Proteins of the Human 40S Ribosomal Subunit Involved in Hepatitis C IRES Binding as Revealed from Fluorescent Labeling
A. A. Malygin1, I. N. Shatsky2, and G. G. Karpova1*
1Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, pr. Lavrentieva 8, 630090 Novosibirsk, Russia; fax: (383) 363-5153; E-mail: karpova@niboch.nsc.ru2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119899 Moscow, Russia; fax: (495) 939-0338; E-mail: shatsky@genebee.msu.su
* To whom correspondence should be addressed.
Received July 24, 2012; Revision received September 7, 2012
Initiation of translation of genomic RNA (gRNA) of hepatitis C virus (HCV) is provided by a highly structured fragment in its 5′-untranslated region, the so-called Internal Ribosome Entry Site (IRES). In this work, the exposed NH2-groups of proteins in the 40S subunit of the human ribosome and in its binary complexes with RNA transcripts corresponding to the full-size HCV IRES or its fragments were probed using the N-hydroxysuccinimide derivative of the fluorescent dye Cy3. Comparison of efficiencies of modification of ribosomal proteins in free subunits and in their binary complexes with the RNA transcripts revealed ribosomal proteins involved in the HCV IRES binding. It was found that binding of the 40S subunits with the RNA transcript corresponding to full-size HCV IRES results in a decrease in modification levels of ribosomal protein (rp) S27 and, to a lesser extent of rpS10; also, a noticeable decrease in the efficiency of labeling of proteins RACK1/S2/S3a was observed. When a fragment of HCV IRES containing the initial part of the open reading frame (ORF) of the viral gRNA was deleted, the level of rpS10 modification became the same as in free subunits, whereas the levels of modification of rpS27 and the RACK1/S2/S3a group remained virtually unchanged compared to those observed in the complex of 40S subunit with the full-size HCV IRES. Binding of 40S subunits to a fragment of the HCV IRES lacking an ORF and domain II increased the modification level of the RACK1/S2/S3a proteins, while the efficiencies of labeling of rpS10 and rpS27 remained the same as upon the deletion of the ORF fragment. Comparison of these results with known structural and biochemical data on the organization of 40S subunit and the location of the HCV IRES on it revealed structural elements of the IRES contacting exposed lysine residues of the above-mentioned ribosomal proteins. Thus, it was found that the majority of exposed lysine residues of rpS27 are involved in the binding of the HCV IRES region formed by the junction of subdomains IIIa, IIIb, and IIIc with the central stalk of domain III, and that several lysine residues of rpS10 participate in the binding of the HCV IRES region corresponding to the initial part of the ORF of the viral gRNA. In addition, we concluded that lysine residues of rpS3a are involved in the binding of domains II and III of HCV IRES.
KEY WORDS: HCV IRES, human ribosome, fluorescent labeling, ribosomal protein S3a, ribosomal protein S10, ribosomal protein S27DOI: 10.1134/S0006297913010069
Abbreviations: cryo-EM, cryoelectron microscopy; HCV, hepatitis C virus; IRES, internal ribosome entry site; NHS-Cy3, 1,1′-(3-dihydroxypropyl)-3,3,3′,3′-tetramethylindocarbocyanine N-hydroxysuccinimide ester; ORF, open reading frame; rp, ribosomal protein; 5′-UTR, 5′-untranslated region.
The initiation of translation in eukaryotes occurs with the
participation of a large set of protein factors involved in the search
for the start codon AUG of mRNA by the 40S ribosomal subunit associated
with the ternary complex
eIF2•Met-tRNAiMet•GTP (see [1] for review). A characteristic feature of eukaryotic
mRNAs is the presence of a specific structural element at their
5′-ends named the cap, which is directly involved in this process
[1, 2]. However, many viruses,
whose genomes are represented by a single-stranded RNA lacking a
5′ cap, are able to realize their genetic information without the
participation of a number of translation initiation factors. The
functional role of these factors is performed by a highly structured
fragment in the 5′-untranslated region (5′-UTR) of the
genomic RNA (gRNA) called IRES (Internal Ribosome Entry Site) [3-5].
Hepatitis C virus (HCV), belonging to the Flaviviridae family, is one of the most dangerous pathogens; it infects more than 180 million people worldwide [6] and results in more than 350,000 deaths annually. The gRNA of this virus contains an IRES comprising a conserved stretch of about 330 nt with rigid secondary structure, which consists of several helices, hairpins, and pseudoknots forming separate domains and sub-domains [5]. The HCV IRES is able to bind to the 40S ribosomal subunit in the absence of translation initiation factors providing such location of the coding part of the gRNA virus in the mRNA-binding center that the initiation codon AUG occurs in the subunit area corresponding to the P-site [7].
To study the HCV IRES binding site on the 40S ribosomal subunit, approaches based primarily on chemical cross-linking, as well as the method of cryoelectron microscopy (cryo-EM) have been used. With the latter method, as far as we know, only two works at low resolution (about 15 Å) have been performed [8, 9], and these provided a general overview of the location of the HCV IRES on the 40S subunit. As for studies with the use of chemical cross-linking, in the early works it was shown that the HCV IRES forms a UV cross-link with ribosomal protein (rp) S5 during irradiation in its complex with 40S subunits [7, 10]. Later, a large number of 40S subunit proteins cross-linked to HCV IRES derivatives containing 4-thiouridines randomly replacing uridine residues were identified [11]. Relatively recently, with the use of HCV IRES derivatives bearing photoactivated groups in definite locations, ribosomal proteins neighboring subdomains IIId [12], IIIe [13], and IIb [12] of IRES complexed with the 40S subunit were established.
In this work we have used an approach based on fluorescent labeling of exposed lysine residues in proteins with N-hydroxysuccinimide derivative of the dye Cy3 (NHS-Cy3) to identify human 40S ribosomal subunit proteins involved in binding of the HCV IRES and its deletion mutants; by comparing the results obtained with the available data on the structure of 40S subunit and location of HCV IRES on the subunit, the regions of the IRES contacting these ribosomal proteins were established.
MATERIALS AND METHODS
Materials. In this work we used ribonucleoside triphosphates, spermidine, and DTT from Sigma (USA), acrylamide, N,N′-methylene-bis-acrylamide, KCl, SDS, and HEPES from AppliChem (Germany), and NHS-Cy3 from BioDye (Russia). Other reagents were of chemical grade from Russian suppliers. RNA polymerase of phage T7 was kindly provided by I. Eperon (University of Leicester, UK), ribonuclease inhibitor was from Fermentas (Lithuania) and Promega (USA), [α-32P]GTP (1000 Ci/mmol) was synthesized in the Laboratory of Biotechnology at the Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences (ICBFM SB RAS), and oligodeoxyribonucleotides were synthesized at the Group of Oligonucleotide Synthesis in the Laboratory of Medical Chemistry of ICBFM SB RAS.
Synthesis of RNA transcripts corresponding to HCV IRES and its deletion forms. DNA template for the synthesis of an RNA transcript corresponding to the full length HCV IRES (IRES40-372) – fragment 40-372 of HCV gRNA [4] was obtained by linearization of plasmid pXL40-372.NS′ [4] using endonuclease BamHI (Fermentas). DNA templates for RNA transcripts corresponding to the deletion forms of HCV IRES were synthesized by PCR using plasmid pXL40-372.NS′ as a template. For the synthesis of DNA template corresponding to the RNA transcript IRESΔORF, which lacked the sequence 345-372 of HCV IRES, the following oligodeoxyribonucleotides were used as primers: 5′-aaattaatacgactcactatagggagactcccctgtgaggaactac-3′ and 5′-catggtgcacggtctacg-3′ (primer R). For the synthesis of the DNA template corresponding to the RNA transcript IRESΔDIIΔORF, where both the sequence 345-372 and domain II of HCV IRES were absent, oligodeoxyribonucleotide 5′-aattaatacgactcactatagggagaccctcccgggagagcc-3′ and primer R were used. 32P-labeled RNA transcripts were synthesized using T7 transcription as described [14].
Binding of HCV IRES and its deletion forms to 40S ribosomal subunits. Ribosomal subunits were isolated from full-term placenta according to the method described in [15]. The 40S subunits were bound with HCV IRES and its deletion forms by incubation of 4 pmol subunits and the indicated amounts of RNA transcript in 10 µl of 20 mM HEPES-KOH, pH 7.5, containing 100 mM KCl and 2.5 mM MgCl2 (buffer A) at 30°C for 30 min (with deletion forms of HCV IRES) or at 37°C for 2 h (with RNA transcript IRES40-372). The extent of binding of RNA transcripts to subunits was determined by filtration through nitrocellulose filters as described [16].
Probing of exposed lysine residues in 40S subunit ribosomal proteins by NHS-Cy3. For fluorescent labeling of ribosomal proteins in free 40S subunits or their complexes with RNA transcripts, to 10 pmol of 40S subunits in 25 µl of buffer A, or to a mixture of 10 pmol of subunits and 40 pmol of an RNA transcript in 25 µl of buffer A (after preincubation of the mixture of the components under binding conditions, see above), 0.5 µl of 1 mM solution of NHS-Cy3 in DMSO was added. The reaction mixtures were incubated at 25°C for 4 h and then dialyzed through a VSWP 14250 membrane (Millipore, USA) against 10 ml of 20 mM Tris-HCl, pH 7.5, containing 100 mM KCl and 10 mM MgCl2 for 16 h. Ribosomal proteins were extracted from the 40S subunits with two volumes of acetic acid and precipitated with five volumes of acetone as described [17]. Pellets of ribosomal proteins were dried and dissolved in 10 µl of 125 mM Tris-HCl, pH 6.8, containing 2% SDS, 10% glycerol, 0.2% 2-mercaptoethanol, and 0.1% bromophenol blue, and then they were separated by 15% SDS-PAGE as described [13]. The gel was scanned on a Molecular Imager Pro FX (BioRad, USA) using the program QuantityOne, and the densitograms were normalized using as internal control the fluorescent label in the lane corresponding to the isolated 40S subunits.
Mass spectrometric analysis of proteins. Protein bands stained with Coomassie BB G250 were excised from the gel, and after removal of the dye by washing sulfhydryl groups of the proteins were reduced by DTT and blocked by iodoacetamide. Proteins in the gel were digested with trypsin, and the resulting peptides were extracted and purified on a ZipTip column (Millipore) according to the manufacturer’s recommendations. Mass spectra were recorded on a tandem TOF mass spectrometry MALDI-TOF Autoflex speed series LIFT (Bruker Daltonics, Germany) using 4-hydroxycyanocinnamic acid as a template.
RESULTS
To modify the primary amino groups of ribosomal proteins (amino groups in the side chains of lysine residues and the non-blocked N-terminal amino groups) exposed in human 40S ribosomal subunits, free or in complexes with the HCV IRES and its deletion mutants, fluorescent reagent NHS-Cy3 was used, and the labeled proteins were then analyzed by SDS-PAGE. This reagent readily reacts with deprotonated amino groups; therefore the modification was carried out in slightly alkaline buffer (pH 8.5) in the absence of components containing primary amino groups, and 40S subunits and NHS-Cy3 were taken in relatively low concentrations to avoid the effect of residues of dye cross-linked to proteins on their mobilities in the SDS-PAGE. To identify 40S ribosomal subunit proteins involved in the binding of HCV IRES, we compared the values of the efficiencies of 40S subunit modification with NHS-Cy3 in the absence and in the presence of HCV IRES or its deletion mutants. This approach is methodologically similar to footprinting of RNA in ribonucleoprotein complexes by chemical probes, so it can be called protein footprinting.
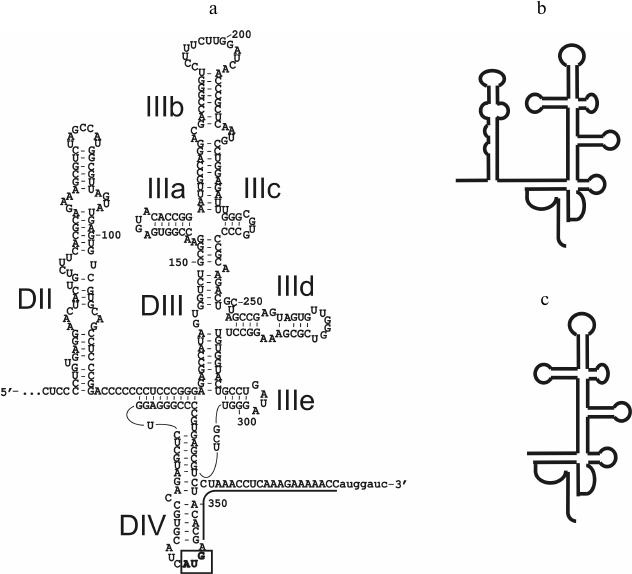
To determine the proteins of 40S subunits involved in the binding of HCV IRES, we used three RNA transcripts, one of which corresponded to HCV IRES described in [4], and the other two were its shortened forms. In particular, RNA transcript IRES40-372 (hereinafter referred to as full-length IRES) contained a portion of HCV gRNA from the 40th to the 372nd nt, which includes a large fragment of the 5′-UTR and the initial part of the open reading frame (ORF) (sequence 345-372) immediately after the AUG codon (positions 342-344) (Fig. 1). RNA transcript IRESΔORF corresponded to HCV IRES lacking the sequence 345-372 that includes a part of the ORF (Fig. 1). This sequence, while it does not contribute significantly to the binding of HCV IRES to 40S subunits [18], is required for its functional activity [4]. Finally, the RNA transcript IRESΔDIIΔORF corresponded to HCV IRES where both the ORF and domain II (Fig. 1), whose contribution to the affinity of the HCV IRES to 40S subunits is also insignificant [18], were lacking. It should be noted that domain IV of HCV IRES contains a part of the ORF and that only the 5′-part of this domain is present in the RNA transcripts that do not contain the ORF (Fig. 1).
Fig. 1. RNA transcripts corresponding to HCV IRES and its deletion mutant forms: a) secondary structure of RNA transcript IRES40-372, according to [25]. Domains and sub-domains are indicated, the initiator codon is boxed, the fragment of the open reading frame is underlined, nucleotide residues present in the RNA transcript but not related to HCV IRES are marked by lower case letters; b) diagram of the secondary structure of RNA transcript of IRESΔORF; c) diagram of the secondary structure of RNA transcript of IRESΔDIIΔORF.
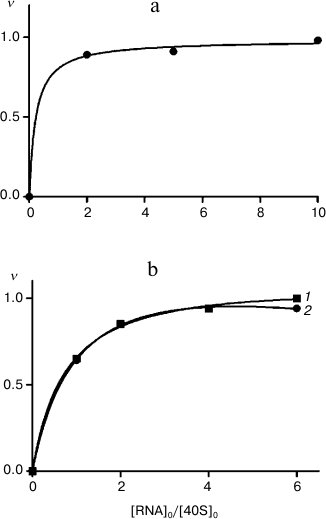
It is worth mentioning that participation of ribosomal protein in binding of 40S subunits with HCV IRES does not mean that all exposed lysine residues in this protein are also involved in the binding. Therefore, differences in modification levels of the protein in free 40S subunits and in their complexes with RNA transcripts may be in the range of the error of measurements. Hence, to maximize the difference in the levels of modification, it is necessary to use 40S subunits saturated with RNA transcripts up to the stoichiometric ratio. The 40S ribosomal subunits preparations used here were able to be saturated with RNA transcripts IRESΔORF and IRESΔDIIΔORF after incubation for 30 min at 30°C (Fig. 2). To saturate 40S subunits with RNA transcript IRES40-372, a longer incubation at a higher temperature (37°C, 2 h) was required (Fig. 2), probably because domain IV in HCV IRES (Fig. 1) should be melted for the subsequent binding of the AUG codon and a part of the ORF in the ribosomal mRNA-binding center.
Fig. 2. Binding of RNA transcripts IRES40-372, IRESΔORF, and IRESΔDIIΔORF to human 40S ribosomal subunits. Adsorption isotherms of IRES40-372 (a), IRESΔORF (b, 1), and IRESΔDIIΔORF (b, 2) are presented; ν, extent of binding of RNA, mol/mol 40S subunits. The relative error of measurement was 10%.
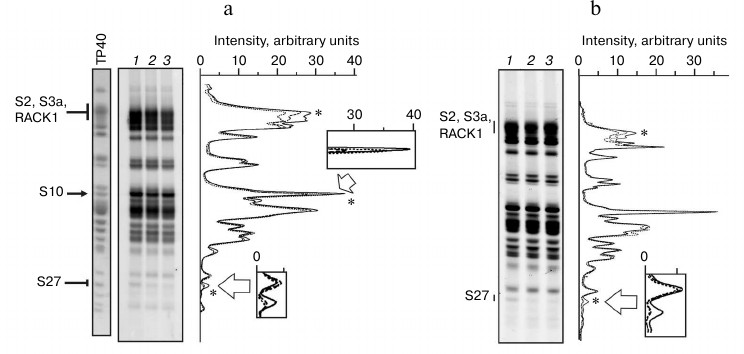
The results of probing of exposed primary amino groups of ribosomal proteins with NHS-Cy3 in isolated 40S subunits and their complexes with IRES40-372, IRESΔORF, and IRESΔDIIΔORF are shown in Fig. 3. Analysis of the fluorescence densitograms obtained by scanning of the gels after SDS-PAGE separation of the labeled ribosomal proteins showed that almost all the 40S subunit proteins can be modified by NHS-Cy3. This is consistent with the general view on the structure of the 40S ribosomal subunit [19] according to which proteins hold peripheral positions in the subunit. Different intensities of fluorescence in the protein bands observed in the gels reflect different numbers of exposed lysine residues in the corresponding proteins and their different accessibilities to NHS-Cy3. One can see that the binding of RNA transcripts to 40S subunits results in a significant reduction in the fluorescence intensities in some protein bands (Fig. 3), indicating the involvement of lysine residues of the corresponding proteins in binding to HCV IRES. However, fluorescence intensity in only three protein bands decreased more than by 10% as compared to the intensity of the corresponding bands observed in the experiments with the isolated subunits (relative measurement error was less than 5%).
So, the binding of all three RNA transcripts resulted in a decrease in the fluorescence intensity of the band at the top of the gel (Fig. 3), which was most pronounced with IRESΔORF (about 15%). Mass spectrometric analysis of the protein composition of this band revealed the presence of ribosomal proteins S2, S3a, and RACK1 (data not shown), which, in general, is consistent with previously published data [20]. In addition, in the complex of IRES40-372 with the 40S subunits, a 22% decrease in the fluorescence intensity of the protein band in the middle of the gel was observed, whereas with the complexes containing other RNA transcripts lacking the fragment corresponding to the ORF, the intensity of this band was unchanged (Fig. 3). Earlier it was established that this band corresponds to rpS10 [20]. Finally, there was a protein band at the bottom of the gel whose fluorescence intensity with free 40S subunits was small, but the presence of each of the RNA transcripts reduced by more than 70% (Fig. 3). Mass spectrometric analysis of proteins in this gel band showed that it contained only rpS27 (data not shown). Strong protection of this protein against the modification in the presence of each of the three RNA transcripts indicates that most of the exposed lysine residues in the protein are involved in the binding of HCV IRES. Protection from fluorescent labeling by the shortest RNA transcript (IRESΔDIIΔORF) indicates that rpS27 is not involved in the binding of domain II or the fragment of the ORF of HCV IRES, i.e. in fact, it binds only to domain III.
Fig. 3. Fluorescent labeling of 40S subunits in complexes with HCV IRES or its fragments. a) Fluorogram of 14% SDS-PAGE after separation of ribosomal proteins extracted from modified 40S subunits, free or complexed with IRES40-372 or IRESΔORF (lanes 1-3, respectively). Lane TP40, 40S subunit proteins separated in the same gel and stained with Coomassie Brilliant Blue R250. Positions of proteins in the gel are marked on the left. Right, densitometric profiles of lanes 1 (solid line), 2 (long dashed line), and 3 (short dashed lines). Asterisks indicate peaks whose intensity varies. In separated sections, areas corresponding to proteins S10 and S27 are zoomed. b) Fluorogram of 14% SDS-PAGE after separation of ribosomal proteins extracted from modified 40S subunits, free or complexed with IRESΔORF or IRESΔDIIΔORF (lanes 1-3, respectively). Right, densitometric profiles of lanes 1 (solid line), 2 (long dashed line), and 3 (short dashed lines). Designations are the same as on panel (a).
DISCUSSION
In this work, using a method of fluorescent labeling of proteins of human 40S ribosomal subunit with NHS-Cy3 and a set of RNA transcripts corresponding to the HCV IRES and its fragments lacking either the ORF or domain II and the ORF simultaneously, ribosomal proteins participating in the binding of HCV IRES were identified and IRES regions involved in their binding were established. It turned out that rpS27 contacts domain III, and rpS10 interacts with part of the ORF present in HCV IRES. In addition, binding of 40S subunits with domains II and III of HCV IRES involves a protein (or proteins) from the group of proteins RACK1/S2/S3a, which could not be resolved by SDS-PAGE.
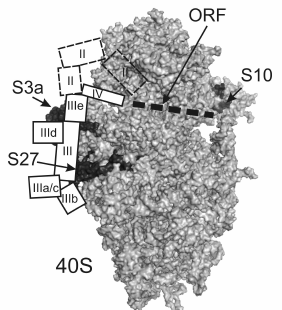
According to X-ray analysis of ribosomes of yeast Saccharomyces cerevisiae and of ciliate Tetrahymena thermophila, rpS27 is located under the subunit platform, between proteins S3a and S13 [19, 21] (Fig. 4). This protein is specific to eukaryotes and archaea, and its part exposed on the surface of the 40S subunit forms a zinc finger class C4 structure [22]. Assuming that rpS27 structure in the human 40S ribosomal subunit forms the same fold as that of the homologous proteins in 40S subunits of yeast and ciliates, and using crystal structures of these subunits, one can determine how many lysine residues in human rpS27 are exposed on the 40S subunit surface. It turned out that three of 10 lysine residues in the protein are accessible to NHS-Cy3 modification. Our data (decrease in the modification level of rpS27 by 70% in the presence of HCV IRES or its fragments) suggest that at least two of these three residues are involved in the binding of domain III of HCV IRES. To compare the results obtained with cryo-EM data, the electron density maps presented in [8], which provide more detailed information on the location of HCV IRES on the 40S subunit than the maps presented in [9], were used together with the crystal structure of the 40S ribosomal subunit of yeast [19]. Indeed, it appears that the site of domain III in HCV IRES formed by the junction of four helices (IIIa, IIIb, IIIc, and the central stem of domain III) is located near rpS27, and probably contacts it (Fig. 4). Interestingly, the deletion of this region in HCV IRES deprives it of the ability to bind to 40S subunits [18], which points to the critical role of this region in the functional activity of HCV IRES. The results presented here are consistent with data on cross-linking of rpS27 in 40S subunits to 4-thiouridine-containing derivatives of HCV IRES and its deletion mutant lacking domain II [11].
Fig. 4. Comparison of results on fluorescent labeling of human 40S ribosomal subunits in complex with HCV IRES with X-ray crystallographic data on the structure of the 40S ribosomal subunits of yeast [19] and with cryo-EM data on the location of the HCV IRES on 40S subunits [8]. The 40S subunit is presented in the standard orientation, the solvent side of the subunit is shown, and ribosomal proteins S3a, S27, and S10 are marked in black and indicated. Locations of HCV IRES domains are presented by white numbered rectangles. The mRNA binding channel occupied by the ORF is marked by the dotted line.
Eukaryote-specific rpS10, according to X-ray analysis of ribosomes from lower eucaryotes [19, 21], is located at the “beak” region of the 40S subunit near the mRNA entry site (Fig. 4). Since partial protection of exposed lysine residues in this protein from modification by NHS-Cy3 was observed only in the presence of IRES40-372 (Fig. 3), which in contrast to IRESΔORF contains 28 nt of the ORF after the AUG-codon, we assume that it is this part of the ORF that protects lysine residues in rpS10 from modification. Therefore, rpS10 probably participates rather in formation of the mRNA binding center than in the binding of the 5′-UTR of the HCV gRNA. According to the X-ray analysis, rpS10 is located in the 40S subunit near rpS3, which is one of the ribosomal components forming the mRNA entry site [19]. In addition, according to the data on affinity modification of human ribosomes with mRNA analogs, rpS3 neighbors the mRNA region 3′ to the A-site codon (nucleotides at positions from +7 to +12 relatively to the first nucleotide of a codon in the P-site) [23, 24]. Thus, there is every reason to assume that rpS10 contacts mRNA nucleotides located 3′ of the nucleotide at position +12, and that, according to our data, several of its 13 lysine residues participate in this interaction. All this also explains the previously observed cross-link of rpS10 in 40S subunits with HCV IRES carrying 4-thiouridine residues scattered randomly over the molecule [11], because the uridine residues at positions +15 and +21 of the HCV IRES ORF could be substituted for 4-thiouridine.
Proteins S2, S3a, and RACK1, the largest in the 40S subunit, form a broad band at the top of the gel after SDS-PAGE (Fig. 3), and therefore one cannot accurately judge which of these proteins was protected by RNA transcripts from modification by NHS-Cy3. However, comparing cryo-EM data on location of the HCV IRES on the 40S subunit [8] and X-ray crystallographic data on the structure of the 40S subunit [19], we conclude that of these three proteins only rpS3a, which is specific to eukaryotes and archaea, is located close to the binding site of HCV IRES near subdomains IIId and IIIe and the lower part of domain II (Fig. 4). It is noteworthy that rpS3a was detected earlier among proteins cross-linked to HCV IRES derivatives bearing reactive groups at the nucleotides in the subdomains IIIe [13] and IIId in their complexes with human 40S ribosomal subunits [12]. Thus, the decreasing of the fluorescent labeling intensity of the proteins of the 40S subunit migrating at the top of the gel observed with RNA transcripts IRES40-372, IRESΔORF, and IRESΔDIIΔORF is most likely caused by the protection of rpS3a from the modification. This protein seems to contact not only domain III of HCV IRES, but also domain II, because the efficacy of the modification of the protein in the presence of IRESΔDIIΔORF is higher than that in the complex of the 40S subunits with IRESΔORF.
Thus, applying a protein footprinting method with the use of a fluorescent reagent specific for NH2-groups, we were able to establish that the lysine residues of the ribosomal proteins S3a, S10, and S27, which have no homologs in eubacteria, are directly involved in the formation of the HCV IRES binding site on the human 40S ribosomal subunit. This information together with the structural and biochemical data on the structure of 40S subunits and the HCV IRES location on 40S subunit allowed us to determine the structural elements of the IRES contacting these proteins, expanding significantly our knowledge on the structural basis of the molecular interactions providing the binding of HCV IRES to 40S subunit.
This work was supported by the Russian Foundation for Basic Research (grant 11-04-00672-a to A.M.) and by the program of the Presidium of the Russian Academy of Sciences “Molecular and Cellular Biology” (G.K.).
REFERENCES
1.Jackson, R. J., Hellen, C. U., and Pestova, T. V.
(2010) Nat. Rev. Mol. Cell. Biol., 11, 113-127.
2.Merrick, W. C. (2004) Gene, 332,
1-11.
3.Rijnbrand, R., Bredenbeek, P., van der Straaten,
T., Whetter, L., Inchauspe, G., Lemon, S., and Spaan, W. (1995) FEBS
Lett., 365, 115-119.
4.Reynolds, J. E., Kaminski, A., Kettinen, H. J.,
Grace, K., Clarke, B. E., Carroll, A. R., Rowlands, D. J., and Jackson,
R. J. (1995) EMBO J., 14, 6010-6020.
5.Honda, M., Beard, M. R., Ping, L. H., and Lemon, S.
M. (1999) J. Virol., 73, 1165-1174.
6.Brocard, M., Paulous, S., Komarova, A. V., Deveaux,
V., and Kean, K. M. (2007) Virus Genes, 35, 5-15.
7.Pestova, T. V., Shatsky, I. N., Fletcher, S. P.,
Jackson, R. J., and Hellen, C. U. (1998) Genes Dev., 12,
67-83.
8.Boehringer, D., Thermann, R., Ostareck-Lederer, A.,
Lewis, J. D., and Stark, H. (2005) Structure, 13,
1695-1706.
9.Spahn, C. M., Kieft, J. S., Grassucci, R. A.,
Penczek, P. A., Zhou, K., Doudna, J. A., and Frank, J. (2001)
Science, 291, 1959-1962.
10.Fukushi, S., Okada, M., Stahl, J., Kageyama, T.,
Hoshino, F. B., and Katayama, K. (2001) J. Biol. Chem.,
276, 20824-20826.
11.Otto, G. A., Lukavsky, P. J., Lancaster, A. M.,
Sarnow, P., and Puglisi, J. D. (2002) RNA, 8,
913-923.
12.Babaylova, E., Graifer, D., Malygin, A., Stahl,
J., Shatsky, I., and Karpova, G. (2009) Nucleic Acids Res.,
37, 1141-1151.
13.Laletina, E., Graifer, D., Malygin, A., Ivanov,
A., Shatsky, I., and Karpova, G. (2006) Nucleic Acids Res.,
34, 2027-2036.
14.Malygin, A. A., Graifer, D. M., Laletina, E. S.,
Shatsky, I. N., and Karpova, G. G. (2003) Mol. Biol. (Moscow),
37, 1027-1034.
15.Matasova, N. B., Myltseva, S. V., Zenkova, M. A.,
Graifer, D. M., Vladimirov, S. N., and Karpova, G. G. (1991) Anal.
Biochem., 198, 219-223.
16.Ilin, A. A., Malygin, A. A., and Karpova, G. G.
(2011) Biochim. Biophys. Acta, 1814, 505-512.
17.Vladimirov, S. N., Ivanov, A. V., Karpova, G. G.,
Musolyamov, A. K., Egorov, T. A., Thiede, B., Wittmann-Liebold, B., and
Otto, A. (1996) Eur. J. Biochem., 239, 144-149.
18.Kieft, J. S., Zhou, K., Jubin, R., and Doudna, J.
A. (2001) RNA, 7, 194-206.
19.Ben-Shem, A., Garreau de Loubresse, N., Melnikov,
S., Jenner, L., Yusupova, G., and Yusupov, M. (2011) Science,
334, 1524-1529.
20.Malygin, A. A., Shaulo, D. D., and Karpova, G. G.
(2000) Biochim. Biophys. Acta, 1494, 213-216.
21.Rabl, J., Leibundgut, M., Ataide, S. F., Haag,
A., and Ban, N. (2011) Science, 331, 730-736.
22.Razin, S. V., Borunova, V. V., Maksimenko, O. V.,
and Kantidze, O. L. (2012) Biochemistry (Moscow), 77,
217-226.
23.Graifer, D., Molotkov, M., Styazhkina, V.,
Demeshkina, N., Bulygin, K., Eremina, A., Ivanov, A., Laletina, E.,
Ven’yaminova, A., and Karpova, G. (2004) Nucleic Acids
Res., 32, 3282-3293.
24.Pisarev, A. V., Kolupaeva, V. G., Yusupov, M. M.,
Hellen, C. U., and Pestova, T. V. (2008) EMBO J., 27,
1609-1621.
25.Zhao, W. D., and Wimmer, E. (2001) J.
Virol., 75, 3719-3730.