176.MacCallum, R. M., Martin, A. C., and Thornton,
J. M. (1996) J. Mol. Biol., 262, 732-745.
177.Bruccoleri, R. E. (2000) Meth. Mol.
Biol., 143, 247-264.
178.Martin, A. C., Cheetham, J. C., and Rees, A. R.
(1991) Meth. Enzymol., 203, 121-153.
179.Whitelegg, N. R., and Rees, A. R. (2000)
Protein Eng., 13, 819-824.
180.Whitelegg, N., and Rees, A. R. (2004) Meth.
Mol. Biol., 248, 51-91.
181.Marcatili, P., Rosi, A., and Tramontano, A.
(2008) Bioinformatics, 24, 1953-1954.
182.Sircar, A., Kim, E. T., and Gray, J. J. (2009)
Nucleic Acids Res., 37, W474-479.
183.Wiehe, K., Peterson, M. W., Pierce, B.,
Mintseris, J., and Weng, Z. (2008) Meth. Mol. Biol., 413,
283-314.
184.Comeau, S. R., Gatchell, D. W., Vajda, S., and
Camacho, C. J. (2004) Bioinformatics, 20, 45-50.
185.Sternberg, M. J., Gabb, H. A., Jackson, R. M.,
and Moont, G. (2000) Meth. Mol. Biol., 143, 399-415.
186.Chaudhury, S., and Gray, J. J. (2008) J.
Mol. Biol., 381, 1068-1087.
187.Goodsell, D. S., Morris, G. M., and Olson, A.
J. (1996) J. Mol. Recognit., 9, 1-5.
188.Ewing, T. J., Makino, S., Skillman, A. G., and
Kuntz, I. D. (2001) J. Comput. Aided Mol. Des., 15,
411-428.
189.Rarey, M., Kramer, B., Lengauer, T., and Klebe,
G. (1996) J. Mol. Biol., 261, 470-489.
190.Halgren, T. A., Murphy, R. B., Friesner, R. A.,
Beard, H. S., Frye, L. L., Pollard, W. T., and Banks, J. L. (2004)
J. Med. Chem., 47, 1750-1759.
191.Verdonk, M. L., Cole, J. C., Hartshorn, M. J.,
Murray, C. W., and Taylor, R. D. (2003) Proteins, 52,
609-623.
192.Dominguez, C., Boelens, R., and Bonvin, A. M.
(2003) J. Am. Chem. Soc., 125, 1731-1737.
193.Totrov, M., and Abagyan, R. (1997)
Proteins, Suppl. 1, 215-220.
194.Wriggers, W., and Birmanns, S. (2001) J.
Struct. Biol., 133, 193-202.
195.Sousa, S. F., Fernandes, P. A., and Ramos, M.
J. (2006) Proteins, 65, 15-26.
196.Skerra, A. (2000) J. Mol. Recognit.,
13, 167-187.
197.Sidhu, S. S., Li, B., Chen, Y., Fellouse, F.
A., Eigenbrot, C., and Fuh, G. (2004) J. Mol. Biol., 338,
299-310.
198.Bostrom, J., Lee, C. V., Haber, L., and Fuh, G.
(2009) Meth. Mol. Biol., 525, 353-376.
199.Lee, C. V., Liang, W. C., Dennis, M. S.,
Eigenbrot, C., Sidhu, S. S., and Fuh, G. (2004) J. Mol. Biol.,
340, 1073-1093.
200.Rajpal, A., Beyaz, N., Haber, L., Cappuccilli,
G., Yee, H., Bhatt, R. R., Takeuchi, T., Lerner, R. A., and Crea, R.
(2005) Proc. Natl. Acad. Sci. USA, 102, 8466-8471.
201.Schier, R., Balint, R. F., McCall, A., Apell,
G., Larrick, J. W., and Marks, J. D. (1996) Gene, 169,
147-155.
202.Brorson, K., Thompson, C., Wei, G.,
Krasnokutsky, M., and Stein, K. E. (1999) J. Immunol.,
163, 6694-6701.
203.Jackson, T., Morris, B. A., Martin, A. C.,
Lewis, D. F., and Sanders, P. G. (1992) Protein Eng., 5,
343-350.
204.Clark, L. A., Boriack-Sjodin, P. A., Eldredge,
J., Fitch, C., Friedman, B., Hanf, K. J., Jarpe, M., Liparoto, S. F.,
Li, Y., Lugovskoy, A., Miller, S., Rushe, M., Sherman, W., Simon, K.,
and van Vlijmen, H. (2006) Protein Sci., 15, 949-960.
205.Liu, Y., and Kuhlman, B. (2006) Nucleic
Acids Res., 34, W235-238.
206.Yuan, L., Kurek, I., English, J., and Keenan,
R. (2005) Microbiol. Mol. Biol. Rev., 69, 373-392.
207.Wells, J. A. (1990) Biochemistry,
29, 8509-8517.
208.Riechmann, L., and Weill, M. (1993)
Biochemistry, 32, 8848-8855.
209.Foote, J., and Winter, G. (1992) J.
Mol. Biol., 224, 487-499.
210.Schier, R., McCall, A., Adams, G. P., Marshall,
K. W., Merritt, H., Yim, M., Crawford, R. S., Weiner, L. M., Marks, C.,
and Marks, J. D. (1996) J. Mol. Biol., 263, 551-567.
211.Chen, C., Roberts, V. A., and Rittenberg, M. B.
(1992) J. Exp. Med., 176, 855-866.
212.Gustchina, E., Louis, J. M., Frisch, C., Ylera,
F., Lechner, A., Bewley, C. A., and Clore, G. M. (2009)
Virology, 393, 112-119.
213.Hanes, J., Schaffitzel, C., Knappik, A., and
Pluckthun, A. (2000) Nat. Biotechnol., 18,
1287-1292.