REVIEW: Poly(A)-Binding Proteins: Structure, Domain Organization, and Activity Regulation
I. A. Eliseeva, D. N. Lyabin, and L. P. Ovchinnikov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: yeliseeva@vega.protres.ru; lyabin@vega.protres.ru; ovchinn@vega.protres.ru* To whom correspondence should be addressed.
Received May 6, 2013
RNA-binding proteins are of vital importance for mRNA functioning. Among these, poly(A)-binding proteins (PABPs) are of special interest due to their participation in virtually all mRNA-dependent events that is caused by their high affinity for A-rich mRNA sequences. Apart from mRNAs, PABPs interact with many proteins, thus promoting their involvement in cellular events. In the nucleus, PABPs play a role in polyadenylation, determine the length of the poly(A) tail, and may be involved in mRNA export. In the cytoplasm, they participate in regulation of translation initiation and either protect mRNAs from decay through binding to their poly(A) tails or stimulate this decay by promoting mRNA interactions with deadenylase complex proteins. This review presents modern notions of the role of PABPs in mRNA-dependent events; peculiarities of regulation of PABP amount in the cell and activities are also discussed.
KEY WORDS: PABPs, mRNA, translation, poly(A) tail, polyadenylation, mRNA decayDOI: 10.1134/S0006297913130014
Abbreviations: ARE, AU-rich element; ARS, adenine-rich autoregulatory sequence; Cp, ceruloplasmin; CPE, cytoplasmic polyadenylation element; CPEBP, cytoplasmic polyadenylation element binding protein; CPSF, cleavage and polyadenylation specificity factor; eIF, eukaryotic initiation factor; EJC, exon junction complex; eRF, eukaryotic release factor; GAIT-element, interferon γ-activated inhibitor of translation; hnRNP, heterogeneous nuclear ribonucleoprotein; IFN-γ, interferon γ; IRES, internal ribosome entry site; MLLE, C-terminal domain of PABP; Msi1, Musashi 1; NMD, nonsense mediated mRNA decay; PABPs, poly(A)-binding proteins; Paip1/2, PABP-interacting protein 1/2; PAM, PABP-interacting motif; PAN2/3, PABP-dependent poly(A)-specific ribonuclease 2/3; PAP, poly(A) polymerase; PARN, poly(A)-specific ribonuclease; PTC, premature termination codon; RRM, RNA recognition motif; TOP, terminal oligopyrimidine track; UNR, upstream of N-ras; UTR, untranslated region.
Poly(A)-binding proteins (PABPs) are a highly conserved class of
eukaryotic RNA-binding proteins that specifically recognize the
polyadenylic acid sequence typically present at the 3′ end of the
majority of eukaryotic mRNAs. Members of this class lack their own
catalytic activity but are capable of binding many protein partners and
virtually all mRNAs in the cell. This capability makes PABPs key
participants in a diversity of cellular events.
Historically, the first poly(A)-binding protein was discovered as a major protein of cytoplasmic mRNPs [1] showing an elevated affinity for a poly(A) sequence [2]. Its basic function was believed to be protection of mRNA against decay, though eventually PABPs were proved to participate in many cellular mRNA-dependent events. PABPs accompany mRNAs during their entire life from synthesis to decay. Bound to the growing poly(A) tail, PABPs determine the tail length and assist in mRNA transportation to the cytoplasm, where they contribute to “sorting out” of mRNAs into those for decay and translation according to the presence or absence of premature termination codons. PABPs are involved in mRNA lifespan control and set a term to mRNA degradation as dictated by cell growth conditions and mRNA regulatory sequences. In association with viral RNAs, PABPs participate in translation and replication of some viruses.
The set of PABPs varies from organism to organism. For example, Xenopus laevis has three of them, Mus musculus – two, while Arabidopsis thaliana – as many as eight. There are seven human PABPs that can be grouped as follows: cytoplasmic (PABPC1, PABPC3, PABPC4, PABPC4L), embryonic (ePABP, also termed PABPC1L), nuclear (PABPN1), and the X chromosome-encoded protein PABPC5. Among these, the best studied are PABPC1 and PABPN1 [3, 4].
Here, we summarize the available data on structure and functions of PABPs, as well as on the regulatory mechanisms of their expression and activity; the best known proteins PABPC1 and PABPN1 are the principal subjects of this review.
STRUCTURE, DOMAIN ORGANIZATION, AND INTERACTION WITH OTHER
MOLECULES
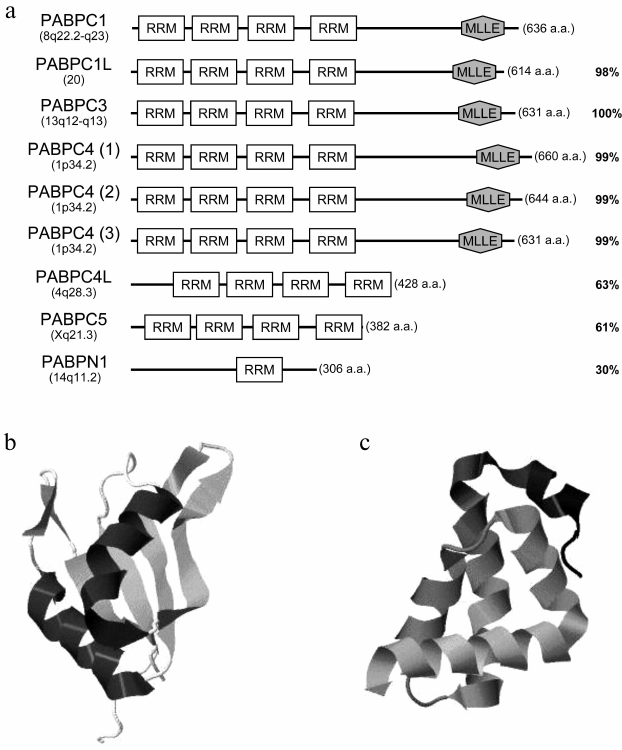
Members of the class of poly(A)-binding proteins differ from one another in length, domain organization, and amino acid sequence homology (Fig. 1a). A distinguishing feature of cytoplasmic PABPs is the presence of four RNA-binding domains termed RRM (RNA recognition motif) in their N-termini. RRMs are typical for many RNA-binding proteins. An RRM (~90-100 a.a.) has two highly conserved consensuses (K/R)G(F/Y)(G/A)FVX(F/Y) (RNP-1) and (L/I)(F/Y)( V/I)(G/K)(N/G)(L/M) (RNP-2) of eight and six amino acids, respectively. Structurally, it is composed of four β-strands and two α-helices assembled in a globular domain shaped as a four-stranded antiparallel β-sheet flanked with two α-helices (Fig. 1b) [5]. RNP-1 and RNP-2 are located on the two central β-strands.
Fig. 1. a) Domain organization of human PABPs. The gene positions on the chromosome, the lengths of amino acid sequences, and homology with PABPC1 are indicated. For PABPC4, its three known isoforms are shown. RRM, RNA-binding domain; MLLE, the α-helical C-terminal domain. b) PABPC1 RRM2 crystal structure (pdb # 4F25) [6]. c) PABPC1 MLLE crystal structure (pdb # 3KUR) [1].
The C-terminal part of some cytoplasmic PABPs contains a “helical” domain (PABPC or MLLE) consisting of five α-helices (Fig. 1c). The domain MLLE is named after its highly conserved amino acid motif Met-Leu-Leu-Glu. It is present only in PABPs, and it is linked with the RRM cluster through a disordered Pro/Met-rich linker.
The nuclear poly(A)-binding protein PABPN1, unlike cytoplasmic ones, has only one RRM. Its N-terminus contains a cluster of negatively charged amino acids, while its rather short C-terminus has an Arg-rich sequence (Fig. 1a) [7].
The poly(A)-binding proteins specifically interact with poly(A) sequences and exhibit a lower affinity for other homo- and heterogeneous sequences in vitro [8]. For oligo(A) and (A)-rich sequences, the PABP dissociation constants (Kd) amount to 1-20 nM and depend on the poly(A) length. For other sequences, Kd = 10-50 nM, i.e. a 3-5 times lower binding level [9-11]. The minimal oligo(A) length required for interaction with PABP is about 12 nucleotides. However, PABP bound to a long poly(A) tail can protect as many as about 27 nucleotides from the action of ribonucleases [7, 12]. Similarly to the majority of RNA-binding proteins, PABP employs its RRM for interaction with RNA. The efficient poly(A) binding is provided by any two RRMs, while interaction with other sequences requires three RRMs [7, 10]. It has been reported that two initial RRMs of PABP from X. laevis are poly(A)-specific, while the third and the last ones are used for nonspecific RNA binding [7]. But for human and rat PABPs, the last two RRMs have been shown to exhibit the highest affinity for both poly(A) and other heterogeneous sequences [11, 13], while the first and the second ones displayed neither specificity nor high RNA-binding activity. This was explained by species-dependent specificity and experimental conditions. However, subsequent studies by Khanam and Patel showed good agreement between the data on human PABP and that from X. laevis. They reported that in both cases the two initial RRMs displayed a higher affinity for poly(A) than the two others [9, 10].
Apart from RNA, PABP RRMs bind to some proteins, such as Paip1 (PABP-interacting protein 1), Paip2, eIF4G, GW182, and MKRN1 (table). Of note, the interaction of some proteins with an RRM can influence the affinity of PABP for poly(A). For example, RRM-to-eIF4G binding increases PABP affinity for poly(A) by an order of magnitude [14, 15]. According to X-ray data, eIF4G allosterically regulates PABP binding to poly(A) sequences. As a result of either RRM-2-to-eIF4G or RRM-1-2-to-poly(A) binding, these RNA-binding domains undergo some conformational changes. The expanded inter-RRM regions cause transition of PABP into a more “open” state, which facilitates binding to another partner [6]. In contrast, the Paip2-to-PABP binding decreases PABP affinity for poly(A), because in the PABP molecule the binding sites of Paip2 and poly(A) overlap [16].
Some PABP-interacting proteins
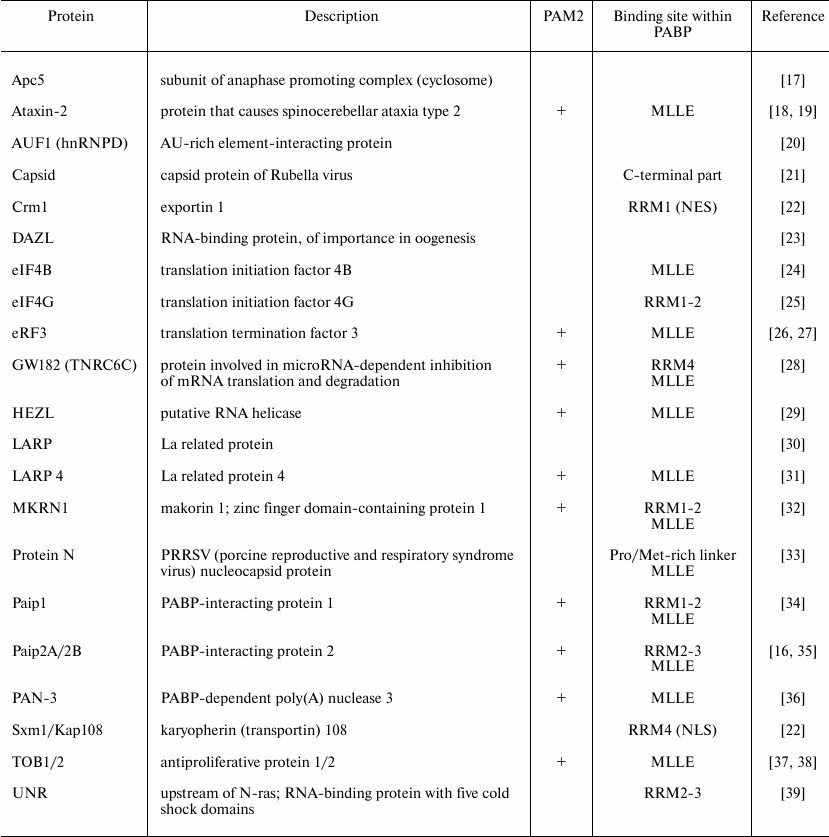
Notice that RRMs contain the nuclear localization signal (NLS) and the nuclear export signal (NES) that interact with importin Sxm1/Kap108 and exportin Crm1, respectively [22].
The Pro/Met-rich linker between the RRM cluster and MLLE is responsible for multimerization of PABP and for its cooperative binding to a poly(A) sequence [40, 41]. As shown, this linker contains proteolytic cleavage sites for proteases of polioviruses, lentiviruses, and caliciviruses [42]. Interestingly, unlike completely viral protease-processed eIF4G, PABP undergoes processing in poliovirus-infected cells only partially (30-60% of its total amount), which probably is explained by its proline-rich composition that allows a few conformations, including viral protease-resistant ones [43, 44]. Specifically, the ribosome-associated fraction of PABP has been found to be most sensitive to protease activity. This is supposed to be functionally important [44, 45].
The C-terminal domain MLLE is responsible for the interaction of PABP with other proteins [3, 46] whose typical feature is the presence of PABP-interacting motif 2 (PAM2) comprising 12 amino acids (those at positions 3, 5, 7, 10, and 12 are hydrophobic). The key role is played by the Phe/Tyr stacking interaction at position 10 of PAM2 and at Phe/Tyr567 of the PABP molecule [46, 47]. Most frequently, PAM2 is localized to disordered regions near protein phosphorylation sites, which may be indicative of regulation of the interaction of PABP with its partners [48]. Of note, some proteins have two PAMs, one of which is responsible for specific PABP recognition, while the other stabilizes such complexes [28, 49].
FUNCTIONS OF PABPs
PABPs play an important role in many cellular events, such as mRNA polyadenylation, translation, degradation, and nuclear export. These are discussed in detail below.
Polyadenylation
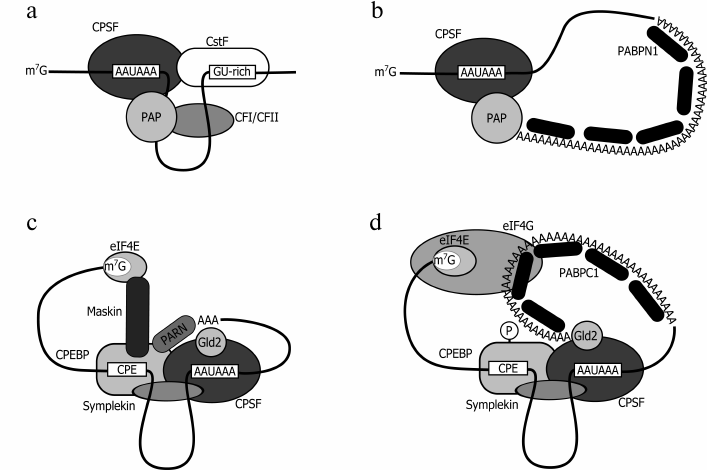
Unlike prokaryotes where protein synthesis is coupled with mRNA synthesis, eukaryotes have their initial transcript processed prior to translation. One of the processing steps consists in cotranscriptional addition of ~200-250 adenyl residues (for mammals) or ~70-90 (for yeast) to the 3′ terminus [3, 50]. This process includes two steps. First, RNA undergoes endonuclease-induced cleavage by a large protein complex, where the key role is played by the cleavage and polyadenylation specificity factor (CPSF) that recognizes the motif AAUAAA or AUUAAA located 20-30 nt upstream of the cleavage site (Fig. 2a). After the cleavage, the 3′-terminal fragment becomes degraded, while the rest of this mRNA undergoes polyadenylation catalyzed by poly(A) polymerase (PAP) [51]. It was found that synthesis can be performed by PAP alone, but then the processing is very slow; a more efficient activity requires involvement of two factors, CPSF and PABPN1, whose effect is synergic. PABPN1 stimulates poly(A) synthesis by poly(A) polymerase and protects the growing poly(A) tail from degradation. The consecutive binding of PABPN1 results in formation of filaments of 7 nm in diameter and, finally, of a spherical particle of 21 nm in diameter, that is a “molecular ruler” showing that the required length of the poly(A) sequence has been achieved (Fig. 2b). Thus, defining the length of the growing poly(A) tail is yet another function of PABPN1 [50-52].
Fig. 2. Models for nuclear (a, b) and cytoplasmic (c, d) polyadenylation. a) The cleavage complex model (CPSF, cleavage and polyadenylation specificity factor; PAP, poly(A) polymerase; CstF, cleavage stimulating factor; CF, cleavage factor). b) The polyadenylation complex model. c) The masked mRNA model (CPEBP, cytoplasmic polyadenylation element binding protein; eIF4E, cap-binding protein; Gld2, poly(A) polymerase; Maskin, mRNA-masking protein; PARN, poly(A) specific ribonuclease; Symplekin, scaffold protein). d) The cytoplasmic polyadenylation complex model: after CPEBP phosphorylation, the complex loses PARN and Maskin. PABPC1 that interacts with the growing poly(A) tail stabilizes the interaction between eIF4G and the mRNA 5′ end.
However, some researchers believe that polyadenylation is terminated at a later step and connected with the rearrangement of the mRNA–protein complex and mRNA export to the cytoplasm [53]. Interestingly, this event may be controlled by PABPC1 but not PABPN1 [54]. It was found that yeasts that express PABPC1 with a mutation in the nuclear localization signal accumulate hyperadenylated mRNA in the nucleus [22, 55]. Besides, mammalian PABPC1 was detected in a complex with poly(A) polymerase and unspliced mRNA [56]. It can be thought that in the course of rearrangement of the mRNA complexes on nuclear pore proteins PABPN1 is completely or partially substituted by PABPC1, thereby causing polyadenylation termination and mRNA export.
Both PABPN1 and PABPC1 are proteins shuttling between the nucleus and the cytoplasm. PABPC1 is supposed to displace PABPN1 from the poly(A) tail in the nucleus and to be exported to the cytoplasm in association with mRNA [50, 56]. Besides, both proteins were detected in a complex with CBP80/CBP20-bound mRNA and mRNA having the premature termination codon (PTC)1. In the cytoplasm, the imported mRNA is believed to be checked for the presence of the PTC during a pioneer round of translation, and the presence of PTC leads to decay of this mRNA [57]. In the course of the pioneer round, the rest of PABPN1 is substituted by PABPC1 in a translation-dependent manner [56, 58].
1 CBP80/CBP20 (cap-binding protein) is a heterodimeric nuclear cap-binding protein that is associated with mRNA only during a pioneer round of translation. After the absence of the mRNA PTC has been verified, eIF4E displaces CBP80/CBP20 from the complex. It has been recently reported that PABPN1-depleted cells showed no disturbed transcription, processing, polyadenylation, or nuclear export of mRNA. In the absence of PABPN1, its functions were probably performed by its cytoplasmic homologs. As shown, PABPN1 depletion was accompanied by nuclear translocation of PABPC4, while the amount of PABPC5 increased in the cytoplasm. Most probably, PABPC4 played the functional role of PABPN1 in the nucleus, while PABPC5 functionally replaced PABPC4 in the cytoplasm. Of note, the number of apoptotic cells was higher upon PABPN1 depletion, which is indicative of its possible antiapoptotic functions [59].
These results suggest that cytoplasmic PABPs that have been transferred to the nucleus can participate in maturation of mRNA, its verification for the presence of the premature termination codon with possible subsequent degradation, and in mRNA export to the cytoplasm.
It should be noted that polyadenylation can occur in the cytoplasm during mRNA maturation in the period of early embryonal development, and also in neurons [60, 61]. The key role in regulation of cytoplasmic polyadenylation is played by the cytoplasmic polyadenylation element binding protein (CPEBP) that recognizes the cytoplasmic polyadenylation element (CPE) in the target mRNA 3′ UTR (untranslated region). It binds to the processed mRNA in the nucleus. Upon export of the target mRNA to the cytoplasm, CPEBP promotes its association with some proteins, such as CPSF, noncharacteristic poly(A) polymerase Gld-2, deadenylase PARN, the eIF4E-binding protein Maskin, and the scaffold Symplekin [62]. It was shown that in this complex the deadenylase PARN dominates in activity over the polymerase, thus shortening the poly(A) tail from 100-250 nt down to 20-30 nt (Fig. 2c). Besides, non-phosphorylated CPEBP showed a high level of binding to the protein Maskin displaying a high affinity for eIF4E; it displaces eIF4G from its complex with eIF4E, thereby triggering translation inhibition. Of note, PABPs are not associated with short poly(A) tails of these mRNAs [63, 64]. After oocyte stimulation or activation in neurons of the N-methyl-D-aspartate receptor (NMDAR), CPEBP phosphorylation enhances its affinity for the CPSF and contributes to dissociation of PARN [65-67]. In such a complex, addition of a poly(A) tail is not accompanied by deadenylase-induced truncation (Fig. 2d). The newly synthesized poly(A) sequence interacts with PABPC1 (or ePABP) that, in turn, through binding to eIF4G enhances its affinity for eIF4E. This succession of events leads to displacement of Maskin from its complex with eIF4E and formation of a classical closed-loop structure including PABP, eIF4G, and eIF4E, thus ensuring efficient translation [63, 67-69].
Translation
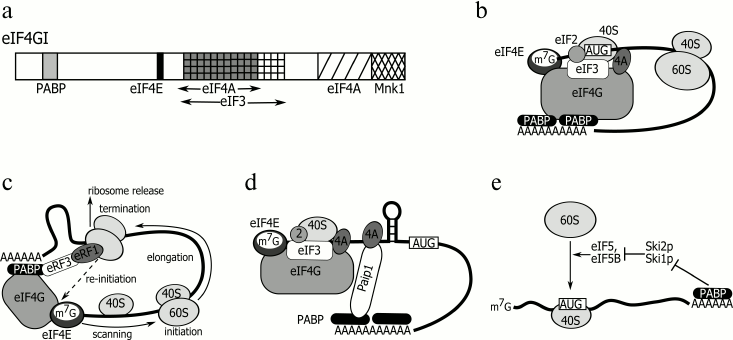
The canonical stimulators of protein synthesis are the 3′-terminal poly(A) tail and the 5′-terminal cap structure. It is believed that the cytoplasmic poly(A)-binding protein PABPC1 (hereinafter referred to as PABP) associated with the poly(A) tail stimulates translation initiation. Some reported data suggest a detailed mechanism of this process. As found, PABP can interact with the initiation factor eIF4G through its two initial RNA-recognizing motifs [25, 70, 71]. The protein eIF4G, a member of the cap-binding initiation complex eIF4F, acts as a scaffold that brings into association other members of this complex, i.e. the cap-binding protein eIF4E and the ATP-dependent RNA helicase eIF4A. It also interacts with the multisubunit translation initiation factor eIF3 that, in turn, recruits the 40S ribosome subunit. The mutually spaced positions of the PABP- and eIF4E-binding sites within the eIF4G molecule (Fig. 3a) suggests a mechanism in which initiation of translation can be stimulated by a PABP molecule located at the 3′ end. The poly(A) tail-bound PABP interacts with eIF4F through eIF4G, thereby enhancing its affinity for the cap by about an order of magnitude [14, 15] and contributing to formation of a closed-loop structure (Fig. 3b) [72] that is probably additionally supported by interaction between the termination factor eRF3 and PABP [26, 27]. The N-terminus of PABP binds to the poly(A) tail and eIF4G, while its C-terminus interacts with eRF3, which brings together AUG (initiation codon) and the stop codon (Fig. 3c). Such a closed-loop structure is believed to facilitate translation initiation and ribosome recycling [26, 73].
Fig. 3. a) eIF4G: position of domains interacting with other proteins. b) “Closed loop” model. c) Ribosome recycling model. d) Paip1-induced translation stimulation model. e) Model of PABP effect on 60S subunit recruitment stimulation. For details, see text.
An extensive literature has evolved that traces the effect of PABP on translation by other mechanisms [74]. For example, it was shown that mutant PABP unable to stimulate poly(A)-dependent translation and bind to eIF4G retains its ability to promote cap-dependent translation [71]. Moreover, separate PABP domains can stimulate translation in an independent manner, and each tandem of these RNA-binding domains shows an activity level comparable with that of the full-length PABP [70]. As known, RRM1-2 stimulates translation via its interaction with eIF4G. Surprisingly, considerable translation stimulation activity is displayed by RRM3-4 that is unable to selectively bind to poly(A) and is involved in no interaction with any translation factor. This probably means that PABP can have at least two different activities and stimulate the translation of a capped and polyadenylated mRNA either in cis (on the poly(A) tail) or in trans through its direct interaction with translation initiation factors or other regulatory proteins [71]. Specifically, the PABP-to-eIF4B binding adds to helicase activity of the eIF4A–eIF4B complex that unwinds the secondary structure of the mRNA 5′ UTR, which facilitates scanning of this 5′ untranslated region by the 43S pre-initiation complex [24]. Of note, exogenous poly(A) can in trans stimulate the cap-dependent translation in a rabbit reticulocyte cell-free system, and therefore, no closed-loop structure is required in this case [75].
The translation regulation can also be mediated by interaction of PABP with Paip1, Paip2A, and Paip2B. These proteins associate with PABP in a similar way, that is, through their two PAM motifs, with PAM1 binding to the N-terminal part of the PABP molecule (RRM1 and 2) and PAM2 to its MLLE domain. At both termini, the interaction between PABP and eIF4G is blocked. Yet, Paip1-bound PABP keeps stimulating translation because it acquires affinity for eIF4A, thus promoting unwinding of the 5′ UTR secondary structure (Fig. 3d) [34, 76]. The translation can also be stimulated in the case that a PABP–Paip1–eIF3 complex has been formed [77]. Unlike Paip1, PABP-bound Paip2A and Paip2B reduce the ability of PABP to bind to poly(A), eIF4G, and Paip1, which results in inhibition of translation initiation [16]. There are reports indicating that the amount of Paip2 in the cell is regulated in response to varying PABP concentration. With its decreased level, Paip2 undergoes polyubiquitination with ubiquitin ligase (HYD) followed by 26S proteasome-induced degradation [35]. Interestingly, the PABP C-terminal domain MLLE that binds to Paip2, Paip1, and some other proteins shows high homology to one of the HYD substrate-recognizing domains. The mechanism of Paip2-to-HYD binding is believed to be analogous to that of the Paip2-to-PABP binding. Presumably, other PABP C-terminus-binding proteins are subject to a similar regulation [78, 79].
As shown recently, excess PABP inhibits translation in vitro. A mechanism has been proposed according to which excess PABP competes with poly(A)-bound PABP for binding to eIF4F. When free PABP interacts with eIF4F, the poly(A)–PABP–eIF4G complex is destabilized and the cap-recognizing activity of eIF4E goes down, thereby provoking translation inhibition [80]. Added Paip2 neutralizes the inhibiting effect of excess PABP. Most probably, a similar situation is observed in vivo in the final steps of spermatogenesis. Then the amount of Paip2A grows, and translation of mRNAs of regulatory proteins, such as Prm (protamines) and Tps (transition proteins), is activated. Paip2A-null mice show disturbed fertility and spermatogenesis in its final steps characterized by an increased amount of PABP [80]. A comparison of the proteomic profiles of elongated spermatids from wild-type and Paip2A-null mice demonstrated changes in expression of only a small number of genes coding for proteins involved in metabolism, reproduction, translation, and cytoskeleton organization. This suggests that the discovered mechanism is functional only for a limited number of mRNAs whose translation is most sensitive to the amount of PABP and/or eIF4E [81].
It was found that PABP can stimulate translation initiation at the stage of 60S subunit recruitment [74, 82]. Studies of yeast with deleted pabp revealed suppressor mutations in a number of genes. One of these occurring in the gene encoding ribosomal protein L46 may be indicative of the direct interaction between PABP and the large ribosome subunit [83]. Besides, as shown for S. cerevisiae, PABP contributes to activity of the fifth group of initiation factors (eIF5, eIF5B) by suppressing their inhibitors Ski1p and Ski2p, which promotes recruitment of the 60S subunit (Fig. 3e) [84].
The importance of interaction between PABP and translation initiation factors is emphasized by the fact that some viruses can use PABP and eIF4G as targets for suppression of synthesis of the host cell proteins [42], e.g. the rotavirus protein NSP3A binds to eIF4G, thus displacing PABP from its complex with the cap-binding proteins [85, 86]; picornavirus proteases cleave PABP, thereby inducing its inactivation [45, 87, 88]; some viral proteases deprive eIF4G of the ability to bind to PABP by cutting off its N-terminus [24].
The majority of viral RNAs comprising the internal ribosome entry site (IRES) do not use PABP for their efficient translation. The exceptions are not numerous; such viral RNAs as coxackievirus 3 (CBV3) [89], some polioviruses [43], human cytomegalovirus (HCMV) [90, 91], etc. contain IRES but their translation is stimulated by PABP. Virus translation and replication can be “switched over” depending on the accessibility/amount of PABP [42]. For example, in the course of hepatitis A virus infection, PABP cleavage inhibits the IRES-dependent translation and stimulates replication. As reported, an N-terminal fragment of PABP is responsible for this event [92]. Another mechanism is used in the case of Rubella virus infection. The viral protein Capsid withdraws PABP from translation by binding to its C-terminal domain. Of note, PABP interacts solely with the pool of free Capsid protein [21].
Upon specific regulation of translation of some mRNAs, PABP may be either directly bound to the poly(A) tail/(A)-rich segments or interact with other sequences via its protein partners.
PABP binding to the mRNA poly(A) tail. It was found that inhibition of translation initiation of some mRNAs, such as msl-2 mRNA [93] or Cp (ceruloplasmin) mRNA [94], occurs only in the presence of the poly(A) tail and PABP.
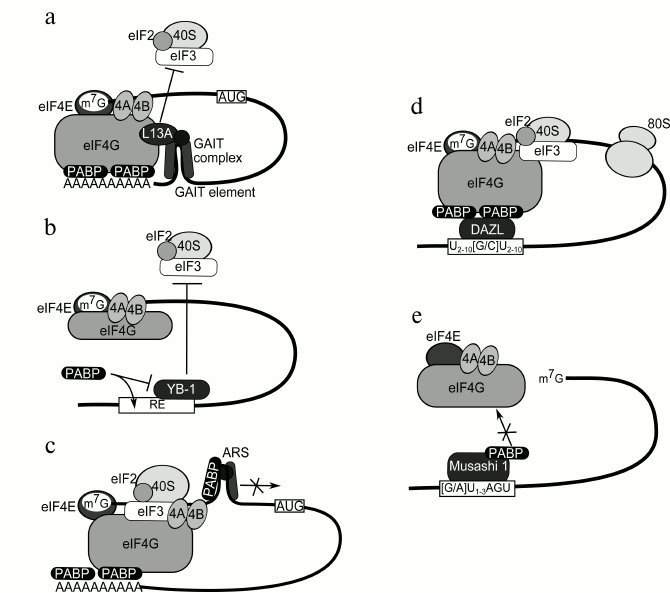
The inhibitory mechanism of Cp mRNA was studied in detail to find that the Cp mRNA 3′ UTR contains a 29 nt regulatory element GAIT (interferon γ-activated inhibitor of translation) in association with a large multiprotein complex. The key member of this complex is the ribosomal protein L13a. The synthesis of ceruloplasmin starts 2-4 h after cell stimulation with IFN-γ. During this time non-phosphorylated L13a is located on the ribosome. Its phosphorylation occurs after 16-24 h and is followed by its release from the ribosome. Free L13a interacts with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and with a pre-GAIT complex comprising phosphorylated Glu-Pro-tRNA synthetase and hnRNPQ1 (Fig. 4a). The resultant 450 kDa complex interacts with GAIT within the Cp mRNA 3′ UTR [94-96]. With the GAIT element and the 5′ UTR brought together by the PABP–poly(A)–eIF4F association, L13a binds to the C-terminal part of eIF4G that is responsible for eIF3 binding. This prevents recruitment of the 43S pre-initiation complex, and hence, causes inhibition of translation initiation [97]. A number of mRNAs were afterwards shown to be regulated in a similar way, including mRNAs of VEGF, APOL2, CCL22, CCR4, etc. [98].
Fig. 4. a) Model of inhibition of ceruloplasmin mRNA translation. The PABP–eIF4G interaction brings together the GAIT element (that blocks recruitment of the 43S pre-initiation complex) and the mRNA 5′ end. The GAIT-element is interferon γ-activated inhibitor of translation. b) Model of stimulation of YB-1 mRNA translation. PABP displaces the negative regulator YB-1 from the regulatory element (RE) within the 3′ UTR. c) Model of autoinhibition of PABP mRNA translation. PABP associated with other proteins binds to the A-rich autoregulatory sequence (ARS) within its own mRNA 5′ UTR and blocks its further scanning. d) Model of DAZL-mediated translation stimulation. Upon its binding to a specific sequence within poly(A)– mRNA 3′ UTR, the protein DAZL brings PABP into interaction with eIF4G, thereby stimulating translation. e) Model of Musashi 1-mediated translation inhibition. PABP is brought to Msi1 through its interaction with a specific sequence, which prevents its interaction with eIF4G.
PABP binding to A-rich segments within mRNA UTRs. Most frequently, PABP interacts with A-rich segments within regulatory regions of mRNAs. PABP bound to such a region in the YB-1 mRNA 3′ UTR (Fig. 4b) stimulates translation of this mRNA due to displacement of the negative regulator (YB-1) from this region [99-101]. The presence of the poly(A) tail in the mRNA molecule neutralizes the stimulating effect of PABP because of formation of a 3′ “mini-loop” structure [102]. Association of PABP with A-rich segments in the 3′ UTRs of RNAs of the red clover necrotic mosaic virus (RCNMV) [103], the Dengue virus [104], and others contributes to their translation by an unknown mechanism. However, bound PABP may sometimes cause translation inhibition, e.g. in the case of PABP mRNA (see Fig. 4c and the chapter “Regulation of Synthesis and Activity of PABPs”).
Mediated binding of PABP to A-unenriched segments within mRNA 3′UTRs. Stimulation of PABP-induced translation initiation is also observed in the absence of the poly(A) tail for mRNAs having in their 3′ UTRs the element U(2-10)[G/C]U(2-10) that is the binding site of the PABP-interacting protein DAZL [105]. The known DAZL targets are Sycp3, HuB, and Mvh mRNAs. It was shown in an in vitro system that DAZL can associate with 3′ UTRs and recruit PABP (Fig. 4d) [23].
In some cases interaction of PABP with 3′ UTR-associated proteins results in translation inhibition. For example, mRNAs of numb, p21WAF1, c-mos, doublecortin, and Adenomatous Polyposis Coli contain the element [G/A]U(1-3)AGU that is the binding site of the protein Msi1 (Musashi 1) whose interaction with PABP prevents binding of the latter to eIF4G, and hence, provokes translation inhibition. Of note, the Msi1-to-PABP binding was observed solely in neurons and can take place in the absence of RNA [106, 107].
Also, PABP is a member of regulatory complexes of some mRNAs, including those of PRE, Tim [108], caudal [109], insulin [110], and gurken [111]. But its role there is still obscure.
Degradation of mRNA
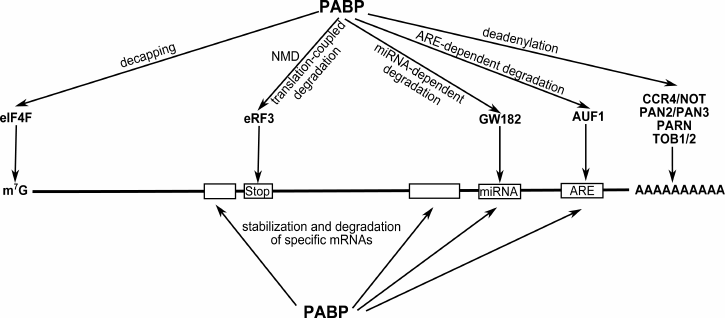
PABP is involved in a variety of mRNA stabilization and degradation events (Fig. 5). Its interaction with the termination factor eRF3 is important both for the efficient translation (as discussed above) and regulation of mRNA stability. The interaction of PABP with eRF3 reduces the number of PABP molecules bound to the poly(A) tail [26]. Each round of translation causes further truncation of the unprotected poly(A) tail. Its shortening down to 10-15 nt triggers the mechanisms of mRNA degradation [3]. This is why the eRF3–PABP interaction may be called a “molecular watch” showing the lifespan of an mRNA in the cell. This interaction might also be of importance for regulation and triggering of the nonsense-mediated mRNA decay (NMD). The key NMD proteins are UPF1 and the exon junction complex (EJC). PABP was shown to compete with UPF1 for binding to eRF3. This competition enhanced by PABP overexpression causes a higher level of termination and a decreased NMD-type mRNA degradation. Besides, as found, a closer proximity of the PABP binding site to the stop codon than that of the EJC binding site hampers mRNA degradation [112, 113].
Fig. 5. Involvement of PABP in a variety of mRNA stabilization/degradation events. The PABP–eIF4G interaction inhibits decapping and stabilizes mRNA. Through its interaction with CCR4-NOT, PARN, TOB1/2, and PAN2/3, PABP can influence the deadenylation reaction and the choice of degradation type. By binding to GW182, PABP can participate in microRNA-mediated inhibition of target mRNA translation and degradation. Besides, PABP is involved in stabilization and degradation of mRNAs that are controlled by specific sequences within these mRNAs, e.g. ARE (the decay-mediating AU-rich element).
There exist several pathways of mRNA degradation in the cell [114]. The most common one includes mRNA deadenylation, decapping, and subsequent exonuclease-induced cleavage. PABP plays a part in a variety of steps of this process. At the step of deadenylation, PABP can exhibit either an inhibitory or a stimulating effect. It inhibits the CCR4–NOT complex (a multiprotein complex in charge of deadenylation of the majority of mRNAs) and PARN; it stimulates the complex PAN2/3 responsible for truncation of the newly synthesized poly(A) tail and specific degradation of RAD5 mRNA [114-116]. Some researchers believe that PABP has no effect on the enzyme activity shown by these deadenylases. Its role is supposed to be choosing between the two degradation pathways, CCR4/POP2/NOT or PAN2/3. As known, the deadenylases PAN2/PAN3 can be recruited through a direct interaction between PABP and PAN3. The CCR4–NOT complex is recruited indirectly via protein adapters TOB1/2. It was shown that PABP can associate with either of them [37, 117].
The decapping step is controlled predominantly by stabilization of the cap-binding complex through the PABP–eIF4G interaction. It is conceived that PABP alone can sterically hamper binding of proteins that activate decapping enzymes (e.g. Lsm1p-7p) [114].
Lately, much attention has been paid to the involvement of PABP in microRNA-dependent repression of translation and mRNA degradation [118]. As reported, mRNAs are transcribed as precursors with a well-developed secondary structure; then, using Drosha (in the nucleus) and Dicer (in the cytoplasm) they are cut into 21-22 nt double-stranded fragments with protruding ends. One of their strands is in part complementary to the target mRNA 3′ UTR, while the other is subject to degradation [119]. By binding to their targets, microRNAs inhibit translation and/or trigger mRNA degradation.
Supposedly, in mammalian cells, microRNAs regulate about 60% of all cell mRNAs [120]. The microRNA-dependent deadenylation, translation repression, and mRNA degradation are widely studied [121], but no detailed mechanism of the microRNA effect on translation has been proposed so far. According to the recent full genome data, translation inhibition precedes deadenylation that in most cases ends with mRNA decapping and degradation [122, 123]. Different experimental systems (different organisms, tissues, or conditions) give different kinetics of repression and deadenylation. In some of them, repression and degradation are hardly separable [124], while in others deadenylation cannot be observed [125]. Presumably, different organisms or different events show some difference in the microRNA-dependent regulation. Regardless of a few reports on reactivation of repressed mRNAs [125], it still remains unclear whether translation repression and mRNA deadenylation are mostly reversible or not.
The major participant of a microRNA-dependent event is the RISC complex composed of a microRNA, the protein Ago (Argonaut), and the scaffold GW182 (in Drosophila melanogaster) or its paralogs (in other organisms). Due to its partial complementarity to the target mRNA, the Ago-associated microRNA delivers RISC to this mRNA. In turn, the scaffold GW182 recruits additional components of this complex, i.e. PABP, deadenylase complexes CCR4–NOT and PAN2/3, and E3-ubiquitin ligase EDD [121]. In this process, GW182 or TNRC6 (human paralog) is known to be the main PABP partner, but its role has not been determined yet. Some researchers believe that PABP plays a role in microRNA-mediated translation repression [28, 126], while others think it important for mRNA degradation [127, 128]. It was shown that GW182 may compete with eIF4G, and probably with the poly(A) tail too, for binding to PABP, which can lead to translation inhibition and mRNA degradation [118, 128]. According to one of the hypotheses, the GW182-to-mRNA binding results in a lower level of mRNA-associated eIF4E (mates with decapping), eIF4G (mates with deadenylation), and PABP (presumably, mates with translation inhibition); of note, the latter may occur in the absence of deadenylation [129]. PABP is also supposed to be able to recruit the deadenylase complex CCR4–NOT through its interaction with the adapter TOB1/2 [117, 127]. Another hypothesis discussed in the literature is that PABP brings to mRNA proteins from the RISC complex through its interaction with GW182. The RISC-to-mRNA binding leads to PABP displacement, thus triggering the mechanisms of translation repression and mRNA degradation [126]. Some recent studies show that PABP is not directly involved either in mRNA degradation or translation inhibition under the action of microRNAs [130, 131]. Because translation regulation mediated by microRNAs is a most complicated and still poorly studied process, it is difficult to say which of these hypotheses is the closest to the truth. It can be believed that all these scenarios may be good for the cell, and its choice depends on conditions, the particular microRNA, or the target mRNA.
Participation of PABP in mRNA stabilization/degradation controlled by specific 3′ UTR sequences is also worth mentioning. The presence of the AU-rich element (ARE) causes a rapid degradation of mRNAs. The protein AUF1 playing the key role in the ARE-dependent degradation was shown to associate with the poly(A) tail, thereby displacing PABP. Hence, this interaction may contribute much to translation inhibition and mRNA degradation [20, 132].
As found, PABP may also be a component of ARE-independent mechanisms of stabilization of specific mRNAs. For example, PABP inhibits erythroid cell-enriched endonuclease (ErEN) whose target is the cytosine-rich element (CRE) located within the 3′ UTR of α-globin mRNA. The poly(A) tail-bound PABP interacts with the poly(C)-binding proteins (αCP), thereby enhancing their affinity for the CRE and, in turn, protecting this element against the action of the endonuclease ErEN [133].
Another example is regulation of stability of the neurofilament NF-L mRNA. By binding to the decay-mediating element within this mRNA, PABP protects it from aldolase C-induced degradation. Free PABP binding to dimeric (active) aldolase prevents the aldolase–mRNA association [134].
In the case of the transcription factor c-fos mRNA, the element termed the major protein-coding region determinant (mCRD) is located within the 3′ UTR and covers a part of the coding region. The mCRD is protected by the mRNA-associated protein complex composed of UNR (comprising five cold-shock domains), PABP, Paip1, hnRNPD, and hnRNPQ. Having reached this complex, the ribosome displaces it from the mCRD, thereby initiating mRNA deadenylation followed by degradation [135].
There are a few other reports on PABP involvement in regulation of mRNA stability. This protein is reported to be able both to stabilize mRNAs, e.g. those of oscar [136] or TIMP1, iNOS [137] and to destabilize them, e.g. in the case of MKK2, GAPDH, etc. [138], although the mechanism of both is unknown.
REGULATION OF SYNTHESIS AND ACTIVITY OF PABPs
As mentioned above, regulation of PABP activity may be mediated by PABP-binding proteins (Paip1, Paip2, DAZL, GW182, etc.) due to their modification and degradation [48, 139]. Similarly, PABP functioning can be influenced by other poly(A)-interacting proteins. For example, competition between PABP and hnRNPQ2 for poly(A) binding results in repression of global translation and let7 miRNA-dependent degradation of target mRNAs [140]. Since PABP is a nucleocytoplasmic protein, its involvement in a certain event may be dictated by its subcellular localization [54]. Accumulation of this protein in the nucleus has been reported to result from some types of stress, such as oxidative stress [141], UV irradiation [142], heat shock [143], and some viral infections (rotavirus, herpes simplex virus) [144, 145]. It is known that PABP translocation into the nucleus is mediated by karyopherin 108, but the regulatory mechanism of its accumulation there is unclear [22]. It was found recently that the carrier of the yeast PABN1 (Pab2) is karyopherin β2. The Rmt1-induced arginine methylation within the nuclear localization signal results in a lower Pab2 import into the nucleus. Of note, this mechanism functions only for the yeast protein, whereas no analogy for the human protein has been reported [146]. In some cases, instead of nuclear import activation, disturbance of export from the nucleus was observed. The export of PABP can be either Crm1-dependent or mRNA-dependent [22]. Under some stress conditions, splicing and polyadenylation appear to be blocked, and hence, the mRNA transport ceases [142, 147]. Then, the idle PABP accumulates in the nucleus.
PABP synthesis can be regulated at the stage of translation initiation. The PABP mRNA contains in its 5′ UTR two regulatory sequences: the terminal oligopyrimidine track (TOP) and the adenine-rich autoregulatory sequence (ARS).
It is believed that ARS performs a constitutive control of PABP synthesis in the cell. Previously, PABP was suggested to specifically inhibit translation of its own mRNA (Fig. 4c) by binding to the A-rich region within the 5′ UTR [148]. However, later studies showed that the PABP-to-ARS binding was a few times inferior to that of PABP to the poly(A) sequence. Also, with the poly(A) sequence substituted for ARS, the translation inhibition reduced 3-fold. This suggests another, more complicated mechanism of translation regulation. The ARS consists of 4-6 oligo(A) segments interlinked with conserved pyrimidine-rich linkers of a certain length. It is this arrangement that presumably contributes to a more efficient inhibition of translation. As found, apart from PABP, ARS partners are UNR and the insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1). These three proteins form a complex that binds to ARS and blocks mRNA scanning by the 43S pre-initiation complex, thereby inhibiting translation of the PABP mRNA [10, 149, 150].
The presence of the TOP sequence is typical for mRNAs whose translation is regulated in the course of cell growth and development (e.g. those of ribosomal proteins, elongation factors). The regulatory mechanism of translation of TOP-containing mRNAs is obscure, although translation activation is known to be accompanied by activation of the mTORC1 signaling pathway [151]. The targets of mTOR kinase are eIF4E binding proteins (4EBP) and S6 kinase. A model based on mTOR-activating cytomegalovirus infection showed that activation of PABP translation requires only hyperphosphorylation of 4EBP but not S6 kinase [90, 91]. The regulation of translation of TOP-containing mRNAs can additionally involve other proteins. For example, myotonic dystrophy type 2 is characterized by a typically decreased amount of ZNF9 and a lower level of translation of TOP mRNAs, including PABP mRNA. The mechanism of the ZNF9 effect on translation remains to be elucidated [152]. The stress granule proteins TIA-1 and TIAR may also participate in TOP mRNA translation regulation. Under amino acid starvation, TIA-1 and TIAR interact with TOP sequences and transfer mRNAs from polysomes to stress granules [153]. Thus, PABP synthesis is synchronized with synthesis of other components of the translation apparatus.
It should be noted that the amount of PABP increases after cessation of heat shock action [143] and decreases as a result of knockdown of the gene LARP4 coding for La related protein 4 [31]. But the mechanism of this regulation has not been studied.
Supposedly, the functional activity of PABP can be regulated by phosphorylation. It was reported that in plants the hypophosphorylated PABP shows low activity in binding to poly(A) and eIF4G [154]. In extracts of PABP-hyperexpressing HeLa cells PABP phosphorylation was shown to be possibly performed by the ERK1/2 signaling pathway, thus stimulating translation [138]. As shown recently, ePABP, an embryo homolog of PABP, undergoes phosphorylation upon oocyte maturation. All these modifications are localized to the proline-rich linker. The hyperphosphorylated ePABP has been shown to interact in vivo with the mRNA poly(A) tail, the cap, polysomes, and cytoplasmic polyadenylation proteins. Moreover, only hyperphosphorylated PABP can bind to the regulator Paip2 [155]. Thus, phosphorylation may be another important modification in functioning of other members of the family of poly(A) binding proteins.
Apart from phosphorylation, other posttranslational modifications of PABP have been reported recently, such as methylation of glutamine and asparagine residues, and methylation and acetylation of lysine residues. The latter is of most interest. As found, acetylation of lysine residues changes in accordance with the cell cycle, i.e. the amount of non-acetylated protein gradually decreases to disappear by the beginning of mitosis. Presumably, acetylation of some PABP residues may decrease eRF3 binding and increase Paip2 binding [156].
CONCLUSION
Poly(A) binding proteins have long been in the focus of studies primarily due to their unique ability to interact with mRNA poly(A) tails. Eventually, this allowed discovering their role in mRNA translation and stability, as well as in other events where the mRNA poly(A) tail is involved. Their importance for translation is emphasized by the fact that many researchers believe them to be noncanonical translation factors. However, as it often happens, intensive studies of PABPs properties and functions gave ambiguous interpretations of data on PABP-involving regulatory mechanisms. In spite of the abundance of information, it is still unclear how PABPs participate in microRNA-mediated inhibition of mRNA translation and/or degradation. The mechanism of replacement of nuclear PABP by its cytoplasmic form also needs to be made clearer.
Besides, new challenges are to be addressed. The numerous recent data on covalent modification-mediated regulation of PABPs activity may give a stimulus for investigating more mechanisms of PABPs functioning in the cell. Numerous PABPs homologs and their functional implications are also of great interest. Recent reports on proteins that, like PABPs, exhibit a high affinity for poly(A) sequences (e.g. hnRNPQ) suggest their importance for the mechanisms of specific regulation of mRNA translation and stability. Specificity of PABPs not only to homopoly(A) ribonucleotides but also to AU-rich and A-rich sequences suggests existence of their still unknown functions in mRNA-dependent events.
Thus, despite the long-term studies of PABPs, their functioning in the cell is still obscure in many respects. With the advance of cell studying techniques novel results may be expected that will allow a different view on the role of PABPs in the life of a cell and the whole organism.
The authors thank E. V. Serebrova for help in manuscript preparation.
This study was supported by the Russian Foundation for Basic Research (grant No. 11-04-00267) and Programs on “Molecular and Cell Biology” and “Basic Sciences to Medicine” from the Russian Academy of Sciences.
REFERENCES
1.Blobel, G. (1972) Biochem. Biophys. Res.
Commun., 47, 88-95.
2.Blobel, G. (1973) Proc. Natl. Acad. Sci.
USA, 70, 924-928.
3.Mangus, D. A., Evans, M. C., and Jacobson, A.
(2003) Genome Biol., 4, 223.
4.Goss, D. J., and Kleiman, F. E. (2013) Wiley
Interdiscip. Rev. RNA, 4, 167-179.
5.Deo, R. C., Bonanno, J. B., Sonenberg, N., and
Burley, S. K. (1999) Cell, 98, 835-845.
6.Safaee, N., Kozlov, G., Noronha, A. M., Xie, J.,
Wilds, C. J., and Gehring, K. (2012) Mol. Cell, 48,
375-386.
7.Kuhn, U., and Pieler, T. (1996) J. Mol.
Biol., 256, 20-30.
8.Burd, C. G., Matunis, E. L., and Dreyfuss, G.
(1991) Mol. Cell Biol., 11, 3419-3424.
9.Khanam, T., Muddashetty, R. S., Kahvejian, A.,
Sonenberg, N., and Brosius, J. (2006) RNA Biol., 3,
170-177.
10.Patel, G. P., and Bag, J. (2006) FEBS J.,
273, 5678-5690.
11.Sladic, R. T., Lagnado, C. A., Bagley, C. J., and
Goodall, G. J. (2004) Eur. J. Biochem., 271, 450-457.
12.Baer, B. W., and Kornberg, R. D. (1983) J.
Cell Biol., 96, 717-721.
13.Mullin, C., Duning, K., Barnekow, A., Richter,
D., Kremerskothen, J., and Mohr, E. (2004) FEBS Lett.,
576, 437-441.
14.Von Der Haar, T., Ball, P. D., and McCarthy, J.
E. (2000) J. Biol. Chem., 275, 30551-30555.
15.Wei, C. C., Balasta, M. L., Ren, J., and Goss, D.
J. (1998) Biochemistry, 37, 1910-1916.
16.Khaleghpour, K., Kahvejian, A., De Crescenzo, G.,
Roy, G., Svitkin, Y. V., Imataka, H., O’Connor-McCourt, M.,
and Sonenberg, N. (2001) Mol. Cell Biol., 21,
5200-5213.
17.Koloteva-Levine, N., Pinchasi, D., Pereman, I.,
Zur, A., Brandeis, M., and Elroy-Stein, O. (2004) Mol. Cell
Biol., 24, 3577-3587.
18.Kozlov, G., Safaee, N., Rosenauer, A., and
Gehring, K. (2010) J. Biol. Chem., 285, 13599-13606.
19.Lessing, D., and Bonini, N. M. (2008) PLoS
Biol., 6, e29.
20.Lu, J.-Y., Bergman, N., Sadri, N., and Schneider,
R. J. (2006) RNA, 12, 883-893.
21.Ilkow, C. S., Mancinelli, V., Beatch, M. D., and
Hobman, T. C. (2008) J. Virol., 82, 4284-4294.
22.Brune, C., Munchel, S. E., Fischer, N.,
Podtelejnikov, A. V., and Weis, K. (2005) RNA, 11,
517-531.
23.Collier, B., Gorgoni, B., Loveridge, C., Cooke,
H. J., and Gray, N. K. (2005) EMBO J., 24, 2656-2666.
24.Bushell, M., Wood, W., Carpenter, G., Pain, V.
M., Morley, S. J., and Clemens, M. J. (2001) J. Biol.
Chem., 276, 23922-23928.
25.Imataka, H., Gradi, A., and Sonenberg, N. (1998)
EMBO J., 17, 7480-7489.
26.Hoshino, S., Imai, M., Kobayashi, T., Uchida, N.,
and Katada, T. (1999) J. Biol. Chem., 274,
16677-16680.
27.Uchida, N., Hoshino, S.-I., Imataka, H.,
Sonenberg, N., and Katada, T. (2002) J. Biol. Chem., 277,
50286-50292.
28.Huntzinger, E., Braun, J. E., Heimstadt, S.,
Zekri, L., and Izaurralde, E. (2010) EMBO J., 29,
4146-4160.
29.Hasgall, P. A., Hoogewijs, D., Faza, M. B.,
Panse, V. G., Wenger, R. H., and Camenisch, G. (2011) PLoS One,
6, e22107.
30.Blagden, S. P., Gatt, M. K., Archambault, V.,
Lada, K., Ichihara, K., Lilley, K. S., Inoue, Y. H., and Glover, D. M.
(2009) Dev. Biol., 334, 186-197.
31.Yang, R., Gaidamakov, S. A., Xie, J., Lee, J.,
Martino, L., Kozlov, G., Crawford, A. K., Russo, A. N., Conte, M. R.,
Gehring, K., and Maraia, R. J. (2011) Mol. Cell Biol.,
31, 542-556.
32.Miroci, H., Schob, C., Kindler, S.,
Olschlager-Schutt, J., Fehr, S., Jungenitz, T., Schwarzacher, S. W.,
Bagni, C., and Mohr, E. (2012) J. Biol. Chem., 287,
1322-1334.
33.Wang, X., Bai, J., Zhang, L., Wang, X., Li, Y.,
and Jiang, P. (2012) Antiviral Res., 96, 315-323.
34.Roy, G., De Crescenzo, G., Khaleghpour, K.,
Kahvejian, A., O’Connor-McCourt, M., and Sonenberg, N. (2002)
Mol. Cell Biol., 22, 3769-3782.
35.Berlanga, J. J., Baass, A., and Sonenberg, N.
(2006) RNA, 12, 1556-1568.
36.Siddiqui, N., Mangus, D. A., Chang, T.-C.,
Palermino, J.-M., Shyu, A.-B., and Gehring, K. (2007) J. Biol.
Chem., 282, 25067-25075.
37.Ezzeddine, N., Chang, T.-C., Zhu, W., Yamashita,
A., Chen, C.-Y. A., Zhong, Z., Yamashita, Y., Zheng, D., and Shyu,
A.-B. (2007) Mol. Cell Biol., 27, 7791-7801.
38.Okochi, K., Suzuki, T., Inoue, J., Matsuda, S.,
and Yamamoto, T. (2005) Genes Cells, 10, 151-163.
39.Chang, T.-C., Yamashita, A., Chen, C.-Y. A.,
Yamashita, Y., Zhu, W., Durdan, S., Kahvejian, A., Sonenberg, N., and
Shyu, A.-B. (2004) Genes Dev., 18, 2010-2023.
40.Lin, J., Fabian, M., Sonenberg, N., and Meller,
A. (2012) Biophys. J., 102, 1427-1434.
41.Melo, E. O., Dhalia, R., Martins de Sa, C.,
Standart, N., and De Melo Neto, O. P. (2003) J. Biol. Chem.,
278, 46357-46368.
42.Smith, R. W. P., and Gray, N. K. (2010)
Biochem. J., 426, 1-12.
43.Bonderoff, J. M., Larey, J. L., and Lloyd, R. E.
(2008) J. Virol., 82, 9389-9399.
44.Rivera, C. I., and Lloyd, R. E. (2008)
Virology, 375, 59-72.
45.Kuyumcu-Martinez, N. M., Joachims, M., and Lloyd,
R. E. (2002) J. Virol., 76, 2062-2074.
46.Kozlov, G., De Crescenzo, G., Lim, N. S.,
Siddiqui, N., Fantus, D., Kahvejian, A., Trempe, J.-F., Elias, D.,
Ekiel, I., Sonenberg, N., O’Connor-McCourt, M., and Gehring, K.
(2004) EMBO J., 23, 272-281.
47.Kozlov, G., Menade, M., Rosenauer, A., Nguyen,
L., and Gehring, K. (2010) J. Mol. Biol., 397,
397-407.
48.Huang, K.-L., Chadee, A. B., Chen, C.-Y. A.,
Zhang, Y., and Shyu, A.-B. (2013) RNA, 19, 295-305.
49.Kononenko, A. V., Mitkevich, V. A.,
Atkinson, G. C., Tenson, T., Dubovaya, V. I., Frolova, L. Y., Makarov,
A. A., and Hauryliuk, V. (2010) Nucleic Acids Res., 38,
548-558.
50.Keller, R. W., Kuhn, U., Aragon, M., Bornikova,
L., Wahle, E., and Bear, D. G. (2000) J. Mol. Biol., 297,
569-583.
51.Wahle, E., and Ruegsegger, U. (1999) FEMS
Microbiol. Rev., 23, 277-295.
52.Kuhn, U., Gundel, M., Knoth, A., Kerwitz, Y.,
Rudel, S., and Wahle, E. (2009) J. Biol. Chem., 284,
22803-22814.
53.Chekanova, J. A., and Belostotsky, D. A. (2003)
RNA, 9, 1476-1490.
54.Lemay, J.-F., Lemieux, C., St-Andre, O., and
Bachand, F. (2010) RNA Biol., 7, 291-295.
55.Dunn, E. F., Hammell, C. M., Hodge, C. A., and
Cole, C. N. (2005) Genes Dev., 19, 90-103.
56.Hosoda, N., Lejeune, F., and Maquat, L. E. (2006)
Mol. Cell Biol., 26, 3085-3097.
57.Maquat, L. E., Hwang, J., Sato, H., and Tang, Y.
(2010) Cold Spring Harb. Symp. Quant. Biol., 75,
127-134.
58.Sato, H., and Maquat, L. E. (2009) Genes
Dev., 23, 2537-2550.
59.Bhattacharjee, R. B., and Bag, J. (2012) PLoS
One, 7, e53036.
60.Richter, J. D., and Lasko, P. (2011) Cold
Spring Harb. Perspect. Biol., 3, a002758.
61.Darnell, J. C., and Richter, J. D. (2012) Cold
Spring Harb. Perspect. Biol., 4, a012344.
62.Richter, J. D. (2007) Trends Biochem.
Sci., 32, 279-285.
63.Kim, J. H., and Richter, J. D. (2006) Mol.
Cell, 24, 173-183.
64.Kuersten, S., and Goodwin, E. B. (2003) Nat.
Rev. Genet., 4, 626-637.
65.Tay, J., Hodgman, R., Sarkissian, M., and
Richter, J. D. (2003) Genes Dev., 17, 1457-1462.
66.Atkins, C. M., Nozaki, N., Shigeri, Y., and
Soderling, T. R. (2004) J. Neurosci., 24, 5193-5201.
67.Udagawa, T., Swanger, S. A., Takeuchi, K., Kim,
J. H., Nalavadi, V., Shin, J., Lorenz, L. J., Zukin, R. S., Bassell, G.
J., and Richter, J. D. (2012) Mol. Cell, 47, 253-266.
68.De Moor, C. H., and Richter, J. D. (1999) EMBO
J., 18, 2294-2303.
69.Groisman, I., Jung, M.-Y., Sarkissian, M., Cao,
Q., and Richter, J. D. (2002) Cell, 109, 473-483.
70.Gray, N. K., Coller, J. M., Dickson, K. S., and
Wickens, M. (2000) EMBO J., 19, 4723-4733.
71.Otero, L. J., Ashe, M. P., and Sachs, A. B.
(1999) EMBO J., 18, 3153-3163.
72.Wells, S. E., Hillner, P. E., Vale, R. D., and
Sachs, A. B. (1998) Mol. Cell, 2, 135-140.
73.Rajkowitsch, L., Vilela, C., Berthelot, K.,
Ramirez, C. V., and McCarthy, J. E. G. (2004) J. Mol. Biol.,
335, 71-85.
74.Kahvejian, A., Svitkin, Y. V., Sukarieh,
R., M’Boutchou, M.-N., and Sonenberg, N. (2005) Genes
& Development, 19, 104-113.
75.Borman, A. M., Michel, Y. M., Malnou, C. E., and
Kean, K. M. (2002) J. Biol. Chem., 277, 36818-36824.
76.Craig, A. W., Haghighat, A., Yu, A. T., and
Sonenberg, N. (1998) Nature, 392, 520-523.
77.Martineau, Y., Derry, M. C., Wang, X., Yanagiya,
A., Berlanga, J. J., Shyu, A.-B., Imataka, H., Gehring, K., and
Sonenberg, N. (2008) Mol. Cell Biol., 28, 6658-6667.
78.Lim, N. S., Kozlov, G., Chang, T.-C., Groover,
O., Siddiqui, N., Volpon, L., De Crescenzo, G., Shyu, A.-B., and
Gehring, K. (2006) J. Biol. Chem., 281, 14376-14382.
79.Yoshida, M., Yoshida, K., Kozlov, G., Lim, N. S.,
De Crescenzo, G., Pang, Z., Berlanga, J. J., Kahvejian, A., Gehring,
K., Wing, S. S., and Sonenberg, N. (2006) EMBO J., 25,
1934-1944.
80.Yanagiya, A., Delbes, G., Svitkin, Y. V.,
Robaire, B., and Sonenberg, N. (2010) J. Clin. Invest.,
120, 3389-3400.
81.Delbes, G., Yanagiya, A., Sonenberg, N., and
Robaire, B. (2012) Biol. Reprod., 86, 95.
82.Jacobson, A. (1996) Translational Control,
Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 30,
451-480.
83.Proweller, A., and Butler, J. S. (1996) J.
Biol. Chem., 271, 10859-10865.
84.Searfoss, A., Dever, T. E., and Wickner, R.
(2001) Mol. Cell Biol., 21, 4900-4908.
85.Padilla-Noriega, L., Paniagua, O., and
Guzman-Leon, S. (2002) Virology, 298, 1-7.
86.Piron, M., Vende, P., Cohen, J., and Poncet, D.
(1998) EMBO J., 17, 5811-5821.
87.Joachims, M., Van Breugel, P. C., and Lloyd, R.
E. (1999) J. Virol., 73, 718-727.
88.Kerekatte, V., Keiper, B. D., Badorff, C., Cai,
A., Knowlton, K. U., and Rhoads, R. E. (1999) J. Virol.,
73, 709-717.
89.Bradrick, S. S., Dobrikova, E. Y., Kaiser, C.,
Shveygert, M., and Gromeier, M. (2007) RNA, 13,
1582-1593.
90.Perez, C., McKinney, C., Chulunbaatar, U., and
Mohr, I. (2011) J. Virol., 85, 156-164.
91.McKinney, C., Perez, C., and Mohr, I. (2012)
Proc. Natl. Acad. Sci. USA, 109, 5627-5632.
92.Zhang, B., Morace, G., Gauss-Muller, V., and
Kusov, Y. (2007) Nucleic Acids Res., 35, 5975-5984.
93.Duncan, K. E., Strein, C., and Hentze, M. W.
(2009) Mol. Cell, 36, 571-582.
94.Mazumder, B., Seshadri, V., Imataka, H.,
Sonenberg, N., and Fox, P. L. (2001) Mol. Cell Biol., 21,
6440-6449.
95.Mazumder, B., Sampath, P., Seshadri, V., Maitra,
R. K., DiCorleto, P. E., and Fox, P. L. (2003) Cell, 115,
187-198.
96.Sampath, P., Mazumder, B., Seshadri, V., Gerber,
C. A., Chavatte, L., Kinter, M., Ting, S. M., Dignam, J. D., Kim, S.,
Driscoll, D. M., and Fox, P. L. (2004) Cell, 119,
195-208.
97.Kapasi, P., Chaudhuri, S., Vyas, K., Baus, D.,
Komar, A. A., Fox, P. L., Merrick, W. C., and Mazumder, B. (2007)
Mol. Cell, 25, 113-126.
98.Vyas, K., Chaudhuri, S., Leaman, D. W., Komar, A.
A., Musiyenko, A., Barik, S., and Mazumder, B. (2009) Mol. Cell
Biol., 29, 458-470.
99.Skabkina, O. V., Skabkin, M. A., Popova,
N. V., Lyabin, D. N., Penalva, L. O., and Ovchinnikov, L. P.
(2003) J. Biol. Chem., 278, 18191-18198.
100.Skabkina, O. V., Lyabin, D. N., Skabkin,
M. A., and Ovchinnikov, L. P. (2005) Mol. Cell Biol., 25,
3317-3323.
101.Lyabin, D. N., Eliseeva, I. A., Skabkina,
O. V., and Ovchinnikov, L. P. (2011) RNA Biol., 8,
883-892.
102.Eliseeva, I. A., Ovchinnikov, L. P., and
Lyabin, D. N. (2012) RNA Biol., 9, 1473-1487.
103.Iwakawa, H.-O., Tajima, Y., Taniguchi, T.,
Kaido, M., Mise, K., Tomari, Y., Taniguchi, H., and Okuno, T. (2012)
J. Virol., 86, 7836-7849.
104.Polacek, C., Friebe, P., and Harris, E. (2009)
J. Gen. Virol., 90, 687-692.
105.Reynolds, N., Collier, B., Maratou, K.,
Bingham, V., Speed, R. M., Taggart, M., Semple, C. A., Gray, N. K., and
Cooke, H. J. (2005) Hum. Mol. Genet., 14, 3899-3909.
106.Kawahara, H., Imai, T., Imataka, H., Tsujimoto,
M., Matsumoto, K., and Okano, H. (2008) J. Cell Biol.,
181, 639-653.
107.Ohyama, T., Nagata, T., Tsuda, K., Kobayashi,
N., Imai, T., Okano, H., Yamazaki, T., and Katahira, M. (2011)
Nucleic Acids Res., 40, 3218-3231.
108.Lim, C., Lee, J., Choi, C., Kilman, V. L., Kim,
J., Park, S. M., Jang, S. K., Allada, R., and Choe, J. (2011)
Nature, 470, 399-403.
109.Singh, N., Morlock, H., and Hanes, S. D. (2011)
Dev. Biol., 352, 104-115.
110.Kulkarni, S. D., Muralidharan, B., Panda, A.
C., Bakthavachalu, B., Vindu, A., and Seshadri, V. (2011) J. Biol.
Chem., 286, 14146-14156.
111.Clouse, K. N., Ferguson, S. B., and Schupbach,
T. (2008) Dev. Biol., 313, 713-724.
112.Ivanov, P. V., Gehring, N. H., Kunz, J.
B., Hentze, M. W., and Kulozik, A. E. (2008) EMBO J., 27,
736-747.
113.Singh, G., Rebbapragada, I., and
Lykke-Andersen, J. (2008) PLoS Biol., 6, e111.
114.Parker, R., and Song, H. (2004) Nat. Struct.
Mol. Biol., 11, 121-127.
115.Brown, C. E., and Sachs, A. B. (1998) Mol.
Cell Biol., 18, 6548-6559.
116.Kuhn, U., and Wahle, E. (2004) Biochim.
Biophys. Acta, 1678, 67-84.
117.Ezzeddine, N., Chen, C.-Y. A., and Shyu, A.-B.
(2012) Mol. Cell Biol., 32, 1089-1098.
118.Tritschler, F., Huntzinger, E., and Izaurralde,
E. (2010) Nat. Rev. Mol. Cell Biol., 11, 379-384.
119.Lai, E. C. (2003) Curr. Biol.,
13, r925-936.
120.Friedman, R. C., Farh, K. K.-H., Burge, C. B.,
and Bartel, D. P. (2009) Genome Res., 19, 92-105.
121.Fabian, M. R., and Sonenberg, N. (2012) Nat.
Struct. Mol. Biol., 19, 586-593.
122.Bazzini, A. A., Lee, M. T., and Giraldez, A. J.
(2012) Science, 336, 233-237.
123.Djuranovic, S., Nahvi, A., and Green, R. (2012)
Science, 336, 237-240.
124.Eulalio, A., Huntzinger, E., Nishihara, T.,
Rehwinkel, J., Fauser, M., and Izaurralde, E. (2009) RNA,
15, 21-32.
125.Bhattacharyya, S. N., Habermacher, R., Martine,
U., Closs, E. I., and Filipowicz, W. (2006) Cell, 125,
1111-1124.
126.Moretti, F., Kaiser, C., Zdanowicz-Specht, A.,
and Hentze, M. W. (2012) Nat. Struct. Mol. Biol., 19,
603-608.
127.Fabian, M. R., Mathonnet, G., Sundermeier, T.,
Mathys, H., Zipprich, J. T., Svitkin, Y. V., Rivas, F., Jinek, M.,
Wohlschlegel, J., Doudna, J. A., Chen, C.-Y. A., Shyu, A.-B., Yates, J.
R., Hannon, G. J., Filipowicz, W., Duchaine, T. F., and Sonenberg, N.
(2009) Mol. Cell, 35, 868-880.
128.Zekri, L., Huntzinger, E., Heimstadt, S., and
Izaurralde, E. (2009) Mol. Cell Biol., 29, 6220-6231.
129.Zekri, L., Kuzuoglu-Ozturk, D., and Izaurralde,
E. (2013) EMBO J., 32, 1052-1065.
130.Fukaya, T., and Tomari, Y. (2011) EMBO
J., 30, 4998-5009.
131.Mishima, Y., Fukao, A., Kishimoto, T.,
Sakamoto, H., Fujiwara, T., and Inoue, K. (2012) Proc. Natl. Acad.
Sci. USA, 109, 1104-1109.
132.Sagliocco, F., Laloo, B., Cosson, B., Laborde,
L., Castroviejo, M., Rosenbaum, J., Ripoche, J., and Grosset, C. (2006)
Biochem. J., 400, 337-347.
133.Wang, Z., and Kiledjian, M. (2000) Mol. Cell
Biol., 20, 6334-6341.
134.Stefanizzi, I., and Canete-Soler, R. (2007)
Brain Res., 1139, 15-28.
135.Grosset, C., Chen, C. Y., Xu, N., Sonenberg,
N., Jacquemin-Sablon, H., and Shyu, A. B. (2000) Cell,
103, 29-40.
136.Vazquez-Pianzola, P., Urlaub, H., and Suter, B.
(2011) Dev. Biol., 357, 404-418.
137.Casper, I., Nowag, S., Koch, K., Hubrich, T.,
Bollmann, F., Henke, J., Schmitz, K., Kleinert, H., and Pautz, A.
(2013) Nitric Oxide, 33, 6-17.
138.Ma, S., Musa, T., and Bag, J. (2006) J.
Biol. Chem., 281, 3145-3156.
139.Brooks, S. A. (2010) Wiley Interdiscip. Rev.
RNA, 1, 240-252.
140.Svitkin, Y. V., Yanagiya, A., Karetnikov, A.
E., Alain, T., Fabian, M. R., Khoutorsky, A., Perreault, S.,
Topisirovic, I., and Sonenberg, N. (2013) PLoS Biol., 11,
e1001564.
141.Salaun, C., MacDonald, A. I., Larralde, O.,
Howard, L., Lochtie, K., Burgess, H. M., Brook, M., Malik, P., Gray, N.
K., and Graham, S. V. (2010) J. Virol., 84,
8539-8548.
142.Burgess, H. M., Richardson, W. A., Anderson, R.
C., Salaun, C., Graham, S. V., and Gray, N. K. (2011) J. Cell
Sci., 124, 3344-3355.
143.Ma, S., Bhattacharjee, R. B., and Bag, J.
(2009) FEBS J., 276, 552-570.
144.Dobrikova, E., Shveygert, M., Walters, R., and
Gromeier, M. (2010) J. Virol., 84, 270-279.
145.Harb, M., Becker, M. M., Vitour, D., Baron, C.
H., Vende, P., Brown, S. C., Bolte, S., Arold, S. T., and Poncet, D.
(2008) J. Virol., 82, 11283-11293.
146.Mallet, P.-L., and Bachand, F. (2013)
Traffic, 14, 282-294.
147.Arnold, M. M., Brownback, C. S., Taraporewala,
Z. F., and Patton, J. T. (2012) J. Gen. Virol., 93,
1483-1489.
148.Bag, J., and Wu, J. (1996) Eur. J.
Biochem., 237, 143-152.
149.Bag, J. (2001) J. Biol. Chem.,
276, 47352-47360.
150.Patel, G. P., Ma, S., and Bag, J. (2005)
Nucleic Acids Res., 33, 7074-7089.
151.Hornstein, E., Git, A., Braunstein, I., Avni,
D., and Meyuhas, O. (1999) J. Biol. Chem., 274,
1708-1714.
152.Huichalaf, C., Schoser, B., Schneider-Gold, C.,
Jin, B., Sarkar, P., and Timchenko, L. (2009) J. Neurosci.,
29, 9042-9049.
153.Damgaard, C. K., and Lykke-Andersen, J. (2011)
Genes Dev., 25, 2057-2068.
154.Le, H., Browning, K. S., and Gallie, D. R.
(2000) J. Biol. Chem., 275, 17452-17462.
155.Friend, K., Brook, M., Bezirci, F. B., Sheets,
M. D., Gray, N. K., and Seli, E. (2012) Biochem. J., 445,
93-100.
156.Brook, M., McCracken, L., Reddington, J. P.,
Lu, Z.-L., Morrice, N. A., and Gray, N. K. (2012) Biochem. J.,
441, 803-812.