Reply to the Letter of G. Matera, A. Quirino, A. G. Lamberti, A. Foca, and M. C. Liberto
D. S. Kabanov* and I. R. Prokhorenko
Institute of Basic Biological Problems, Russian Academy of Sciences, Institutskaya ul. 2, 142290 Pushchino, Moscow Region, Russia; fax: (4967) 330-532; E-mail: kabanovd1@rambler.ru* To whom correspondence should be addressed.
Received March 18, 2011
DOI: 10.1134/S0006297911090148
Dear authors of Discussion Letter “Bartonella: stealthy pathogens or novel drug factories”, Giovanni Matera, Angela Quirino, Angelo Giuseppe Lamberti, Alfredo Foca, and Maria Carla Liberto.
We are kindly thankful to you for your attention and comments to our review “Structural analysis of lipopolysaccharides from Gram-negative bacteria” published in Biochemistry (Moscow). We apology profoundly for our mistake in quoting data of Prof. Dr. Giovanni Matera et al. [1] and Prof. Dr. Ulrich Zähringer et al. [2] on review p. 390: “In experiments with human whole blood, LPS from B. henselae whose pentaacyl lipid A contains an acyloxyacyl residue 16:0[3-O-(28:0(27-OH))] also did not induce the release of TNF-α [105]” [3]. In fact, these experiments were performed with LPS from Bartonella quintana rather than from Bartonella henselae as we reported [1-3].
At the time of publishing of the review, the structure of lipid A from B. quintana endotoxin, whose biological activity is described in the above-mentioned sentence, was not characterized. Only structures of lipid A from endotoxins of B. henselae, a bacterium related to B. quintana were known [2]. Structural variants of lipid A from B. henselae may include rare acyloxyacyl residues of different composition: 12:0[3-O(26:0(25-OH))], 12:0[3-O(28:0(27-OH))], and 16:0[3-O(26:0(25-OH))] [2]. The authors of this work revealed that the endotoxins from B. henselae were 1000 times less active than those from Salmonella enterica sv. Friedenau in inducing IL-8 synthesis by human embryonic kidney 293 cells (HEK 293) transfected with CD14 and Toll-like receptors [2]. In the early study by the group of M. C. Liberto and A. Foca using human whole blood it was determined that the endotoxins from B. quintana did not induce TNF-α synthesis [1]. Antagonistic activity of LPS from B. quintana was elucidated in the work of C. Popa with coworkers [4].
LPS molecules, which exhibit low endotoxic activity but retain the ability to interact with agonist-specific receptors blocking agonist-mediated cellular responses, have been referred as endotoxin antagonists. For instance, Re-LPS from B. quintana do not cause the stimulation of human mononuclear cells to production of TNF-α, IL-1β, or IL-6 up to tested concentration 1 μg/ml. In addition, the application of tenfold excess of antagonists, Re-LPS from B. quintana, was sufficient for prevention of agonistically highly active S-LPS Escherichia coli-driven mRNA synthesis of the mentioned cytokines [4]. Moreover, TNF-α and IL-6 release from human peripheral blood mononuclear cells activated by SR-LPS from Veillonella parvula ATCC 10790 were significantly inhibited by preliminary exposure of the cells to Re-LPS from B. quintana [5]. LPS with the similar antagonistic properties are produced by phototrophic bacteria such as Rhodobacter sphaeroides and Rhodobacter capsulatus [6] as well as by marine bacterium Marinomonas communis ATCC 27118T [7]. Unlike S-LPS from E. coli O111:B4 triggering already TNF-α production in human whole blood at their concentration 10 ng/ml, LPS from Rb. sphaeroides even at higher concentrations (up to 10 μg/ml) do not elicit such effect [6]. The lipid A from Rb. sphaeroides upon simultaneously addition with agonistically active lipid A from Salmonella minnesota R595 (1 ng/ml) or with LPS from Helicobacter pylori (100 ng/ml) inhibits IL-8 production by human monocytes at antagonist concentrations 100 ng/ml or 10 μg/ml, respectively [8]. The main structural features of lipids A from Rb. sphaeroides and Rb. capsulatus are the presence in their fatty acid residues of rare 3-oxo and unsaturated groups (table). By using lipid A synthetic analogs, the role of cis or trans configurations of double bond in the establishment of peculiar activity and antagonistic property of Rb. sphaeroides lipid A was elucidated [6]. It was shown that the capacity of the synthetic analogs to block TNF-α release in response to S-LPS E. coli O111:B4 (10 ng/ml) decreased in the series: 2′-trans-lipid A > LPS Rb. sphaeroides ≥ 2′-cis-lipid A > 2-cis-lipid A. Thus, the lipid A analog from Rb. sphaeroides with unsaturated bond in trans configuration possessed the most pronounced antagonistic activity [6]. Further, the reduction of 3-oxo and unsaturated groups in fatty acid residues (14:0(3-oxo) and 14:0[3-O(cisΔ714:1)]) of natural lipid A Rb. sphaeroides did not increase its endotoxic activity [12].
Synthetic analogs of natural lipids A
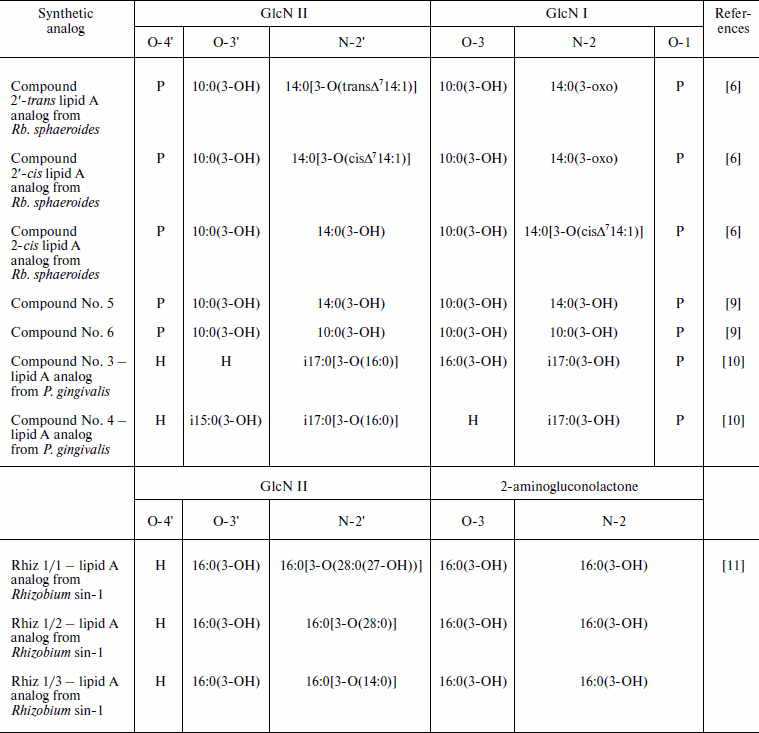
Note: GlcN, glucosamine; i, iso-fatty acids.
Synthetic compounds Rhiz 1/1, Rhiz 1/2 and Rhiz 1/3—analogs of lipid A from Rhizobium sin-1—provide additional examples of lipid A antagonists [11]. They differ from each other by acyloxyacyl residue at N-2′ position of a glucosamine (GlcN) residue (table). The exposure of differentiated Mono Mac 6 cells to these compounds up to their concentration 100 μg/ml (10% serum) led to nearly undetectable TNF-α production. The ability of Rhizobium sin-1 lipid A analogs to inhibit TNF-α release induced by S-LPS E. coli O55:B5 (10 ng/ml) was assessed and decrease in the sequence: LPS Rhizobium sin-1 > Rhiz 1/2 ≥ Rhiz 1/1 > Rhiz 1/3 [11, 13]. These data indicate that the antagonistic activity of lipid A Rhizobium sin-1 is favored by the presence of long chain 27-hydroxyoctacosanoic fatty acid residue. This conclusion is supported by the experiments showing that the substitution of 27-hydroxyoctacosanoic fatty acid residue by the relative shorter tetradecanoic acid residue (C14) devoid of hydroxyl group such in compound Rhiz 1/3 causes significant reduction of antagonistic activity of the studied synthetic analog [11, 13, 14].
The importance of fatty acid residue extension in the maintenance of lipids A antagonistic activity was also reported [9]. Thus, lipid A synthetic analog—tetraacylated compound No. 6 comprised of 3-hydroxydecanoic fatty acid residues only—does not prevent endotoxin-induced IL-6 production. However, another tetraacylated synthetic compound No. 5, differing from compound No. 6 by substitution of two 3-hydroxydecanoic fatty acid residues at N-2′ and N-2 positions of GlcN II and GlcN I by 3-hydroxytetradecanoic acid residues (table), has pronounced antagonistic activity [9].
The role of fatty acid distribution between two glucosamine residues in the manifestation of antagonistic activity by lipids A had been evaluated using tetraacylated synthetic analogs of lipid A from Porphyromonas gingivalis—compound No. 3 and compound No. 4 (table). The latter compound is characterized by asymmetrical [3 + 1] fatty acid residue distribution, whereas the former by a pseudo-symmetrical one [2 + 2]. Comparing the ability of these compounds to inhibit TNF-α release in response to S-LPS E. coli O55:B5 from differentiated Mono Mac 6 cells revealed that compound No. 3 was the most potent antagonist in comparison to compound No. 4 [10].
LPS from Francisella tularensis possessing tetraacylated lipid A with three 3-hydroxyoctadecanoic (C18) fatty acid residues in pseudo-symmetrical distribution have no either agonistic or antagonistic activities [15].
Taking these data into consideration, it can be concluded that the tendency of lipid A to exhibit antagonistic activity is dependent on its composition, with the most pronounced impact of the length and distribution of lipid A fatty acids between two glucosamine residues [10].
In conclusion, we would like to express our gratitude to colleagues of Dr. Giovanni Matera from Catanzaro University (Italy) for broadening the data regarding opportunistic infections caused by B. quintana and biological activity of LPS from this bacterium as well as for the possibility to discuss the dependence of lipid A antagonistic activity on its composition.
REFERENCES
1.Matera, G., Liberto, M. C., Quirino, A., Barreca,
G. S., Lamberti, A. G., Iannone, M., Mancuso, E., Palma E., Cufari, F.
A., Rotiroti, D., and Foca, A. (2003) Int. Immunopharmacol.,
3, 853-864.
2.Zähringer, U., Lindner, B., Knirel, Y. A., van
der Akker, W. M., Hiestand, R., Heine, H., and Dehio, C. (2004) J.
Biol. Chem., 279, 21046-21054.
3.Kabanov, D. S., and Prokhorenko, I. R. (2010)
Biochemistry (Moscow), 75, 383-404.
4.Popa, C., Abdollahi-Roodsan, S., Joosten, L. A.,
Takahashi, N., Sprong, T., Matera, G., Liberto, M. C., Foca, A., van
Deuren, M., Kullberg, B. J., van den Berg, W. B., van der Meer, J. W.,
and Netea, M. G. (2007) Infect. Immun., 75,
4831-4837.
5.Matera, G., Muto, V., Vinci, M., Zicca, E.,
Abdollahi-Roodsaz, S., van de Veerdonk, F. L., Kullberg, B.-J.,
Liberto, M. C., van der Meer, J. W. M., Foca, A., Netea, M. G., and
Joosten, L. A. B. (2009) Clin. Vac. Immunol., 16,
1804-1809.
6.Rose, J. R., Christ, W. J., Bristol, J. R., Kawata,
T., and Rossignol, D. P. (1995) Infect. Immun., 63,
833-839.
7.Vorob’eva, E. V., Krasikova, I. N., and
Solov’eva, T. F. (2006) Biochemistry (Moscow), 71,
759-766.
8.Bliss, Ch. M., Jr., Golenbock, D. T., Keates, S.,
Linevsky, J. K., and Kelly, C. P. (1998) Infect. Immun.,
66, 5357-5363.
9.Fukase, K., Oikawa, M., Suda, Y., Liu, W.-Ch.,
Fukase, Y., Shintaku, T., Sekljic, H., Yoshizaki, H., and Kusumoto, S.
(1999) J. Endotoxin Res., 5, 46-51.
10.Zhang, Y., Gaekwad, J., Wolfert, M. A., and
Boons, G.-J. (2008) Org. Biomol. Chem., 6, 3371-3381.
11.Zhang, Y., Wolfert, M. A., and Boons, G.-J.
(2007) Bioorg. Med. Chem., 15, 4800-4812.
12.Qureshi, N., Takayama, K., Meyer, K. C.,
Kirkland, T. N., Bush, C. A., Chen, L., Wang, R., and Cotter, R. J.
(1991) J. Biol. Chem., 266, 6532-6538.
13.Vasan, M., Wolfert, M. A., and Boons, G.-J.
(2007) Org. Biomol. Chem., 5, 2087-2097.
14.Vandenplas, M. L., Carlson, R. W., Jeyaretnam, B.
S., McNeill, B., Barton, M. H., Norton, N., Murray, T. F., and Moore,
J. N. (2002) J. Biol. Chem., 277, 41811-41816.
15.Hajjar, A. M., Harvey, M. D., Shaffer, S. A.,
Goodlett, D. R., Sjostedt, A., Edebro, H., Forsman, M., Bystrom, M.,
Pelletier, M., Wilson, C. B., Miller, S. I., Skerrett, S. J., and
Ernst, R. K. (2006) Infect. Immun., 74, 6730-6738.