REVIEW: O-Antigens of Bacteria of the Genus Providencia: Structure, Serology, Genetics, and Biosynthesis
O. G. Ovchinnikova1*, A. Rozalski2, B. Liu3, and Y. A. Knirel1
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Leninsky pr. 47, 119991 Moscow, Russia; fax: (499) 137-6148; E-mail: olga.ovchinnikova@gmail.com2Department of Immunobiology of Bacteria, Institute of Microbiology, Biotechnology and Immunology, University of Lodz, PL 90-237 Lodz, Poland
3TEDA School of Biological Sciences and Biotechnology, Nankai University, 23 Hongda Street, TEDA, 300457 Tianjin, P. R. China
* To whom correspondence should be addressed.
Received January 30, 2013; Revision received February 28, 2013
The genus Providencia consists of eight species of opportunistic pathogenic enterobacteria that can cause enteric diseases and urinary tract infections. The existing combined serological classification scheme of three species, P. alcalifaciens, P. stuartii, and P. rustigianii, is based on the specificity of O-antigens (O-polysaccharides) and comprises 63 O-serogroups. Differences between serogroups are related to polymorphism at a specific genome locus, the O-antigen gene cluster, responsible for O-antigen biosynthesis. This review presents data on structures of 36 O-antigens of Providencia, many of which contain unusual monosaccharides and non-carbohydrate components. The structural data correlate with the immunospecificity of the O-antigens and enable substantiation on a molecular level of serological relationships within the genus Providencia and between strains of Providencia and bacteria of the genera Proteus, Escherichia, and Salmonella. Peculiar features of the O-antigen gene cluster organization in 10 Providencia serogroups and biosynthetic pathways of nucleotide precursors of specific monosaccharide components of the O-antigens also are discussed.
KEY WORDS: Providencia, lipopolysaccharide, O-antigen, gene cluster, biosynthesis, serological specificityDOI: 10.1134/S0006297913070110
Abbreviations: Col, colitose; ECA, enterobacterial common antigen; FucNAc4N, 2-acetamido-4-amino-2,4-dideoxyfucose; GalA, galacturonic acid; Kdo, 3-deoxy-d-manno-oct-2-ulosonic acid; LPS, lipopolysaccharide; OGC, O-antigen gene cluster; Qui, quinovose; Qui4N, 4-amino-4-deoxyquinovose; UndP, undecaprenyl phosphate.
Bacteria of the genus Providencia from the Enterobacteriaceae
family reside in soil, wastewater, and polluted water reservoirs; they
also are isolated from a broad range of living organisms. These
opportunistic pathogens can cause acute enteric and urinary tract
diseases, most often in young children and patients whose immune system
has been compromised by surgery or burns. The bacteria first were
described in 1920 by Ornstein, who called them Bacillus
inconstans. In 1943-1946, Stuart isolated bacterial cultures of
what he referred to as “paracolon 29911” from patients
suffering from enteric diseases and characterized their biochemical
properties in detail. In 1944, Gomes described Eberthella
alcalifaciens bacteria, which later would be characterized again
and would become the Providencia type strain. In 1951, Kauffmann
proposed to call Providence strains from the “paracolon
29911” group after the town where Stuart worked at Brown
University, and in 1952, Kauffmann and Edwards reclassified this group
as the genus Providencia. During the following decade, the
position of Providencia bacteria in the taxonomic classification
and their relationships with closely related genera Proteus
and Morganella were revised repeatedly, and species were
moved across these genera (for review see [1]).
In 1962, the genus Providencia finally was accepted as an independent genus and included, together with the genus Proteus, to the Proteeae tribe; later, the genus Morganella was appended to the tribe. Two Providencia biogroups described by Ewing were recognized as the species Providencia alcalifaciens and Providencia stuartii. The most recent changes in the Providencia taxonomy occurred after introduction of DNA–DNA hybridization to classification of bacteria. Based on similarities in the genome, Proteus rettgeri was reclassified as Providencia rettgeri, and Providencia alcalifaciens biogroup 3 became a separate species called Providencia rustigianii. In 1986, Muller, while studying bacteria isolated from penguins’ feces, described a new species, Providencia heimbachae; later, a strain of this species also was found in a patient with idiopathic diarrhea [2]. In 2006, a new species, Providencia vermicola, was proposed for strains that infect juveniles of an entomopathogenic nematode [3]. In 2009, representatives of two new species, Providencia sneebia and Providencia burhodogranariea, were isolated from hemolymph of fruit flies [4]; thereby, the number of Providencia species increased to eight.
In 1954, Ewing et al. developed the first serological classification scheme for P. alcalifaciens, P. stuartii, and P. rustigianii [5]. He collected 631 strains, including “paracolon 29911” cultures, and added strains from Italy and the USA. Based on the serospecificity of the heat-stable somatic O-antigens, heat-labile flagella H-antigens, and capsular K-antigens, 56 O-serogroups, 28 H-serogroups, and two K-serogroups were established, which in various combinations gave rise to 125 serotypes. From 543 smooth strains, ca. 40% were classified into six serogroups (O3, O4, O9, O19, O21, and O23), the O3 being the most common (15.5% strains). Later, Ewing identified six more O-antigen forms, extending the number of O-serogroups to 62 [6].
In 1976, Penner et al. reconstructed Ewing’s scheme [7]. From 62 original strains they received from Ewing, 52 strains showed biochemical reactions typical of Providencia, and each had serological O-specificity different from type strains of all other O-serogroups. Ten strains from Ewing’s collection were replaced by strains from Penner’s personal collection, which could not be classified into any of Ewing’s O-serogroups; the total number of O-serogroups (62) remained thus unchanged. Three original strains (serogroups O17, O18, O56) were excluded based on biochemical reactions typical of Providencia rettgeri (called Proteus rettgeri at that time), three more strains (serogroups O57-O59) displayed atypical positive reaction in the ornithine decarboxylase test. Four strains (O15, O24, O26, O39) were replaced based on serological data. Antisera against serogroup O15 strains could not be used for serotyping as they showed a low titer in the homologous reaction, while cross-reacting with several heterologous strains. Absorption of antisera against three pairs of strains (O20/O26, O24/O42, O35/O39) by heterologous antigens completely abolished the reactivity with both homologous and heterologous strains in each pair, thus suggesting that the O-antigens are pairwise identical.
Later, Penner applied this scheme to serotyping of 829 clinical isolates of P. stuartii, and, as a result, one more serogroup, O63, was added [8]. The most common serogroups were found to be O4, O17, O25, O52, O55, O56, and O63 (combined, they covered 91% isolates). A similar serological screening of 86 clinical isolates of P. alcalifaciens [9] revealed no additional O-antigen forms, and the most common serogroup was O3. Strains of P. stuartii and P. alcalifaciens possessed different O-antigenic determinants, and in most cases did not cross-react between species [8].
Serological relationships within the genus Providencia also were studied by Levina et al. [10]. They used a collection of Providencia strains representing 61 serogroups (O1-O61), which was obtained from Rauss (Hungarian National Collection of Medical Bacteria, National Institute of Hygiene, Budapest) and supposed to be identical to the original Ewing’s collection. Data obtained by Levina et al. confirmed the identity of the O-antigens in the O20/O26, O24/O42, and O35/O39 serogroup pairs and revealed yet another such pair, O55/O57 (Penner has replaced the O57 strain in his collection, and, hence, got no data for this pair). Later, our biochemical testing confirmed that all but two strains from the Hungarian collection belonged to P. alcalifaciens, P. stuartii, or P. rustigianii, whereas strains of serogroups O58 and O59 should be moved to the genus Morganella, the species M. morganii.
The specificity of the immune response to a certain O-serogroup is defined by the fine structure of the O-antigen. From the chemical point of view, the O-antigen or O-specific polysaccharide (O-polysaccharide) is a polysaccharide chain of S-form (“smooth form”) lipopolysaccharide (LPS). It is built up of oligosaccharide repeating units or, less commonly, is a homopolysaccharide. The lipid portion of LPS (lipid A) forms the outer layer of the outer membrane of the bacterial cell wall and anchors the LPS molecule to the cell surface by hydrophobic interactions with inner layer phospholipids. Lipid A also is responsible for manifestation of most biological activities of LPS, including its recognition by specific cell receptors of the host immune system that triggers the innate immunity defense mechanisms. The O-polysaccharide protruding towards the environment is attached to lipid A through a so-called core oligosaccharide. It bears immunodeterminants (epitopes or O-factors), which are binding sites for specific antibodies produced by the adaptive immune system in response to infection and required for efficient phagocytosis. Serological cross-reactions observed within species and genera (sometimes even between genera and families) are substantiated by the occurrence of identical or structurally similar O-antigen epitopes.
The S-form of LPS is typical of the majority of wild-type bacterial strains, which produce smooth colonies. Bacteria that form rough colonies are either natural or laboratory R-mutants whose R-form LPS lacks any O-polysaccharide chain. In addition, there is a so-called SR-form LPS having a short-chain O-antigen consisting of an oligosaccharide with the same structure as the repeating unit of the O-polysaccharide.
Recently, serological methods for typing of bacteria are largely supplanted by more precise molecular detection methods, such as polymerase chain reaction and microarray. Depending on the primers or probes targeting the specific genes, molecular typing can differentiate between genera, species, and serotypes of microorganisms. On the genetic level, differences between O-antigen structures are due to variations in O-antigen gene clusters, which map at different genomic loci in various bacterial genera. The clusters for O-antigens that are synthesized by the Wzx/Wzy-dependent pathway, which is most common for bacterial polysaccharides, include: i) genes necessary for synthesis of nucleotide-activated monosaccharides that are specific components of O-antigens; ii) genes for glycosyltransferases involved in the assembly of a biological repeating unit on an undecaprenyl diphosphate carrier, and iii) O-antigen processing genes, which are responsible for translocation of the repeating unit through the inner membrane and its polymerization. Knowledge of the O-antigen structures combined with DNA sequence data for the O-antigen gene clusters allow assignment of putative functions of O-antigen biosynthesis genes, which, if necessary, can be verified biochemically. These data also enable designing specific markers for molecular typing of strains and development on their basis of methods for molecular diagnostics of infectious diseases.
The present review is devoted to chemical, biological, and genetic characterization of O-antigens of representatives of three Providencia species: P. alcalifaciens, P. stuartii, and P. rustigianii.
CHEMICAL COMPOSITION AND STRUCTURES OF O-ANTIGENS
For structural analysis, LPS were subjected to mild acid or mild alkaline hydrolysis to yield a lipid-free polysaccharide or an O-deacylated LPS, respectively. Studied were LPS from 56 O-serogroups from the Hungarian National Collection of Medical Bacteria that includes strains of 59 Providencia O-serogroups (from 62 original Ewing’s serogroups; O58 and O59 were moved to the genus Morganella, and O62 was absent from the Hungarian collection). Strains from 14 serogroups studied (O1, O10, O13, O15, O17, O37, O41, O42, O50, O53-O56, O61) produced R-form LPS lacking any O-polysaccharide chain, which could result from an S→R dissociation of strains during their storage for a long time. High molecular mass polysaccharides from strains of four serogroups (O11, O24, O38, O45) were not bound to lipid A and were substantially different in structure and composition from the O-antigens of other serogroups. Particularly, the O11 polysaccharide contained derivatives of 2,4-diamino-2,4,6-trideoxyglucose and 2-amino-2-deoxygalacturonic acid, which do not occur in Providencia O-antigens [11]. Strains of serogroups O24, O38, and O45 share a polysaccharide that strikingly resembles the carbohydrate chain of bacterial cell wall peptidoglycan [12]. It has a disaccharide repeating unit that is smaller than repeating units of any Providencia O-antigens (Table 1). These polymers were supposed to be capsular polysaccharides (K-antigens) and are not covered in this review. Therefore, LPS of serogroups O11, O24, O38, and O45 also were present in the R-form.
Table 1. Structures of the O-antigens of
P. alcalifaciens, P. stuartii, and P. rustigianii
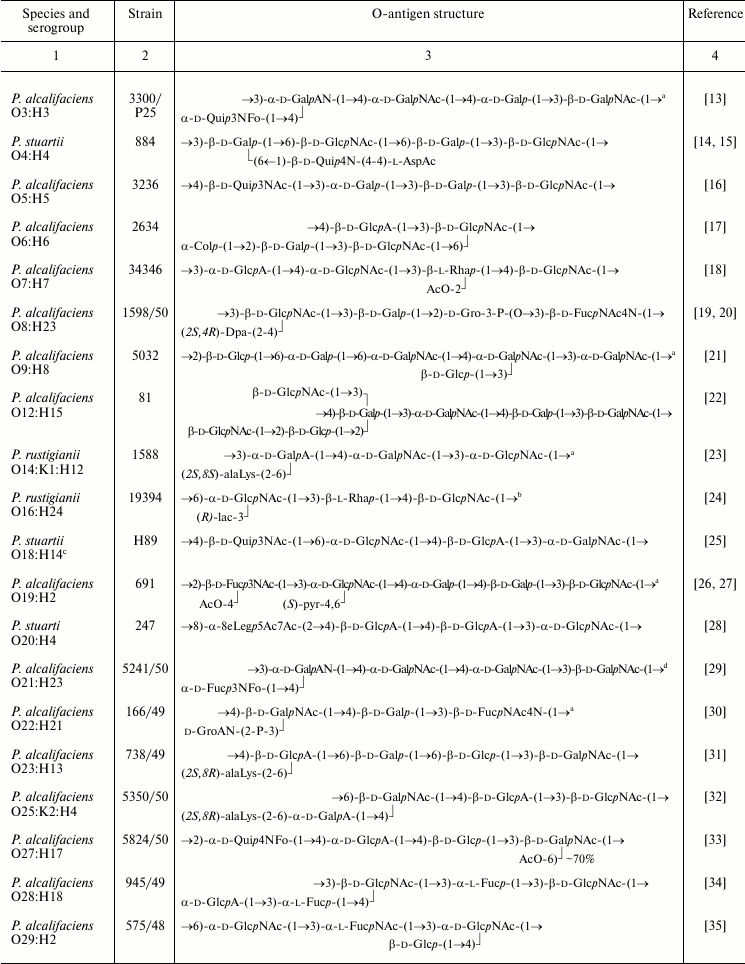
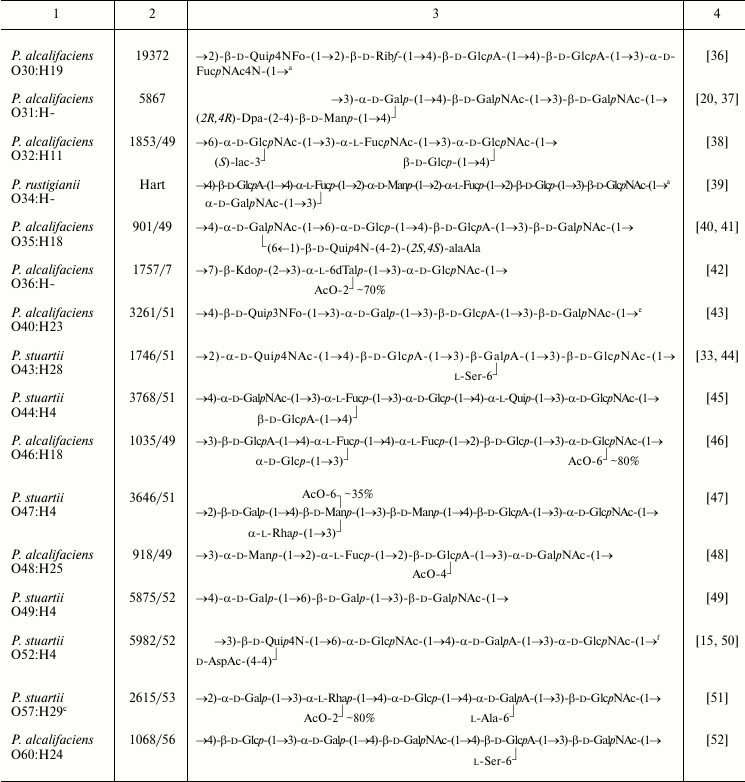
Note: Abbreviations for monosaccharides and non-carbohydrate components
are listed in Table 2.
a The structure represents the biological repeating unit of
the O-antigen.
b Published as the structure of P. alcalifaciens
O16 but later it was found that the strain studied belonged to
P. rustigianii O16.
c In the Penner’s classification scheme, strains from
Ewing’s serogroups O18 and O57 used for structural analysis
were replaced, and it remains unclear which of Penner’s
serogroups the presented O-antigen structures belong to.
d The occurrence of GalA as amide was overlooked in
structural studies of the O-polysaccharide [29]
and demonstrated later by mass spectrometric analysis of an SR-form
LPS-derived oligosaccharide consisting of the core with one O-antigen
repeating unit attached.
e The O-antigen contains a small amount of O-acetyl
groups, whose position(s) was not determined.
f Published as the structure of P. stuartii O33,
but later it was found that the strain studied belonged to P.
stuartii O52.
The O-polysaccharides were obtained from strains representing 38 O-serogroups and studied by chemical methods combined with one- and two-dimensional NMR spectroscopy. In accordance with the serological data (see introductory section), structural analysis confirmed the identity of the O-antigens in the O20/O26 and O35/O39 serogroup pairs. LPS of strains representing the third pair with serologically identical O-antigens, O24/O42, as well as LPS of O55 strain of the O55/O57 pair, occurred in the R-form and possessed no O-polysaccharide chains. The unique structures established for the O-polysaccharides of 36 O-serogroups are shown in Table 1.
All Providencia O-antigens studied are heteropolysaccharides built up of linear or branched oligosaccharide repeating units ranging from tri- to heptasaccharide. A common feature of most O-polysaccharides (~90%) is their acidic or (in case of the presence of FucNAc4N or an amino acid of the opine family) zwitterionic character.
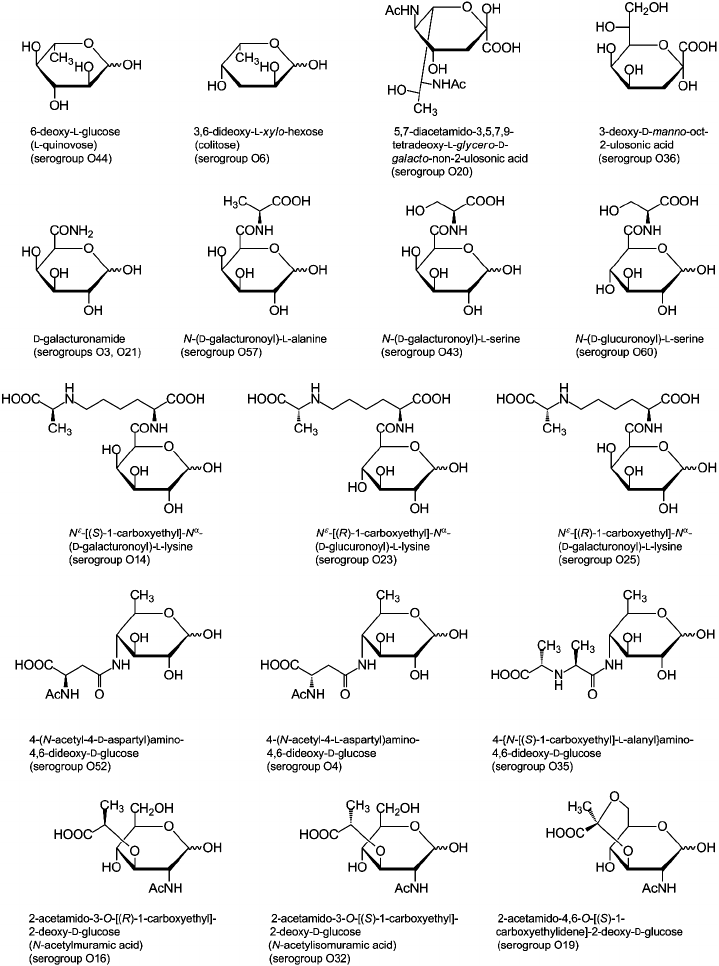
Sugar and non-carbohydrate components found in Providencia O-antigens are listed in Table 2 and are typical of enterobacteria [53, 54]. For example, the most common monosaccharides are 2-acetamido-2-deoxy-d-glucose and 2-acetamido-2-deoxy-d-galactose; d-glucuronic acid, d-galactose, and d-glucose also are fairly abundant. Less common are d-mannose, l-rhamnose, l-fucose, d-galacturonic acid, various 6-deoxyamino sugars, and 4-amino-2-acetamido-2,4-dideoxy-d-fucose. In addition, the following monosaccharides that occur rarely in O-antigens were identified: 6-deoxy-l-glucose (l-quinovose), 6-deoxy-l-talose, 3,6-dideoxy-l-xylo-hexose (colitose), and two aldulosonic acids, 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) and di-N-acetyl-8-epilegionaminic acid (Fig. 1). Kdo is a common component of the LPS core but found in O-polysaccharides of only a few bacteria.
Table 2. Composition of Providencia
O-antigens
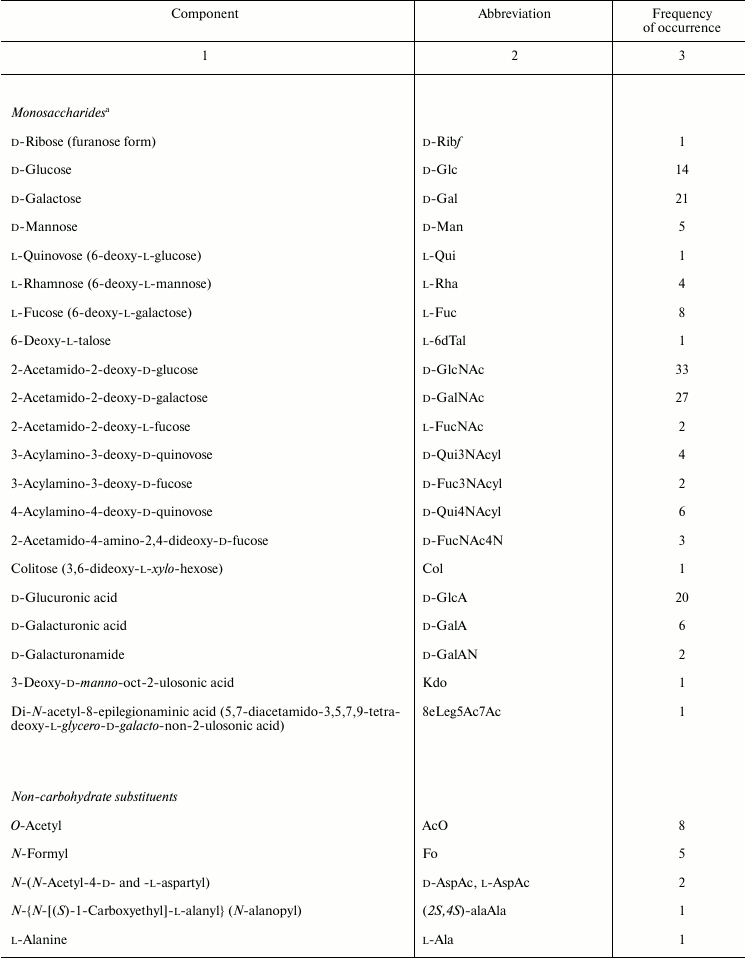

a Monosaccharides occur in the pyranose form unless stated
otherwise.
Fig. 1. Unusual components of Providencia O-antigens.
The carboxyl group of hexuronic acids is present either in the free form or as an amide. The primary amide of d-GalA was found in two O-polysaccharides (serogroups O3 and O21) but more commonly are amides of d-GlcA and d-GalA with amino acids, including l-alanine, l-serine (serogroups O43, O57, and O60), and amino acid derivatives of the opine family: Nε-[(R)-1-carboxyethyl]-l-lysine and Nε-[(S)-1-carboxyethyl]-l-lysine (so-called alaninolysines) (serogroups O14, O23, and O25). Amino groups of amino sugars usually are acetylated, while 6-deoxyamino sugars with an amino group at position 3 or 4 may be N-formylated. Some O-antigens contain other N-acyl substituents, such as N-acetyl-l-aspartyl and N-acetyl-d-aspartyl groups (serogroups O4 and O52) or a residue of another representative of the opine family, N-[(S)-1-carboxyethyl]-l-alanine (alanopine) (serogroup O35). 2-Acetamido-4-amino-2,4-dideoxy-d-fucose (FucNAc4N) is an exception as in the O-antigens of Providencia (serogroups O8, O22, and O30) as well as in all other known FucNAc4N-containing bacterial polysaccharides it has a free amino group at position 4.
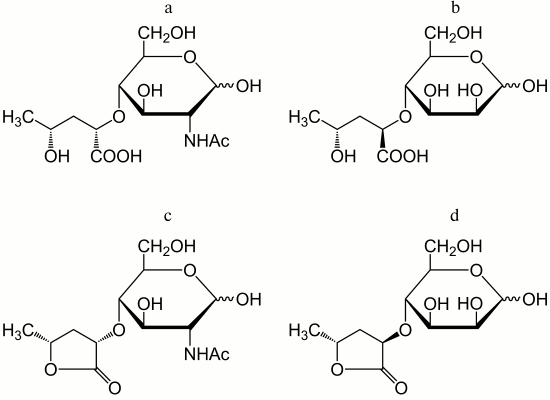
In addition to amino acids, O-polysaccharides may contain oxo acid and hydroxy acids. Pyruvic acid is present as an (S)-acetal attached at positions 4 and 6 of d-GlcNAc (serogroup O19). (R)- and (S)-lactic acids are ether-linked at position 3 of d-GlcNAc giving rise to N-acetylmuramic and N-acetylisomuramic acids, respectively (serogroups O16 and O32). Stereoisomeric (2S,4R)- and (2R,4R)-2,4-dihydroxypentanoic acids (serogroups O8 and O31) are unique components of bacterial polysaccharides. They form ethers between their 2-hydroxy group and the 4-hydroxy group of d-GlcNAc and d-Man (Fig. 2, a and b), partially being present as 1,4-lactones (c and d).
Fig. 2. Ethers of d-GlcNAc and d-Man with 2,4-dihydroxypentanoic acids and their lactones – components of the O-antigens of P. alcalifaciens O8 (a and c) and O31 (b and d).
Although phosphorylation is not typical of Providencia O-antigens, phosphate-containing O-polysaccharides are found in two O-serogroups. In the O8 polysaccharide, glycerol phosphate is present in the main chain so that monomers are linked together not only by glycosidic but also by phosphodiester bonds. This structural feature is characteristic for glycerol teichoic acids of Gram-positive bacteria and some capsular polysaccharides but occurs rarely in LPS of Gram-negative bacteria (among a few examples are O-polysaccharides of Proteus [55], Morganella morganii [56], and Hafnia alvei [57]). The other phosphorylated O-antigen (serogroup O22) contains 2-phospho-d-glyceric acid with the carboxyl group in the amide form. A number of O-antigens are decorated with O-acetyl groups, which in some serogroups occur in non-stoichiometric amounts, thus masking the regularity of the O-polysaccharides.
The O-antigens of Providencia are similar in both sugar and non-carbohydrate composition to O-antigens of Proteus [55]. For example, isomuramic acid, alanopine, and N-acetylaspartic acids hitherto have been found only in these two bacterial genera, and alaninolysine, apart from the O-antigens of Providencia and Proteus, has been identified in the O-polysaccharide of Shewanella fidelis only [58].
Most Providencia strains produce not only S-form LPS but also certain amounts of R- and SR-form LPS. As a result, in addition to an O-polysaccharide, mild acid degradation of LPS afforded oligosaccharides that correspond to the core and the core with one repeating unit attached. The latter was a key to identification of the biological repeating unit of the O-antigen, i.e. the oligosaccharide whose polymerization gives a high molecular mass O-polysaccharide. Its structural analysis allowed determination of the first monosaccharide in the repeating unit that is attached directly to the LPS core and whose transfer to a lipid carrier initiates the O-antigen synthesis. Like in other enterobacteria (Salmonella, Escherichia coli, Shigella, Proteus, Hafnia), in five Providencia O-antigens studied in this respect (serogroups O3, O9, O14, O19, and O34; Table 1), the first monosaccharide is N-acetylhexosamine (d-GlcNAc or d-GalNAc). Notably, formation of the glycosidic bond of HexNAc that connects the O-antigen to the core is catalyzed by ligase, and this linkage has the β-configuration in all LPS studied. However, polymerization of the repeating unit involves O-antigen polymerase and, depending on its specificity, the glycosidic bond of HexNAc connecting the repeating units to each other may have either α- or β-configuration. For instance, in the O-antigen of P. rustigianii O14, the GlcNAc residue is β-linked in the first repeating unit and α-linked in all other repeating units.
Table 1 shows established (for five serogroups) or putative structures of the biological repeating units. Most commonly (in 20 O-serogroups), the reducing end of the polysaccharide chain is occupied by GlcNAc, and in the other O-serogroups (except for O8, O22, and O30) by GalNAc. The O-antigen of P. alcalifaciens O30 contains no HexNAc, while in the O-antigens of P. alcalifaciens O8 and O22, HexNAc residues bear non-carbohydrate substituents, which makes unlikely their involvement in the initiation of the O-antigen synthesis. However, all three O-antigens contain a diamino sugar FucNAc4N, which has been reported as the first monosaccharide in the repeating unit of the O-antigen of Shigella sonnei [59] and some other bacteria. Therefore, it was suggested that FucNAc4N is located at the reducing end in the three exceptional Providencia O-antigens. For serogroups O22 and O30, this suggestion was confirmed by functional analysis of genes responsible for the O-antigen biosynthesis (see below).
Arrangement of the O-antigens structures in accordance with their putative biosynthetic pathways as shown in Table 1 reveals the location of the least common components at the non-reducing end of the polysaccharide chain. These caps are most readily available for interaction with other biological objects, including the immune system. Their uniqueness increases the antigenic diversity of bacteria, which helps pathogens to evade the adaptive immune response of the host.
IMMUNOCHEMISTRY OF O-ANTIGENS
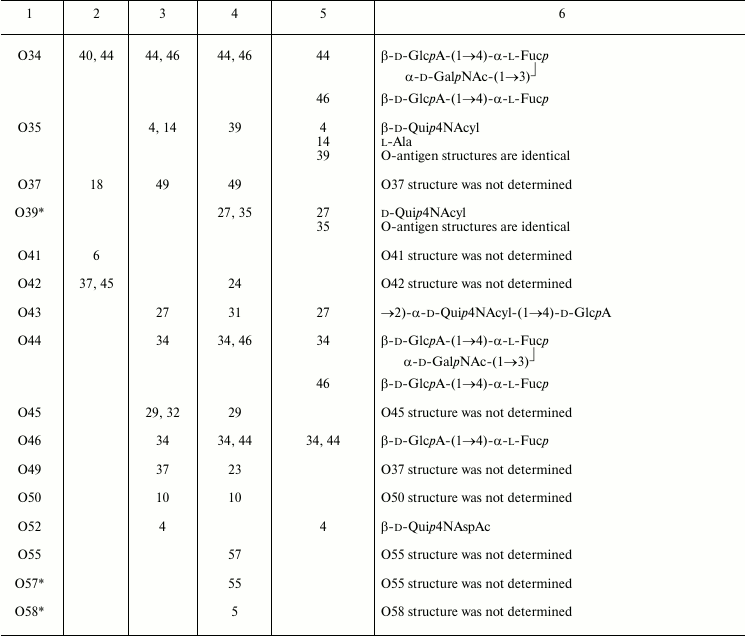
The O-antigenic relationships within the genus Providencia were systematically studied by Ewing et al., Penner et al., and Levina et al. and are summarized in Table 3. A higher sensitivity of the passive hemagglutination test used by Penner et al. and Levina et al. as compared with tube agglutination test used by Ewing et al. evidently accounts for the better agreement between results obtained by the first two research groups. Later, enzyme immunosorbent assay and Western blot were employed, the latter being especially informative as it shows which part of LPS, O-antigen or core or both, binds antibodies.
Table 3. Serological relationships between
Providencia O-serogroups and putative structures within
O-antigens associated with cross-reactive epitopes
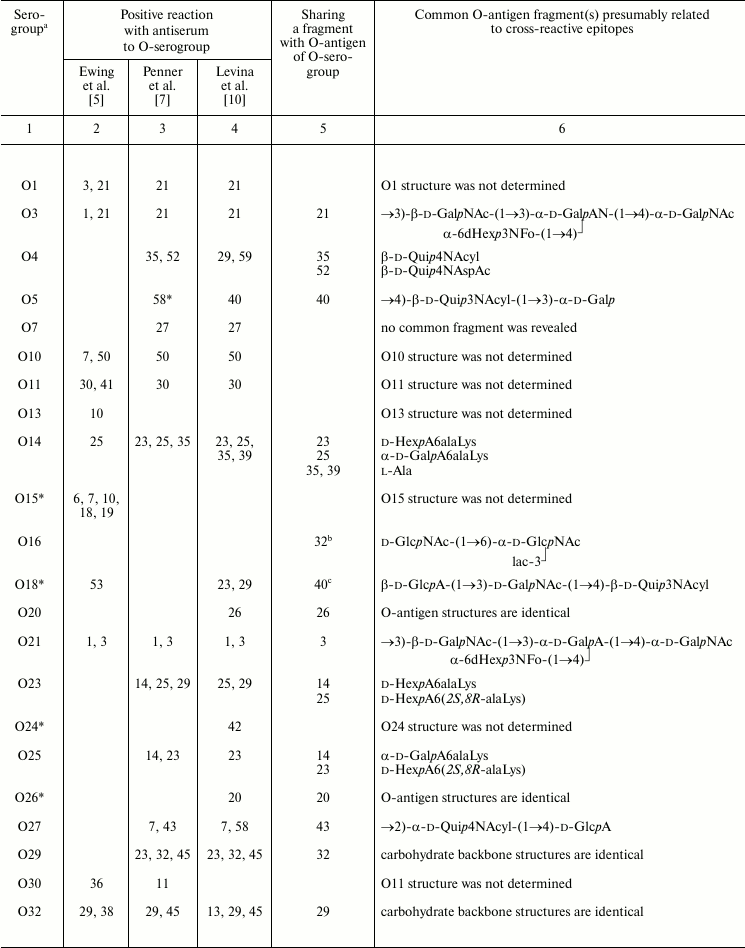
a No serological cross-reactions were observed for serogroups
that are not included in Table 3.
b Positive reaction with antiserum against serogroup O32 was
revealed by authors [38].
c Positive reaction with antiserum against serogroup O40 was
revealed by authors [43].
* Serogroups whose strains were replaced by Penner et al. and differed
from the original Ewing’s collection and the Hungarian
collection, making impossible substantiation of serological
cross-reactions observed by Penner et al. by O-antigen structures.
A comparison of structures of serologically related O-antigens revealed common structural fragments that are presumably associated with cross-reactive epitopes (Table 3). The fragments do not have to be identical; for instance, two-way serological cross-reactions of P. alcalifaciens O3 and O21 could be accounted for by the presence of similar branched tetrasaccharide fragments, whose terminal monosaccharide has either d-gluco or d-galacto configuration, respectively. O-Antiserum against P. alcalifaciens O40 reacted with LPS of P. alcalifaciens O5 and P. stuartii O18 due to sharing of di- and trisaccharide fragments, respectively [43].
As little as one common monosaccharide may be sufficient for cross-reactivity, provided that it occupies the terminal non-reducing end in the O-antigen. For example, cross-reactions of serogroups O4, O35, and O52 evidently are due to the presence of a d-Qui4N derivative at the end of the polysaccharide chain, while its N-acyl substituents are different (N-acetyl-d-aspartyl, N-acetyl-l-aspartyl, or alanopyl).
The O-antigens of serogroups O29 and O32 have the identical carbohydrate backbone and differ in the presence of a non-carbohydrate substituent, (S)-lactic acid, in the latter. LPS of P. alcalifaciens O29 reacted with O-antiserum against P. alcalifaciens O32 significantly weaker than the homologous LPS, thus showing the importance of lactic acid in manifestation of the O32 serospecificity. A weak cross-reaction between O-antiserum against P. alcalifaciens O32 and LPS of P. rustigianii O16 was accounted for by the presence of GlcNAc ethers with (R)- and (S)-lactic acids (N-acetylmuramic and N-acetylisomuramic acids, respectively) [38].
Serological relatedness of serogroups O14, O23, and O25 evidently is due to the occurrence of amides of Nε-(1-carboxyethyl)-l-lysine (alaninolysine) with hexuronic acids in their O-antigens. The pivotal role of the amino acid in manifestation of the O14 serospecificity was confirmed by the loss of the serological reactivity between the homologous LPS and O-antiserum after destruction of alaninolysine by deamination with nitrous acid. The inability of a synthetic antigen, polyacrylamide-linked α-GalA amide with l-lysine, to inhibit O-antiserum against P. rustigianii O14 demonstrated the key role of the 1-carboxyethyl group in alaninolysine. However, its absolute configuration does not seem to be important for recognition of alaninolysine-associated epitopes [23, 60].
Serogroup O35 is serologically related to serogroup O14 but not related to serogroups O23 and O25, which led Penner to a conclusion that the O14 and O35 O-antigens shared an epitope that is absent from the O23 and O25 O-antigens [7]. All four O-antigens contain 1-carboxyethyl group as a part of alanopine (serogroup O35) or alaninolysine (serogroups O14, O23, and O25), but it has the (S)-configuration in cross-reacting serogroups O14 and O35, whereas in serogroups O23 and O25 the configuration is R. One can speculate that in this case, antibodies recognize a smaller common 1-carboxyethyl-associated epitope, and therefore the absolute configuration of the 1-carboxyethyl group is crucial.
Serological cross-reactions have been reported for strains of Providencia and bacteria of other genera, including Proteus, Morganella, Escherichia, Shigella, and Salmonella [6, 61]. Table 4 exemplifies those pairs of cross-reactive strains whose O-antigen structures are known and fragments associated with common epitopes can be putatively identified.
Table 4. Serological cross-reactions between
O-serogroups of Providencia and other enteric bacteria and
putative structures within O-antigens associated with
cross-reactive epitope
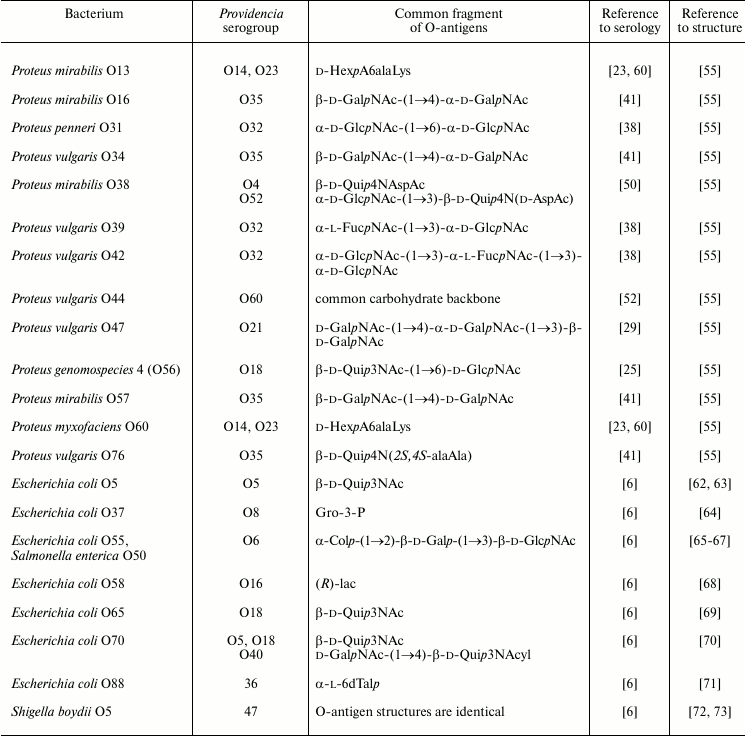
Serological cross-reactions of a group of Providencia and Proteus strains could be accounted for by the presence of common amino acid components, such as amides of uronic acids with N-(1-cabroxyethyl)-l-lysine in the O-antigens of P. rustigianii O14, P. alcalifaciens O23, Proteus mirabilis O13, and Proteus myxofaciens O60. A pronounced serological cross-reactivity was demonstrated for the O-antigens of P. stuartii O52 and Proteus mirabilis O38, which share a disaccharide fragment including an N-acetyl-d-aspartyl derivative of d-Qui4N. However, only a weak serological cross-reaction was observed for the O-antigens of P. stuartii O52 and P. stuartii O4 containing an N-acetyl-l-aspartyl derivative of d-Qui4N, thus showing that also in this case the absolute configuration of aspartic acid is important.
The O-antigens of P. alcalifaciens O21 and Proteus vulgaris O47 share a trisaccharide fragment and, accordingly, O-antiserum against the former cross-reacted with LPS of the latter. The O-antigens of P. alcalifaciens O60 and Proteus vulgaris O44 have the identical carbohydrate backbone and differ only in the nature of the amino acid that amidates glucuronic acid, which is l-serine in P. alcalifaciens O60 and l-alanine in P. vulgaris O44. Antiserum against the former bacterium reacted with both homologous and heterologous LPS to the same titer.
In Western blot, O-antiserum against P. alcalifaciens O35 recognized slowly migrating bands of the LPS of Proteus O16, O34, O57, and O76 as well as rapidly migrating bands of the LPS of Proteus O6 and O57, thus indicating the location of cross-reactive epitopes in the O-polysaccharide or LPS core, respectively. One of the epitopes is evidently associated with the alanopyl derivative of Qui4N, which is common for the O-antigens of P. alcalifaciens O35 and P. vulgaris O76 and core oligosaccharides of P. mirabilis O6 and O57. The cross-reactivity of the LPS of Proteus O16, O34, and O57 could be accounted for by sharing of the β-d-GalpNAc-(1→4)-d-GalpNAc disaccharide fragment of the O-polysaccharides with P. alcalifaciens O35.
Serological relationships between Providencia and Escherichia coli were studied by Ewing. A comparison of O-antigen structures in related strains shows that as little as a glycerol phosphate or lactic acid residue may be sufficient for providing serological cross-reactivity.
Escherichia coli O55 and Salmonella enterica O50 have the identical O-antigens. Their serological cross-reactivity with O-antiserum against P. alcalifaciens O6 could be accounted for by the presence of a common trisaccharide fragment terminated with colitose. This fragment also is shared by the O-antigens of Pseudoalteromonas tetraodonis IAM 14160T [74] and Aeromonas trota [75] and a capsular polysaccharide of Vibrio cholerae O139 [76]. Although no serological investigation has been undertaken, it is likely that P. alcalifaciens O6 and these bacteria also are serologically related. One can speculate that the occurrence of this particular trisaccharide fragment (or a related tetrasaccharide fragment with two colitose residues) is associated with molecular mimicry that allows bacteria to evade the adaptive immune response of the host. Indeed, colitose is a 3-deoxy analog of l-fucose, and therefore the α-Col-(1→2)-β-Gal-(1→3)-β-GlcNAc trisaccharide fragment of the O-antigens simulates the α-l-Fuc-(1→2)-β-Gal-(1→3)-β-GlcNAc H type 1 antigenic determinant.
Serologically related strains of P. stuartii O47 and Shigella boydii O5 share the O-antigen structure, including the pattern and the degree of non-stoichiometric O-acetylation.
The Kdo-containing O-antigen of P. alcalifaciens O36 and an acidic polysaccharide of Pseudoalteromonas flavipulchra NCIMB 2033T [77] have similar structures differing only in one monosaccharide residue, position of O-acetylation, and anomeric configuration of Kdo, but serological relationship of these bacteria has not been examined.
ORGANIZATION OF O-ANTIGEN GENE CLUSTERS
Analysis of full genome sequences of P. alcalifaciens, P. stuartii, and P. rustigianii revealed a polymorphic locus with genes associated with polysaccharide synthesis on a chromosome between two conserved housekeeping genes cpxA and yibK, which encode a two-component system sensor kinase and tRNA/rRNA-methyltransferase, respectively [78]. A tentative assignment of these regions to the O-antigen gene cluster (OGC) was confirmed by further bioinformatic and biochemical studies.
To date, the cpxA-yibK region has been sequenced in 13 Providencia strains, including strains of serogroups O6, O19, O22, O28, O30, O36, O38-O40, O44, O45, and O47 as well as non-typed strain P. rustigianii DSM 4541. As in other enterobacteria studied, OGC of Providencia is distinguished by a lower GC% content compared to the average genome level (31-36% versus ~41%), the lowest GC% content within the cluster being reached by O-antigen processing genes wzx and wzy. However, two sugar synthesis genes, namely ugd and galE, have higher GC content of ~47 and ~43%, respectively. GC% contents that are higher or around the average genome level also is characteristic for wza, wzb, and wzc genes located at the 3′-end of OGC (see below).
Functions of most genes in the Providencia OGC were predicted in silico based on their homology to annotated genes for surface polysaccharide synthesis in other bacteria [43, 78, 79]. Genes responsible for synthesis of nucleotide-activated monosaccharides, genes for glycosyltransferases, and processing genes were identified. The gene clusters of most O-acetylated O-antigens also included genes for O-acetyltransferases. The presence of the wzx and wzy genes in the OGC showed that all Providencia O-antigens studied are synthesized by the Wzx/Wzy-dependent pathway, which is typical of heteropolysaccharides.
The assignment of the glycosyltransferase genes to a certain glycosidic bond based on homology only is complicated by a huge diversity of these enzymes, from which only few have been characterized biochemically. This was not performed in Providencia OGC with the exception of orf5 in the OGC of P. alcalifaciens O36, which was proposed to encode β-Kdo transferase [78]. The orf5 gene is a homolog of kpsS encoding predicted β-Kdo transferase for synthesis and/or addition of the lyso-phosphatidylglycerol-poly-β-Kdo anchor to K-antigens of E. coli and Neisseria meningitidis [80], as well as of wepM in the OGC of Cronobacter sakazakii O6, whose O-antigen contains β-Kdo [81]. Remarkably, all these predicted β-Kdo transferases have no significant homology to well-characterized inverting α-Kdo transferases of the LPS core biosynthetic pathway [82] and to any glycosyltransferase encoded in OGC of C. sakazakii O5 with an α-Kdo-containing O-antigen [81].
Synthesis of O-antigens with GlcNAc or GalNAc at the reducing end of the biological repeating unit is initiated by a transfer of GlcNAc-1-phosphate from UDP-GlcNAc to an undecaprenol phosphate (UndP) lipid acceptor by glycosylphosphatetransferase WecA, which differs from glycosyltransferases in the presence of transmembrane segments. When GalNAc is the first sugar of the repeating unit, the UndPP-GlcNAc is then converted to UndPP-GalNAc by specific 4-epimerase [83]. As both UndPP-GlcNAc 4-epimerase [83] and UDP-GlcNAc 4-epimerase [84, 85] originally have been called Gne, to distinguish between them it was suggested to rename the former Gnu (P. R. Reeves, unpublished data).
In Providencia, like in other enterobacteria, the wecA gene is located in the enterobacterial common antigen (ECA) gene cluster and is involved in synthesis of both O-antigen and ECA. In OGC of serogroups O22 and O30, homologs of the wbgY gene encoding another initiating glycosylphosphatetransferase were found. WbgY of Providencia shares 55% identity with WbgY of Shigella sonnei, whose putative function is to initiate the O-antigen biosynthesis by transfer of FucNAc4N-1-phosphate to UndP [86] (that FucNAc4N occupies the reducing end of the O-antigen of S. sonnei was confirmed chemically [59]). The O-antigens of both P. alcalifaciens O22 and O30 contain FucNAc4N too, and it can be suggested that this diamino sugar also is the first monosaccharide in their repeating units, the more so that the O-antigen of the latter lacks any other 2-acetamido d-sugar that might have acted as the initiating monosaccharide (Table 1).
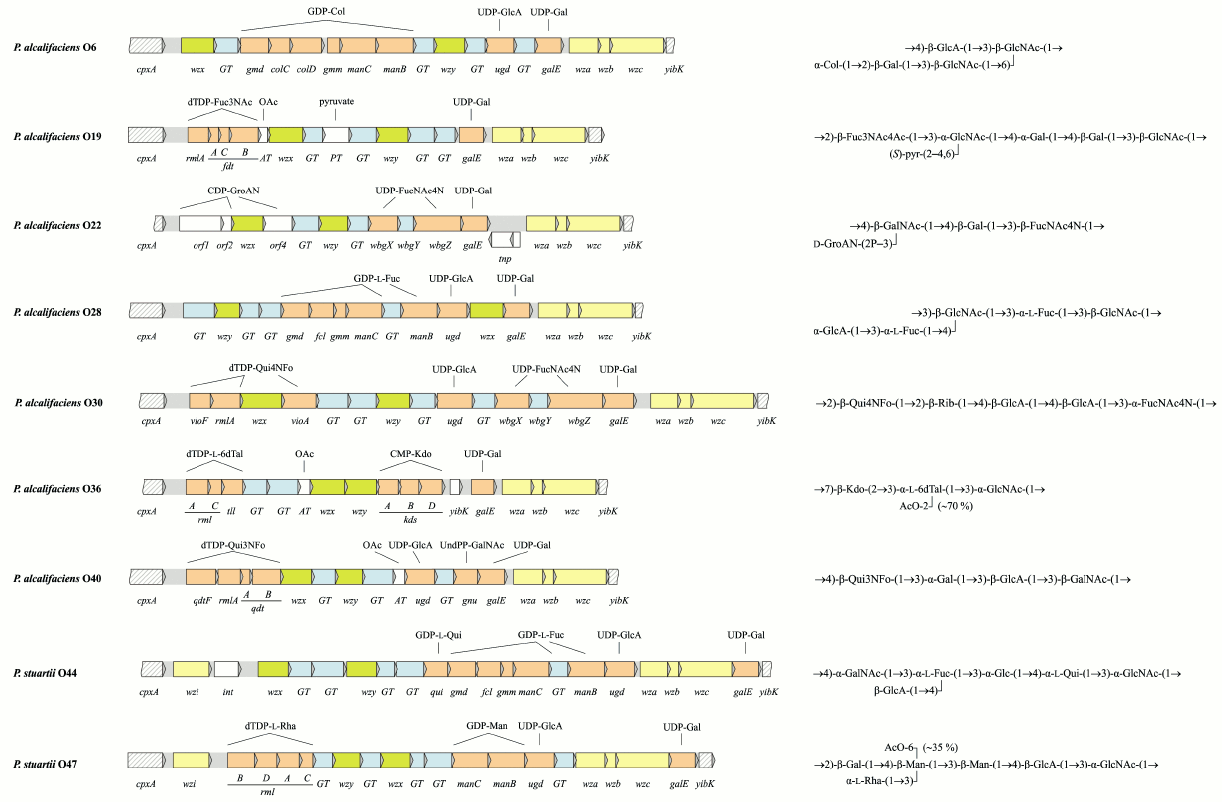
In OGC of 9 from 10 Providencia strains with known O-antigen structures, the putative gene functions are in accordance with the structures of the repeating units [43, 78, 79] (Fig. 3; see color insert). The exceptional OGC of P. alcalifaciens O39 is identical (>99% similarity) to the functional OGC of P. alcalifaciens O6, whose O-antigen is quite different from that of serogroup O39. Therefore, it was suggested that P. alcalifaciens O39 clone was derived from P. alcalifaciens O6 by inactivation of OGC between cpxA and yibK and acquirement of another gene cluster located in another genome locus for synthesis of the existing O-antigen [78].
Fig. 3. Organization of the O-antigen gene clusters and the corresponding O-antigen structures of Providencia. Transferase genes are indicated as follows: GT, glycosyltransferase; AT, acetyltransferase; PT, pyruvate transferase. The DNA sequence of OGC of P. alcalifaciens O22 is reported in this review for the first time and has been deposited in GenBank under accession number JQ417203.
Providencia alcalifaciens O38 and O45 produce R-form LPS and a peptidoglycan-like polysaccharide (presumably a capsule component). They possess different OGC, and neither of the OGC corresponds to the polysaccharide synthesized [12], whereas O-polysaccharides that would correspond to the OGC are not expressed.
An intriguing feature of P. alcalifaciens and P. stuartii is the presence at the 3′-end of the OGC of a conserved set of genes that are homologs of wza, wzb, and wzc [43, 78, 79]. In E. coli, these genes are involved in synthesis and translocation of group 1 and 4 K-antigens, which can be expressed in a lipid A-linked form (so-called K-LPS) [87]. In addition to the three genes, OGC of serogroups O44 and O47 contain a homolog of wzi, which encodes a membrane protein specific for group 1 K-antigens [87, 88]. The occurrence of homologs of these genes in OGC suggests a relationship between the O-antigens and K-antigens in Providencia too, but the exact role of these genes in the expression of surface polysaccharides remains to be determined. Noteworthy, in Proteus, OGC maps to the same locus on the chromosome downstream from cpxA but contains no homologs of wza, wzb, and wzc.
BIOSYNTHESIS OF SUGAR NUCLEOTIDE PRECURSORS
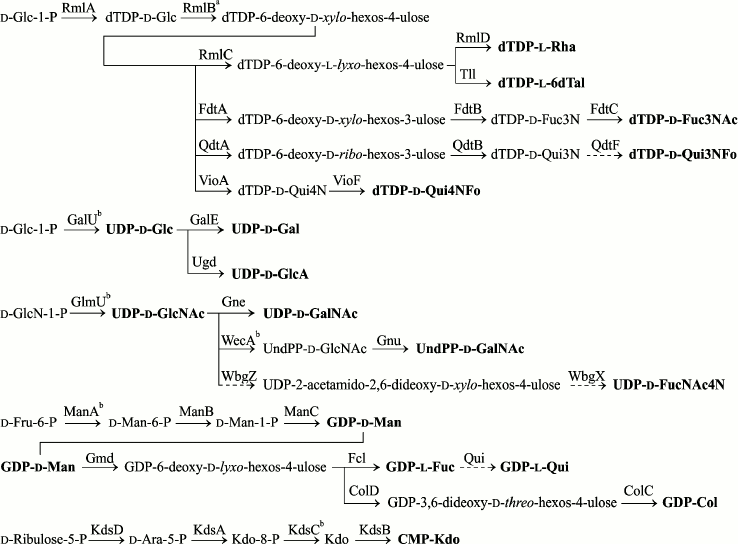
Biosynthetic pathways of specific monosaccharide components of Providencia O-antigens whose gene clusters have been sequenced are presented in Fig. 4. Genes for synthesis of nucleotide precursors of common sugars (UDP-Glc, UDP-GlcNAc, and ADP-Rib) are housekeeping genes and are not duplicated in Providencia OGC. UDP-Gal and UDP-GalNAc are synthesized via reversible 4-epimerization of UDP-Glc and UDP-GlcNAc catalyzed by GalE and Gne, respectively. In bacteria that are capable of utilizing exogenous galactose, the galE gene maps together with galT and galK in the galactose operon. Outside the gal-operon, it is difficult to distinguish between galE and gne based on homology alone. The reason lies in a limited number of the enzymes that are characterized biochemically and the capability of the 4-epimerases to possess both activities with a better specificity towards either UDP-Glc/Gal or UDP-GlcNAc/GalNAc, or the same specificity for both substrates [89].
Fig. 4. Biosynthetic pathways for nucleotide-activated monosaccharides involved in synthesis of Providencia O-antigens. Putative pathways are denoted by dash lines. a The enzyme is encoded by a gene located in the ECA gene cluster and is duplicated in OGC only as a part of the rmlBDAC operon. b The enzyme is encoded by the gene located outside OGC.
Full genome search for homologs of UDP-Glc and UDP-GlcNAc 4-epimerases in P. alcalifaciens, P. stuartii, and P. rustigianii retrieved three genes: galE in the gal-operon (except for P. alcalifaciens), galE in OGC, and a gene with <27% homology to all genes listed in Table 5 (a putative gene for UDP-GlcA 4-epimerase). A homolog of galE is located at the 3′-end of all sequenced OGC next to wza, wzb, wzc in a conservative region, which is distinguished by GC% content close to the average genome level [43, 78, 79]. In P. alcalifaciens O19, whose genome lacks the gal-operon while the O-antigen contains Gal, this gene evidently encodes UDP-Glc 4-epimerase. In serogroups O22 and O44, Gne is necessary for the synthesis of GalNAc-containing O-antigens, and again galE is the only possible candidate in OGC for assignment of the gne function. galE together with the wz genes also retains in serogroups that do not request the UDP precursors of Gal and GalNAc for the O-antigen synthesis, while the rest of OGC varies from serogroup to serogoup.
Table 5. Homology of putative
Providencia Glc/GlcNAc 4-epimerases and related E.
coli proteins with defined functions
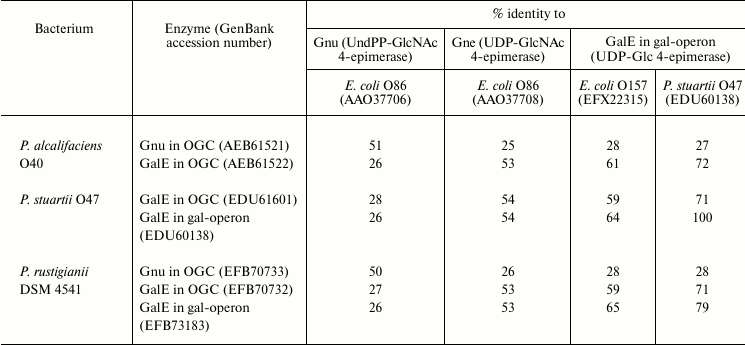
Another GalNAc precursor, namely UndPP-GalNAc, is necessary for the synthesis of the O-antigen of serogroup O40, in which GalNAc is the first monosaccharide in the biological repeating unit. Accordingly, a homolog of the gnu gene responsible for the synthesis of UndPP-GalNAc from UndPP-GlcNAc (see above) was found in OGC of this bacterium [43] as well as in OGC of P. rustigianii DSM 4541, whose O-antigen structure is unknown (Table 5).
The rmlA and rmlB genes encode enzymes for two initial steps of synthesis of dTDP-l-Rha and a number of other 6-deoxy sugars, which are activated as dTDP-monosaccharides. Both genes are located in the ECA gene cluster as RmlA and RmlB are involved in synthesis of an ECA component, d-Fuc4NAc. In Providencia, rmlA is always duplicated in OGC, whereas rmlB is duplicated only as a part of the rmlBDAC operon for synthesis of dTDP-l-Rha [78].
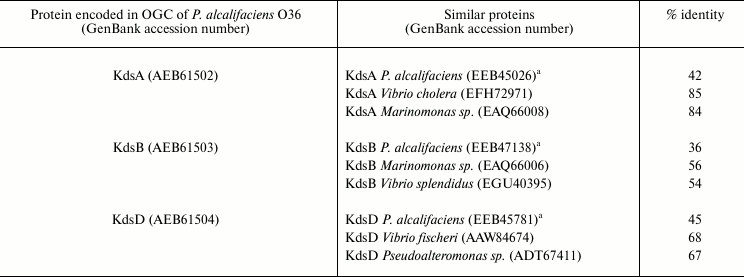
Four genes kdsABCD are necessary for all enterobacteria, including Providencia, for synthesis of CMP-Kdo, a substrate of Kdo-transferase, which participates in the assembly of LPS core. These genes are usually scattered around the chromosome. In P. alcalifaciens O36 that contains Kdo not only in the core but also in the O-antigen, three from the four genes are duplicated in OGC [78]. Notably, kdsA, kdsB, and kdsD in O36 OGC showed a higher degree of amino acid similarity to the homologous genes in marine bacteria (Marinomonas, Pseudoalteromonas, Vibrio sp.) than to those for the Providencia core Kdo synthesis located outside OGC (Table 6). This finding and a close structural similarity between the polysaccharides of P. alcalifaciens O36 and Pseudoalteromonas flavipulchra suggest that kds genes in OGC of Providencia have been acquired by horizontal transfer. Remarkably, in two strains of Cronobacter sakazakii, whose O-antigen structures are not related to that of P. alcalifaciens O36, no gene for synthesis of CMP-Kdo is duplicated in OGC [81, 90].
Table 6. Homology of CMP-Kdo biosynthesis
enzymes encoded in OGC of P. alcalifaciens O36 to those encoded
in P. alcalifaciens genome outside OGC and most similar proteins
in other bacteria
a Encoded outside OGC.
Biosynthetic pathways for 14 from 17 monosaccharides shown in Fig. 4 were verified by biochemical studies [91]. Particularly, the last step of dTDP-d-Qui4NFo synthesis, transfer of a formyl group to the amino group of dTDP-d-Qui4N, was demonstrated in vitro using formyltransferase VioF cloned from OGC of P. alcalifaciens O30 [79]. Cloned enzymes from P. alcalifaciens O39 were used to reproduce the three-step synthesis of GDP-Col from GDP-Man [78], which had been elucidated first in Yersinia pseudotuberculosis [92, 93].
UDP-d-FucNAc4N presumably is synthesized from UDP-GlcNAc in two steps involving UDP-GlcNAc 4,6-dehydratase (WbgV or WbgZ) and aminotransferase WbgX [86, 94]. In two Providencia serogroups that contain d-FucNAc4N (O22 and O30), OGC includes homologs of wbgZ and wbgX. The absence of wbgV supports the suggestion of Xu et al. to revise the UDP-GlcNAc 4,6-dehydratase function from WbgV to WbgZ [94]; however, the final decision on this matter requires biochemical confirmation.
In contrast to many other 6-deoxy sugars, the biosynthetic pathway of l-quinovose (Qui) in bacteria remained unknown until recently, probably, owing to a low abundance of this monosaccharide in polysaccharides. For the first time, l-Qui was reliably identified in bacteria as a component of the O-antigen of P. stuartii O44 (earlier, l-Qui had been tentatively identified in Legionella feeleii [95]). Recent discovery of l-Qui in the O-antigen of Yersinia pseudotuberculosis O12 and analysis of OGC of this bacterium, including its comparison to OGC of P. stuartii O44, enabled suggestion that GDP-l-Qui is synthesized in one step from GDP-l-Fuc by GDP-l-Fuc 4-epimerase called Qui [96]. Earlier, the qui gene in OGC of P. stuartii O44 was identified erroneously as the gne gene for UDP-d-GlcNAc 4-epimerase [78].
Providencia is a genus of ubiquitous opportunistic Gram-negative bacteria from the Enterobacteriaceae family, which may be a part of normal microflora of human intestines. The taxonomic position of Providencia has been revised repeatedly. The genus is closely related to the Proteus and Morganella genera, and the three were considered for a while as members of the Proteaee tribe. Providencia alcalifaciens and P. stuartii were the first Providencia species classified serologically by Ewing. Later, some P. alcalifaciens strains were reclassified as P. rustigianii, and currently the Ewing’s classification scheme includes the three species.
Later studies revealed discrepancies in the original Ewing’s collection of 62 Providencia O-serogroup type strains. Thus, strains of four serogroup pairs O20/O26, O24/O42, O35/O39, and O55/O57 possessed pairwise identical O-antigens, and strains of serogroups O58 and O59 were found to belong to the genus Morganella. The mistakes in classification and serotyping were corrected by Penner, who replaced strains of 10 serogroups and added a new serogroup, O63. However, the original Ewing’s type strains do remain in some other collections, including the Hungarian Collection of Medical Bacteria, which was used in structural and genetic studies of Providencia O-antigens. It is advisable to use in further studies the revised Penner’s typing scheme, which is most comprehensive at present.
The unique O-antigen structures have been established for 36 from 63 Providencia O-serogroups. O-Antigens of 9 serogroups have not been studied, and strains from 18 other serogroups from the Hungarian collection have lost the ability to produce O-polysaccharide-containing S-form LPS. Their replacement by strains from Penner’s collection would allow further structural studies of Providencia O-antigens.
Providencia O-polysaccharides studied to date are distinguished by structural diversity due to variations in constituent monosaccharides (neutral sugars, amino and diamino sugars, their 6-deoxy derivatives, uronic and aldulosonic acids) and non-carbohydrates components (amino and hydroxy acids, pyruvic acid, phosphates of glycerol and glyceramide), the way they are arranged in repeating units, and the mode of the linkage between the repeating units. In some serogroups, a regular O-antigen structure is masked by non-stoichiometric O-acetylation of a sugar residue.
Most O-polysaccharides are acidic due the presence of carbohydrate or non-carbohydrate organic acids and/or phosphate groups. Some O-antigens contain both positive (amino group of FucNAc4N, alaninolysine or alanopine) and negative (carboxylate or phosphate) charged groups, i.e. have zwitterionic character. In contrast to neutral and acidic polysaccharides, which induce a T-independent response and no immune memory, zwitterionic polysaccharides are immunomodulating T-dependent antigens. The unique feature of zwitterionic repetitive patterns useful for design of vaccines first was described for capsular polysaccharides [97] but recently has been demonstrated for O-antigens too [56].
Some Providencia O-antigens display a marked structural similarity to O-antigens of Proteus, Shigella, and Pseudoalteromonas, and O-antigens of P. stuartii O47 and Shigella boydii O5 are even identical. A combination of structural and serological data allowed identification of putative common epitopes responsible for the cross-reactivity of strains within the genus Providencia and between Providencia and other enterobacteria. Most often it is associated with non-carbohydrate substituents and monosaccharides (usually 6-deoxyamino sugars or aldulosonic acids) that occupy the terminal non-reducing end of a polysaccharide chain, where they are most readily available for interactions with cells of the immune system and antibodies.
A polymorphous O-antigen gene cluster was found between the housekeeping genes cpxA and yibK and sequenced in 13 strains of Providencia, including 10 strains with known O-antigen structure. Bioinformatic analysis of genes in OGC has shown the necessity of, and opened the way for, further studies of Providencia O-antigens in various directions. Putative biosynthetic pathways suggested for d-FucNAc4N and l-Qui have to be verified biochemically. In P. alcalifaciens O39, OGC between cpxA and yibK is evidently inactivated, and search for the functional genes necessary for synthesis of the existing O-antigen should be based on the full genome sequencing. Of particular interest is examination of the roles of the wza, wzb, wzc homologs found in all Providencia OGC sequenced so far except for P. rustigianii. These genes are responsible for synthesis of capsular polysaccharides, and their location in OGC has been reported for no other bacteria. A comparison of gene clusters for related O-antigens in various bacteria will provide insight into the evolutionary history of the O-antigen diversification. From a practical point of view, data of OGC are necessary for development of a molecular typing method for Providencia strains based on genes specific for each O-serogroup.
We thank our colleagues who have contributed to the advancement of this field.
This work was supported by the Russian Foundation for Basic Research (project No. 08-04-92221-NNSF), National Key Program for Infectious Diseases of China 2013ZX10004216-001-001, the National Natural Science Foundation of China (NSFC) Key Program Grant 31030002, NSFC General Program Grant 31270003, and Ministry of Science and Higher Education, Poland (core funding for statutory research activity, grant 803 from the Department of Immunobiology of Bacteria, University of Lodz).
REFERENCES
1.O’Hara, C. M., Brenner, F. W., and Miller, J.
M. (2000) Clin. Microbiol. Rev., 13, 534-546.
2.O’Hara, C. M., Steigerwalt, A. G., Green, D.,
Mcdowell, M., Hill, B. C., Brenner, D. J., and Miller, J. M. (1999)
J. Clin. Microbiol., 37, 3048-3050.
3.Somvanshi, V. S., Lang, E., Straubler, B., Sproer,
C., Schumann, P., Ganguly, S., Saxena, A. K., and Stackebrandt, E.
(2006) Int. J. Syst. Evol. Microbiol., 56, 629-633.
4.Juneja, P., and Lazzaro, B. P. (2009) Int. J.
Syst. Evol. Microbiol., 59, 1108-1111.
5.Ewing, W. H., Tanner, K. E., and Dennard, D. A.
(1954) J. Infect. Dis., 94, 134-140.
6.Ewing, W. H. (1986) in Identification of
Enterobacteriaceae (Edwards, P. R., ed.) Elsevier, N. Y., pp.
443-459.
7.Penner, J. L., Hinton, N. A., Hennessy, J. N., and
Whiteley, G. R. (1976) J. Infect. Dis., 133, 283-292.
8.Penner, J. L., Hinton, N. A., Duncan, I. B.,
Hennessy, J. N., and Whiteley, G. R. (1979) J. Clin. Microbiol.,
9, 11-14.
9.Penner, J. L., Fleming, P. C., Whiteley, G. R., and
Hennessy, J. N. (1979) J. Clin. Microbiol., 10,
761-765.
10.Levina, L. A., Kholodkova, E. V., Raginskaia, V.
P., and Kuborina, L. N. (1980) Zh. Mikrobiol. Epidemiol.
Immunobiol., 5, 100-103.
11.Ovchinnikova, O. G., Arbatsky, N. P., Chizhov, A.
O., Kocharova, N. A., Shashkov, A. S., Rozalski, A., and Knirel, Y. A.
(2012) Carbohydr. Res., 349, 95-102.
12.Ovchinnikova, O. G., Liu, B., Kocharova, N. A.,
Shashkov, A. S., Kondakova, A. N., Siwinska, M., Feng, L., Rozalski,
A., Wang, L., and Knirel, Y. A. (2012) Biochemistry (Moscow),
77, 609-615.
13.Ovchinnikova, O. G., Kondakova, A. N., Kocharova,
N. A., Shashkov, A. S., Knirel, Y. A., and Rozalski, A. (2012)
Carbohydr. Res., 361, 27-32.
14.Kocharova, N. A., Torzewska, A., Zatonsky, G. V.,
Blaszczyk, A., Bystrova, O. V., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2004) Carbohydr. Res., 339, 195-200.
15.Kocharova, N. A., Senchenkova, S. N., Kondakova,
A. N., Gremyakov, A. I., Zatonsky, G. V., Shashkov, A. S., Knirel, Y.
A., and Kochetkov, N. K. (2004) Biochemistry (Moscow),
69, 103-107.
16.Zatonsky, G. V., Bystrova, O. V., Kocharova, N.
A., Shashkov, A. S., Knirel, Y. A., Kholodkova, E. V., and
Stanislavsky, E. S. (1999) Biochemistry (Moscow), 64,
523-527.
17.Ovchinnikova, O. G., Kocharova, N. A., Wykrota,
M., Shashkov, A. S., Knirel, Y. A., and Rozalski, A. (2007)
Carbohydr. Res., 342, 2144-2148.
18.Bystrova, O. V., Zatonsky, G. V., Borisova, S.
A., Kocharova, N. A., Shashkov, A. S., Knirel, Y. A., Kholodkova, E.
V., and Stanislavsky, E. S. (2000) Biochemistry (Moscow),
65, 677-684.
19.Toukach, F. V., Kocharova, N. A., Shashkov, A.
S., Knirel, Y. A., and Rozalski, A. (2008) Carbohydr. Res.,
343, 2706-2711.
20.Shashkov, A. S., Kocharova, N. A., Toukach, F.
V., Kachala, V. V., and Knirel, Y. A. (2008) Nat. Prod. Commun.,
3, 1625-1630.
21.Ovchinnikova, O. G., Kocharova, N. A., Shashkov,
A. S., Arbatsky, N. P., Rozalski, A., and Knirel, Y. A. (2011)
Carbohydr. Res., 346, 644-650.
22.Parkhomchuk, A. A., Kocharova, N. A.,
Bialczak-Kokot, M., Shashkov, A. S., Chizhov, A. O., Knirel, Y. A., and
Rozalski, A. (2010) Carbohydr. Res., 345, 1235-1239.
23.Kocharova, N. A., Zatonsky, G. V., Torzewska, A.,
Macieja, Z., Bystrova, O. V., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2003) Carbohydr. Res., 338, 1009-1016.
24.Kocharova, N. A., Zatonsky, G. V., Bystrova, O.
V., Ziolkowski, A., Wykrota, M., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2002) Carbohydr. Res., 337, 1667-1671.
25.Kocharova, N. A., Blaszczyk, A., Zatonsky, G. V.,
Torzewska, A., Bystrova, O. V., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2004) Carbohydr. Res., 339, 409-413.
26.Kocharova, N. A., Maszewska, A., Zatonsky, G. V.,
Torzewska, A., Bystrova, O. V., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A (2004) Carbohydr. Res., 339, 415-419.
27.Kocharova, N. A., Vinogradov, E. V., Kondakova,
A. N., Shashkov, A. S., Rozalski, A., and Knirel, Y. A. (2008) J.
Carbohydr. Chem., 27, 320-331.
28.Shashkov, A. S., Kocharova, N. A., Zatonsky, G.
V., Blaszczyk, A., Knirel, Y. A., and Rozalski, A. (2007) Carbohydr.
Res., 342, 653-658.
29.Kocharova, N. A., Maszewska, A., Zatonsky, G. V.,
Bystrova, O. V., Ziolkowski, A., Torzewska, A., Shashkov, A. S.,
Knirel, Y. A., and Rozalski, A. (2003) Carbohydr. Res.,
338, 1425-1430.
30.Ovchinnikova, O. G., Kocharova, N. A.,
Bialczak-Kokot, M., Shashkov, A. S., Rozalski, A., and Knirel, Y. A.
(2012) Eur. J. Org. Chem., 3500-3506.
31.Kocharova, N. A., Shcherbakova, O. V., Shashkov,
A. S., Knirel, Y. A., Kochetkov, N. K., Kholodkova, E. V., and
Stanislavsky, E. S. (1997) Biochemistry (Moscow), 62,
501-508.
32.Kocharova, N. A., Ovchinnikova, O. G.,
Bialczak-Kokot, M., Shashkov, A. S., Knirel, Y. A., and Rozalski, A.
(2011) Biochemistry (Moscow), 76, 707-712.
33.Ovchinnikova, O. G., Bushmarinov, I. S.,
Kocharova, N. A., Toukach, F. V., Wykrota, M., Shashkov, A. S., Knirel,
Y. A., and Rozalski, A. (2007) Carbohydr. Res., 342,
1116-1121.
34.Ovchinnikova, O. G., Kocharova, N. A., Kondakova,
A. N., Bialczak-Kokot, M., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2011) Carbohydr. Res., 346, 2638-2641.
35.Bushmarinov, I. S., Ovchinnikova, O. G.,
Kocharova, N. A., Toukach, F. V., Torzewska, A., Shashkov, A. S.,
Knirel, Y. A., and Rozalski, A. (2006) Carbohydr. Res.,
341, 1181-1185.
36.Kocharova, N. A., Ovchinnikova, O. G., Torzewska,
A., Shashkov, A. S., Knirel, Y. A., and Rozalski, A. (2006)
Carbohydr. Res., 341, 786-790.
37.Ovchinnikova, O. G., Kocharova, N. A., Shashkov,
A. S., Bialczak-Kokot, M., Knirel, Y. A., and Rozalski, A. (2009)
Carbohydr. Res., 344, 683-686.
38.Bushmarinov, I. S., Ovchinnikova, O. G.,
Kocharova, N. A., Toukach, F. V., Torzewska, A., Shashkov, A. S.,
Knirel, Y. A., and Rozalski, A. (2007) Carbohydr. Res.,
342, 268-273.
39.Kocharova, N. A., Kondakova, A. N., Vinogradov,
E., Ovchinnikova, O. G., Lindner, B., Shashkov, A. S., Rozalski, A.,
and Knirel, Y. A. (2008) Chem. Eur. J., 14,
6184-6191.
40.Kocharova, N. A., Kondakova, A. N., Ovchinnikova,
O. G., Perepelov, A. V., Shashkov, A. S., and Knirel, Y. A. (2009)
Carbohydr. Res., 344, 2060-2062.
41.Ovchinnikova, O. G., Valueva, O. A., Kocharova,
N. A., Arbatsky, N. P., Maszewska, A., Zablotni, A., Shashkov, A. S.,
Rozalski, A., Knirel, Y. A. (2013) Carbohydr. Res., 375,
73-78.
42.Kocharova, N. A., Ovchinnikova, O. G., Torzewska,
A., Shashkov, A. S., Knirel, Y. A., and Rozalski, A. (2007)
Carbohydr. Res., 342, 665-670.
43.Ovchinnikova, O. G., Liu, B., Guo, D., Kocharova,
N. A., Bialczak-Kokot, M., Shashkov, A. S., Feng, L., Rozalski, A.,
Wang, L., and Knirel, Y. A. (2012) FEMS Immunol. Med.
Microbiol., 66, 382-392.
44.Ovchinnikova, O. G., Kocharova, N. A., Torzewska,
A., Blaszczyk, A., Shashkov, A. S., Knirel, Y. A., and Rozalski, A.
(2005) Carbohydr. Res., 340, 1407-1411.
45.Kocharova, N. A., Ovchinnikova, O. G., Toukach,
F. V., Torzewska, A., Shashkov, A. S., Knirel, Y. A., and Rozalski, A.
(2005) Carbohydr. Res., 340, 1419-1423.
46.Ovchinnikova, O. G., Kocharova, N. A., Shashkov,
A. S., Knirel, Y. A., and Rozalski, A. (2009) Russ. J. Bioorg.
Chem., 35, 370-375.
47.Ovchinnikova, O. G., Kocharova, N. A.,
Bakinovskiy, L. V., Torzewska, A., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2004) Carbohydr. Res., 339, 2621-2626.
48.Ovchinnikova, O. G., Kocharova, N. A., Shashkov,
A. S., Knirel, Y. A., and Rozalski, A. (2012) Carbohydr. Res.,
347, 168-171.
49.Bushmarinov, I. S., Ovchinnikova, O. G.,
Kocharova, N. A., Blaszczyk, A., Toukach, F. V., Torzewska, A.,
Shashkov, A. S., Knirel, Y. A., and Rozalski, A. (2004) Carbohydr.
Res., 339, 1557-1560.
50.Torzewska, A., Kocharova, N. A., Zatonsky, G. V.,
Blaszczyk, A., Bystrova, O. V., Shashkov, A. S., Knirel, Y. A., and
Rozalski, A. (2004) FEMS Immunol. Med. Microbiol., 41,
133-139.
51.Kocharova, N. A., Ovchinnikova, O. G.,
Bushmarinov, I. S., Toukach, F. V., Torzewska, A., Shashkov, A. S.,
Knirel, Y. A., and Rozalski, A. (2005) Carbohydr. Res.,
340, 775-780.
52.Ovchinnikova, O. G., Kocharova, N. A.,
Parkhomchuk, A. A., Bialczak-Kokot, M., Shashkov, A. S., Knirel, Y. A.,
and Rozalski, A. (2011) Carbohydr. Res., 346,
377-380.
53.Herget, S., Toukach, P. V., Ranzinger, R., Hull,
W. E., Knirel, Y. A., and von der Lieth, C.-W. (2008) BMC Struct.
Biol., 8, 35.
54.Knirel, Y. A. (2011) in Bacterial
Lipopolysaccharides: Structure, Chemical Synthesis,
Biogenesis and Interaction with Host Cells (Knirel, Y. A., and
Valvano, M. A., eds.) Springer, Wien-N. Y., pp. 41-115.
55.Knirel, Y. A., Perepelov, A. V., Kondakova, A.
N., Senchenkova, S. N., Sidorczyk, Z., Rozalski, A., and Kaca, W.
(2011) Innate Immun., 17, 70-96.
56.Young, N. M., Kreisman, L. S. C., Stupak, J.,
MacLean, L. L., Cobb, B. A., and Richards, J. C. (2011)
Glycobiology, 21, 1266-1276.
57.Romanowska, E. (2000) FEMS Immunol. Med.
Microbiol., 27, 219-225.
58.Kilcoyne, M., Perepelov, A. V., Shashkov, A. S.,
Nazarenko, E. L., Ivanova, E. P., Gorshkova, N. M., Gorshkova, R. P.,
and Savage, A. V. (2004) Carbohydr. Res., 339,
1655-1661.
59.Gamian, A., and Romanowska, T. (1982) Eur. J.
Biochem., 129, 105-109.
60.Torzewska, A., Kocharova, N. A., Maszewska, A.,
Knirel, Y. A., and Rozalski, A. (2004) Arch. Immunol. Ther.
Exp., 52, 43-49.
61.Sedlak, J., and Rische, H. (1986)
Enterobacteriaceae Infektionen, VEB Georg Thieme,
Leipzig, pp. 567-569.
62.MacLean, L. L., and Perry, M. B. (1997)
Biochem. Cell Biol., 75, 199-205.
63.Urbina, F., Nordmark, E.-L., Yang, Z., Weintraub,
A., Scheutz, F., and Widmalm, G. (2005) Carbohydr. Res.,
340, 645-650.
64.Senchenkova, S. N., Knirel, Y. A., Perepelov, A.
V., Ovchinnikova, O. G., Shashkov, A. S., and Wang, Q. (2012) Abst.
26th Int. Carbohydr. Symp., Madrid, Spain, 22-27 July 2012,
P461.
65.Lindberg, B., Lindh, F., Lonngren, J., Lindberg,
A. A., and Svenson, S. B. (1981) Carbohydr. Res., 97,
105-112.
66.Kenne, L., Lindberg, B., Soderholm, E., Bundle,
D. R., and Griffith, D. W. (1983) Carbohydr. Res., 111,
289-296.
67.Senchenkova, S. N., Shashkov, A. S., Knirel, Y.
A., Schwarzmuller, E., and Mayer, H. (1997) Carbohydr. Res.,
301, 61-67.
68.Dmitriev, B. A., Knirel, Y. A., Kochetkov, N. K.,
Jann, B., and Jann, K. (1977) Eur. J. Biochem., 79,
111-115.
69.Perry, M. B., and MacLean, L. L. (1999)
Carbohydr. Res., 322, 57-66.
70.MacLean, L. L., and Perry, M. B. (2010)
Carbohydr. Res., 345, 644-648.
71.Torgov, V. I., Shashkov, A. S., Kochanowski, H.,
Jann, B., and Jann, K. (1996) Carbohydr. Res., 283,
223-227.
72.Albert, M. J., Holme, T., Lindberg, B., Lindberg,
J., Mosihuzzaman, M., Qadri, F., and Rahman, M. M. (1994) Carbohydr.
Res., 265, 121-127.
73.L’vov, V. L., Shashkov, A. S., Knirel, Y.
A., Arifulina, A. E., Senchenkova, S. N., Yakovlev, A. V., and
Dmitriev, B. A. (1995) Carbohydr. Res., 279, 183-192.
74.Muldoon, J., Perepelov, A. V., Shashkov, A. S.,
Gorshkova, R. P., Nazarenko, E. L., Zubkov, V. A., Ivanova, E. P.,
Knirel, Y. A., and Savage, A. V. (2001) Carbohydr. Res.,
333, 41-46.
75.Knirel, Y. A., Senchenkova, S. N., Jansson,
P.-E., Weintraub, A., Ansaruzzaman, M., and Albert, M. J. (1996)
Eur. J. Biochem., 238, 160-165.
76.Knirel, Y. A., Paredes, L., Jansson, P.-E.,
Weintraub, A., Widmalm, G., and Albert, M. J. (1995) Eur. J.
Biochem., 232, 391-396.
77.Muldoon, J., Perepelov, A. V., Shashkov, A. S.,
Nazarenko, E. L., Zubkov, V. A., Gorshkova, R. P., Ivanova, E. P.,
Gorshkova, N. M., Knirel, Y. A., and Savage, A. V. (2003) Carbohydr.
Res., 338, 459-462.
78.Ovchinnikova, O. G., Liu, B., Guo, D., Kocharova,
N. A., Shashkov, A. S., Chen, M., Feng, L., Rozalski, A., Knirel, Y.,
and Wang, L. (2012) Microbiology, 158, 1024-1036.
79.Liu, B., Chen, M., Perepelov, A. V., Liu, J.,
Ovchinnikova, O. G., Zhou, D., Feng, L., Rozalski, A., Knirel, Y. A.,
and Wang, L. (2012) Glycobiology, 22, 1236-1244.
80.Willis, L. M., Stupak, J., Richards, M. R.,
Lowary, T. L., Li, J., and Whitfield, C. (2013) Proc. Natl. Acad.
Sci. USA, 110, 7868-7873.
81.Sun, Y., Wang, M., Wang, Q., Cao, B., He, X., Li,
K., Feng, L., and Wang, L. (2012) Appl. Environ. Microbiol.,
78, 3966-3974.
82.Mamat, U., Skurnik, M., and Bengoechea, J. A.
(2011) in Bacterial Lipopolysaccharides: Structure, Chemical
Synthesis, Biogenesis and Interaction with Host Cells
(Knirel, Y. A., and Valvano, M. A., eds.) Springer, Wien-N. Y., pp.
237-273.
83.Rush, J. S., Alaimo, C., Robbiani, R., Wacker,
M., and Waechter, C. J. (2010) J. Biol. Chem., 285,
1671-1680.
84.Wang, L., Huskic, S., Cisterne, A., Rothemund,
D., and Reeves, P. R. (2002) J. Bacteriol., 184,
2620-2625.
85.Bengoechea, J. A., Pinta, E., Salminen, T.,
Oertelt, C., Holst, O., Radziejewska-Lebrecht, J., Piotrowska-Seget,
Z., Venho, R., and Skurnik, M. (2002) J. Bacteriol.,
184, 4277-4287.
86.Shepherd, J. G., Wang, L., and Reeves, P. R.
(2000) Infect. Immun., 68, 6056-6061.
87.Whitfield, C. (2006) Annu. Rev. Biochem.,
75, 39-68.
88.Rahn, A., Beis, K., Naismith, J. H., and
Whitfield, C. (2003) J. Bacteriol., 185, 5882-5890.
89.Bernatchez, S., Szymanski, C. M., Ishiyama, N.,
Li, J., Jarrell, H. C., Lau, P. C., Berghuis, A. M., Young, N. M., and
Wakarchuk, W. W. (2005) J. Biol. Chem., 280,
4792-802.
90.Shashkov, A. S., Arbatsky, N. P., and Knirel, Y.
A. (2011) Carbohydr. Res., 346, 1924-1929.
91.Hao, Y., and Lam, J. S. (2011) in Bacterial
Lipopolysaccharides: Structure, Chemical Synthesis,
Biogenesis and Interaction with Host Cells (Knirel, Y. A., and
Valvano, M. A., eds.) Springer, Wien-N. Y., pp. 195-235.
92.Beyer, N., Alam, J., Hallis, T. M., Guo, Z., and
Liu, H.-W. (2003) J. Am. Chem. Soc., 125, 5584-5585.
93.Alam, J., Beyer, N., and Liu, H.-W. (2004)
Biochemistry, 43, 16450-16460.
94.Xu, D. Q., Cisar, J. O., Ambulos, J. N. J., Burr,
D. H., and Kopecko, D. J. (2002) Infect. Immun., 70,
4414-4423.
95.Sonesson, A., Jantzen, E., Tangen, T., and
Zahringer, U. (1994) Microbiology, 140, 2663-2671.
96.De Castro, C., Kenyon, J. J., Cunneen, M. M.,
Molinaro, A., Holst, O., Skurnik, M., and Reeves, P. R. (2013)
Glycobiology, 23, 346-353.
97.Tzianabos, A., Wang, J. Y., and Kasper, D. L.
(2003) Carbohydr. Res., 338, 2531-2538.