Pectin Methylesterase-Generated Methanol May Be Involved in Tobacco Leaf Growth
T. V. Komarova1,2, D. V. Pozdyshev1,2, I. V. Petrunia2, E. V. Sheshukova2, and Y. L. Dorokhov1,2*
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; fax: +7 (495) 939-3181; E-mail: dorokhov@genebee.msu.su2Vavilov Institute of General Genetics, Russian Academy of Sciences, ul. Gubkina 3, 119991 Moscow, Russia; fax: +7 (499) 132-8962; E-mail: dorokhov@genebee.msu.ru
* To whom correspondence should be addressed.
Received September 26, 2013; Revision received October 14, 2013
Plant leaves undergo a sink–source modification of intercellular macromolecular transport during the transition from carbon import to carbon export. After assessing the role of metabolite signaling in gene regulation in Nicotiana tabacum sink and source leaves, we observed increased pectin methylesterase (PME)-mediated methanol generation in immature leaves. Using suppression subtractive hybridization (SSH), we identified a number of genes whose activity changes from sink to source leaves. The most abundant SSH-identified genes appeared to be sensitive to methanol. We hypothesize that tobacco leaf maturation and the sink–source transition are accompanied by a change in mRNA levels of genes that function in methanol-dependent cell signaling.
KEY WORDS: pectin methylesterase, methanol, movement, plasmodesmata, Nicotiana tabacum, transportDOI: 10.1134/S0006297914020035
Abbreviations: MIG, methanol-inducible gene; PD, plasmodesmata; PME, pectin methylesterase; SSH, suppression subtractive hybridization.
Plant growth and the transition from an immature apical leaf to a mature
leaf are accompanied not only by an increase in cell size, but also by
two important interconnected processes. First, there is a change in
carbohydrate metabolism when immature leaves, which consume
photosynthetic carbon (sink leaves), are converted into mature leaves
(source leaves) [1]. During the second process,
there is a modification of the intercellular transport of
macromolecules carried out by plasmodesmata (PD) [2]. The sink leaf PD is simple and characterized by
high capacity for providing free transport of macromolecules between
cells, whereas the source leaf PD is branched, with limited
macromolecular transport [2-5].
The sink–source transition occurs under strict genetic control
and is very sensitive to signals far removed from the PD channel
itself. For example, during the sink–source transition in
Arabidopsis thaliana, increased size exclusion limit 1
(ISE1) and ISE2 [6-8] and decreased size exclusion limit 1
(DSE1) [9] up- and down-regulate PD
permeability, respectively. Cell expansion and maturation of the cell
wall are also observed during the leaf sink–source transition.
Because PD are imbedded in the cell wall, the changes in the cell wall
during maturation affect the ability of the PD to increase or decrease
their aperture size. Pectin methylesterase (PME) is an enzymatic factor
involved in cell expansion and cell wall modification [10-12]. PME catalyzes the
demethylesterification of pectin homogalacturonans by converting
methoxyl groups into carboxyl groups, releasing both methanol and
protons [13]. PME-generated methanol is therefore
a byproduct of cell growth, and it is strongly correlated with cell and
leaf expansion. Until recently, methanol was assumed to be a metabolic
waste product of homogalacturonan demethylesterification. Leaf wounding
results in enhanced methanol emission, which increases the accumulation
of methanol-inducible gene (MIG) mRNAs and enhances antibacterial
resistance, as well as cell–cell communication that facilitates
viral spreading in neighboring plants [14].
Methanol probably functions as a signaling molecule that is important
for intra- and inter-plant communication. However, there is also
evidence for a potential role of methanol in the growth and development
of plants [15].
In this study, we investigated whether methanol has a role as a signaling molecule involved in the growth and development of plant leaves. We used tobacco (Nicotiana tabacum) leaves that, in contrast to other plants such as Nicotiana benthamiana, are well-studied with respect to the physiological and anatomical features of sink and source leaves [2-5]. We have identified novel methanol-inducible genes that are probably involved in regulation of the sink–source transformation of leaves.
MATERIALS AND METHODS
Plant growth conditions. Nicotiana tabacum cv. Samsun (nn) plants were grown in soil in a controlled environmental chamber with a 16 h/8 h day/night cycle and 25/18°C temperature. All plants used were 11-12-week-old and had five to six true leaves.
Methanol analysis. To analyze the methanol content in plant sap, 30 mg of leaf tissue was ground in the presence of Celite in microcentrifuge tubes and centrifuged at 16,000g for 10 min. An equal volume of 10% trichloroacetic acid was added to each supernatant aliquot. After 20 min incubation on ice, the samples were centrifuged at 16,000g for 15 min, and 2 µl of the supernatant was used for GC analysis as described previously [14].
Agroinjection experiments. Agrobacterium tumefaciens strain GV3101 was transformed with a binary vector encoding 2×GFP under control of cauliflower mosaic virus 35S promoter (CaMV 35S) and grown at 28°C in LB medium supplemented with 50 mg/liter rifampicin, 25 mg/liter gentamycin, and 50 mg/liter carbenicillin/kanamycin. Agrobacterium cells from an overnight culture (5 ml) were collected by centrifugation (10 min, 4500g), resuspended in 10 mM MES (pH 5.5) buffer supplemented with 10 mM MgSO4, and adjusted to a final OD600 of 0.01. For cell-to-cell movement assays, N. tabacum plant leaves were agroinjected with CaMV 35S 2×GFP and stored for 24 h in a growth chamber at 24°C.
GFP detection. Fluorescent cells containing 2×GFP were counted using an Axiovert 200M fluorescence microscope and the following set of filters: BP470/40 (excitation) and BP525/50 (emission) (Zeiss, Germany).
Suppression subtractive hybridization (SSH). Total RNA was isolated from 3-cm sink and 10-13-cm source tobacco leaves. Amplified double-stranded cDNA was prepared from sink and source leaf RNA with oligonucleotide and CDS primers using the SMART approach [16]. The subtraction was performed in two directions: sink leaf cDNA library was subtracted from source leaf cDNA, and vice versa (http://evrogen.ru/services/cDNA-subtraction/service-cdna-subtraction.shtml).
q-PCR analysis of transcript levels. Total RNA was extracted from plant tissues using TriReagent (MRC, USA) according to the manufacturer’s protocol. After DNase treatment (Fermentas, Lithuania), 2 μg of denatured total RNA was annealed with 0.1 μg of random hexamers and 0.1 μg of Oligo-dT, followed by incubation with 200 units of Superscript II reverse transcriptase (Invitrogen, USA) for 50 min at 43°C to generate cDNA. Real-time qPCR was carried out using an iCycler iQ Real Time PCR detection system (Bio-Rad, USA) as described previously [14]. The thermal profile for EVA Green real-time qPCR included an initial heat-denaturing step at 95°C for 3 min followed by 45 cycles with a denaturation step at 95°C for 15 s, an annealing step for 30 s, and an elongation step at 72°C for 30 s, coupled with measurement of fluorescence. The target gene mRNA levels were normalized to the corresponding reference genes (N. tabacum 18S RNA). The following primers were used: “BG-F1” GATTGTTGTGTCCGAGAGTG; “BG-R1” CCAGTTCAGGGTTCTTGTTG; “NCAPP-F1” CTATGTCTTCAAAGATTAGTCTG; “NCAPP-R1” GGACGGAAATGATTGTGGC; “PME-F1” ATCCTTGGATTCCGGCAAGAACGT; “PME-R1” AAACACTTGCAATTGTAGAGTAAC; “PI-II-F1” ATGTACCTCGCCAAACTTAC; “PI-II-R1” CTCCATCTATTTATAGCAAACG; “MIG-21-F1” AGGAAGGCAGTTTCATGCATAA; “MIG-21-R1” GCCTCTTATTTGTTTGAGTTATGCCTC; “ISE2-F1” ACAGTGGCAAAAGGAAGG; “ISE2-R1” TAAGAGGAGTAGTATAGAATAGTC; “ERP-F1” TGATGGTGGCTATGGTAGTG; “ERP-R1” CTGGCTCTGGCTGTAGTG; “SEO-F1” CTGGAAGCATTGGAAGAGAAGG; “SEO-R1” GGTTGTGAGCGAGTGAAGC; “SABC-F1” TGTCCCTGGTCTTTACTATTCG; “SABC-R1” GCGGTGATTATTGTGATGAGC; “ARP-F1” GGATGATGTTATGGCTGGAC; “ARP-R1” GTGACGGAGTTGTTGGTG; “ST-F1” TCAAGACCAACACTACACTCC; and “ST-R1” CCACCCACCACTGATAATAGG.
Whole plant methanol treatment. Plants were treated with methanol by exposing them to methanol vapors in a single hermetically-sealed 20-liter desiccator. Pots (width, 9.5 cm; depth, 9.5 cm) containing plants (10.0 ± 1.0 g) and soil (198.0 ± 20.0 g) were placed into desiccators with methanol (160 mg) applied to filter paper as described previously [14], and then maintained for 3 h at constant temperature of 24°C. The final concentration of gaseous methanol was 17.5 µg per gram leaf fresh weight. The treated plants were withdrawn from the desiccator, and leaf RNA was isolated as described above.
RESULTS
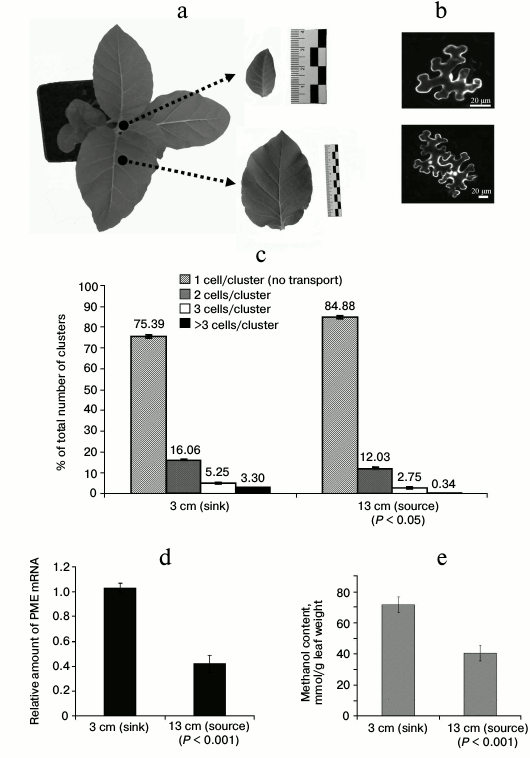
Measurements of PME mRNA and methanol accumulation in tobacco sink and source leaves. We studied the role of PME in the sink–source transition using young tobacco (N. tabacum cv. Samsun nn) plants with 5-6 true leaves (Fig. 1a). For young immature expanding sink leaves, we used small leaves that were 3 cm in length (Fig. 1a, top inset). We chose leaves that were 10-13 cm in length (Fig. 1a, bottom inset) located in the lower tiers of the tobacco plant as mature source leaves. An important characteristic of sink leaves is that dilated PD are open to the movement of protein molecules as large as 47 kDa [3]. To evaluate cell-to-cell communication in sink and source leaves, a reporter macromolecule was used to test movement through PD in different states of dilation. We chose a reporter containing two fused copies of green fluorescent protein (2×GFP). We exploited a previously described “agroinjection strategy” to deliver the CaMV 35S 2×GFP containing plasmid into the cell [14]. To monitor single infection sites, the tobacco plants were agroinjected with a diluted (OD600 ~ 0.01) bacterial suspension, and fluorescent cells were counted after 24 h of incubation in a growth chamber at 24°C. Counting the number of epidermal cells that displayed fluorescence provides a quantitative measure of 2×GFP movement. When the PD were closed, 2×GFP was detected mainly in single cells (Fig. 1b, upper). However, fluorescence signals were distributed in 2- or 4-cell clusters (Fig. 1b, bottom) when the PD were dilated. In the source leaves approximately 15% of the signal was distributed in 2- to 4-cell clusters, whereas in sink leaves approximately 25% of the signal was distributed in 2- to 4-cell clusters (Fig. 1c), thus indicating that the ability to support cell-to-cell movement of 2×GFP was enhanced in sink leaves. An unpaired two-tailed Student’s t-test confirmed the statistical significance of the differences in the cell-to-cell movement of 2×GFP between the source and sink leaves.
Fig. 1. PME mRNA levels and methanol accumulation are increased in tobacco sink leaves. a) A standard Nicotiana tabacum cv. Samsun (nn) plant and the detached sink (upper) and source (bottom) leaves used for analysis. b) Single (upper) and multiple (bottom) epidermal cells containing 2×GFP were observed in sink and source leaves using epifluorescence microscopy. c) Quantification of 2×GFP movement in sink and source leaves. No fewer than 1000 cell clusters were counted for each experiment. The data shown represent five independent experiments. Standard error bars are indicated. The P-value from an unpaired two-tailed Student’s t-test is indicated. d) The relative levels of PME mRNA in sink and source leaves were determined by qRT-PCR. The data represent five independent experiments. Standard error bars and the P-value (unpaired two-tailed Student’s t-test) are indicated. e) Methanol content in the sap of sink and source leaves. Error bars indicate the standard error of the data from six independent samples. The P-value from an unpaired two-tailed Student’s t-test is indicated.
To study leaf PME synthesis, we analyzed PME mRNA content in immature and mature tobacco leaves by quantitative real-time PCR. We found that immature leaves have more than twice greater level of PME mRNA compared to mature tobacco leaves (Fig. 1d). We next expected to observe higher methanol levels in the sap of immature leaves as a result of PME activity. We observed an almost two-fold decrease in methanol in the leaf sap of mature leaves compared to immature leaves (Fig. 1e).
The mRNA accumulation levels in mature and immature leaves were then analyzed for the following genes previously identified as being sensitive to gaseous methanol and involved in plant immunity: β-1,3-glucanase (BG), methanol inducible gene 21 (MIG-21), non-cell-autonomous pathway protein (NCAPP), and proteinase inhibitor II (PI-II) [14]. Although NCAPP mRNA levels were approximately equal in mature and immature leaves, the expression of BG, MIG-21, and PI-II increased significantly during leaf maturation (Table 1). In accordance with a previous study [8], we observed a decrease in ISE2 mRNA expression in mature leaves.
Table 1. Relative quantity of MIGs
and ISE2 mRNAs in N. tabacum leaves
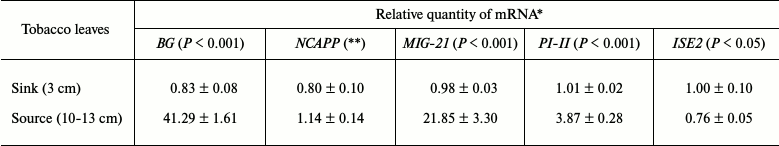
* The data shown represent five independent experiments. The standard
errors and the unpaired two-tailed Student’s t-test
P-values for the statistical significance of the difference
between the sink and source leaves are indicated.
** n.s., not significantly different (Student’s t-test).
Thus, we conclude that the sink–source transition is accompanied by two events. First, there is a change in the transcriptional activity of stress genes, as shown by a decrease in the level of PME mRNA accumulation and an increase in MIG (BG, MIG-21, and PI-II) mRNA levels in source leaf. The second event observed is a reduction in the concentration of methanol in source leaf sap, which correlates with a decrease in the transcriptional activity of PME.
Identification of genes involved in growth and development of tobacco leaves and their sensitivity to methanol. To better understand the mechanisms of gene regulation involved in the sink–source transition, we created a cDNA library from sink and source tobacco leaves and analyzed them using SSH. We identified a total of 92 differentially expressed transcripts, 26 of which were enriched mainly in source leaves and 66 of which were primarily expressed in sink leaves (Table 2).
We selected a few genes to further analyze based on the following properties: first, we selected genes that were represented with a large number of clones in the SSH analysis and second, we selected genes with possible functions in leaf growth and development and/or in the sink–source transformation. We selected five genes: sugar transporter (ST) [17, 18], auxin-repressed protein (ARP) [19], elicitor responsible protein (ERP) [20], salicylic acid binding catalase (SABC) [21, 22], and sieve element occlusion protein (SEOP) [18, 22, 23], the putative functions of which are described in Table 2.
Table 2. List of Nicotiana tabacum
expressed sequence tags (ESTs) identified by SSH
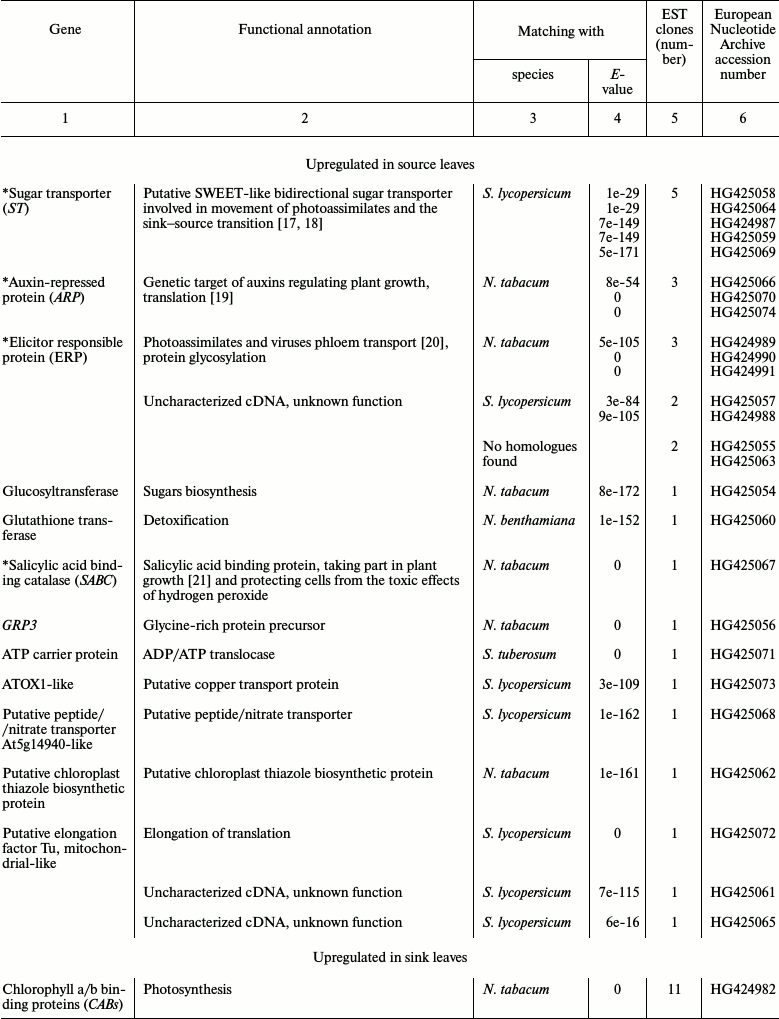
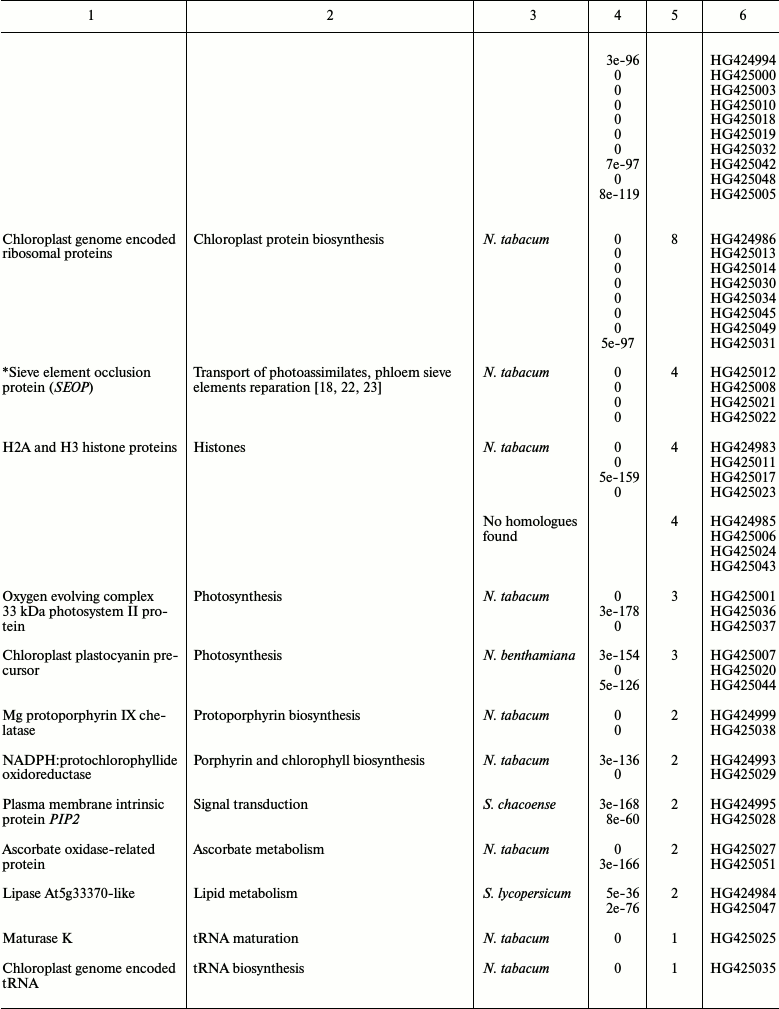
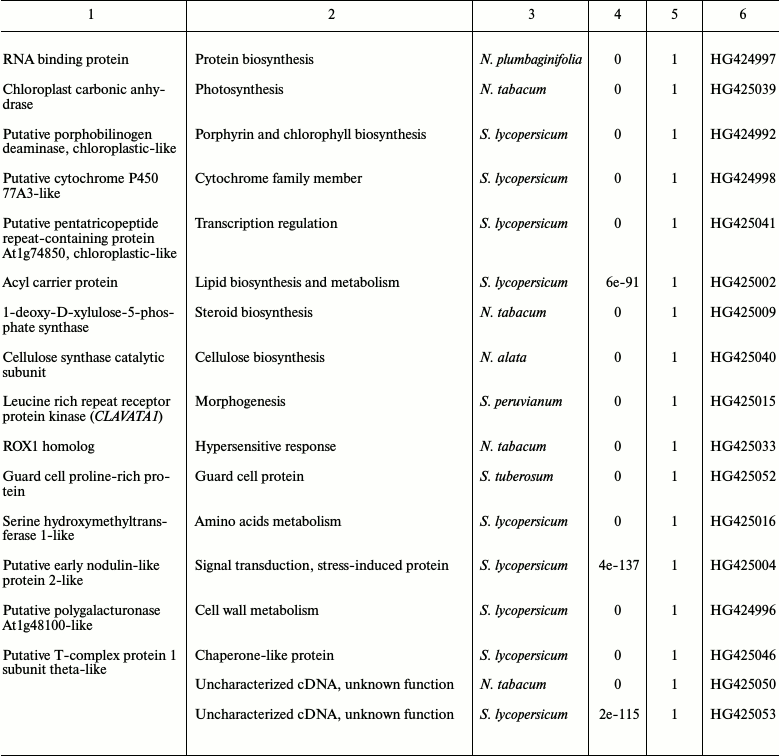
* Genes selected for further analysis.
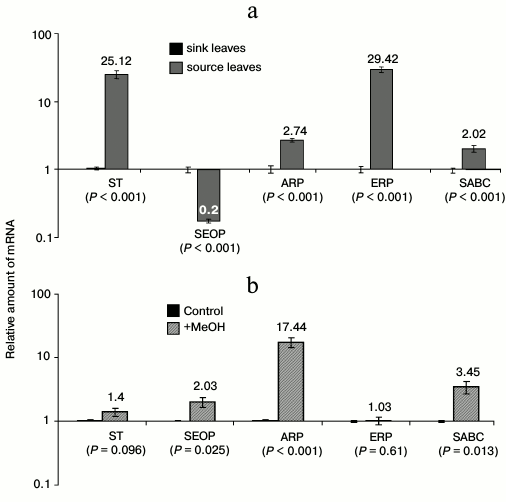
We next validated the changes in gene expression observed by SSH by performing qRT-PCR to determine the mRNA levels in sink and source leaves. We found that leaf maturation was accompanied by an accumulation of ST, ARP, ERP, and SABC mRNA and a decrease in SEOP mRNA levels (Fig. 2a).
Fig. 2. Analysis of mRNA selected by SSH. a) Verification of genes identified by SSH in the cDNA libraries from tobacco leaves. The semi-log plot shows the relative levels of ST, SEOP, ARP, ERP, and SABC mRNA isolated from sink and source leaves. b) Relative levels of ST, SEOP, ARP, ERP, and SABC mRNA in N. tabacum leaves after methanol treatment. The data shown represent five independent experiments. The standard error bars and the P-value (unpaired two-tailed Student’s t-test) are indicated.
The ability of the selected genes to alter their activity under the influence of methanol was tested. We observed an abrupt and significant accumulation of SEOP, ARP, and SABC mRNA when leaves were exposed to gaseous methanol (Fig. 2b).
We hypothesize that the methanol generated by PME can function as a transcriptional regulator of gene activity during the sink–source transition.
DISCUSSION
Until recently, methanol was considered to be a metabolic product of PME activity during growth and cell wall modification [10, 11]. Recent data suggest a role for gaseous methanol in intra- and inter-plant communication [14]. Here we demonstrated that the methanol content in the sap of immature leaves is elevated compared to levels in mature leaves (Fig. 1e). We believe that the methanol detected in each leaf was synthesized by the cells of the individual leaf and not introduced by the phloem sap from other parts of the plant for several reasons. First, the rate of methanol emission (i.e. release of methanol into the air) is correlated with the age of the leaf. Isolated immature leaves emit substantially greater amounts of gaseous methanol than mature leaves [15]. Second, underground methanol production is not essential to the level and emission of methanol in mature and immature leaves [24]. Finally, according to our data, the methanol concentration is correlated with cellular PME mRNA levels in mature and immature leaves (Fig. 1, d and e).
Synthesis of PME is one of the factors in plant growth, leading to pectin demethylation, which accompanies the natural division and maturation of the plant cell. In general, homogalacturonan demethylesterification catalyzed by PME is likely to be a module of plant cell growth control, providing constant remodeling and modification after deposition of the homogalacturonans [25]. Interestingly, overexpression of PME in transgenic plants leads to increased levels of aberrant synthesis of methanol [14], which is accompanied by dwarfism of tobacco [26].
The conversion of homogalacturonan methoxyl groups into carboxyl groups results in methanol release. Methanol is not toxic to plant cells, but rather performs an alarm function and is able to control the activity of genes involved in intercellular transport [14]. Immature leaves are characterized by intense intercellular transport of macromolecules. The PD play an important role in this process, and in immature leaves they are regulated in turn by metabolic signals, among which methanol may be one of the key players. To experimentally confirm this proposed relationship, we have identified mRNAs that differ significantly between mature and immature leaves (Table 2). Two genes, ST [17, 18] and SEOP [18, 22, 23], are directly involved with the operation of phloem transport of carbohydrates in plants, and ARP [19], ERP [20] and SABC [21] are involved in plant morphogenesis and cell signaling. Among the genes identified, at least three (SEOP, ARP, and SABC) are sensitive to methanol (Fig. 2b).
The mechanism of methanol effect on genes involved in sink-source transition remains unclear, but the transcriptional activity of these genes is apparently affected by the specific microenvironment and the concentration of methanol generated during the cell wall modification process.
This work was supported by grants (Nos. 11-04-01152 and 12-04-33016) from the Russian Foundation for Basic Research (to T.V.K.) and stipend of the President of the Russian Federation for young scientists (to T.V.K.).
REFERENCES
1.Ayre, B. G. (2011) Mol. Plant.,
4, 377-394.
2.Burch-Smith, T. M., and Zambryski, P. C. (2012)
Annu. Rev. Plant. Biol., 63, 239-260.
3.Oparka, K. J., Roberts, A. G., Boevink, P., Santa
Cruz, S., Roberts, I., Pradel, K. S., Imlau, A., Kotlizky, G., Sauer,
N., and Epel, B. (1999) Cell, 97, 743-754.
4.Crawford, K., and Zambryski, P. (2000) Curr.
Biol., 10, 1032-1040.
5.Crawford, K., and Zambryski, P. C. (2001) Plant
Physiol., 125, 1802-1812.
6.Burch-Smith, T. M., Brunkard, J. O., Choi, Y. G.,
and Zambryski, P. C. (2011) Proc. Natl. Acad. Sci. USA,
108, E1451-1460.
7.Burch-Smith, T. M., Stonebloom, S., Xu, M., and
Zambryski, P. C. (2011) Protoplasma, 248, 61-74.
8.Burch-Smith, T. M., and Zambryski, P. C. (2010)
Curr. Biol., 20, 989-993.
9.Xu, M., Cho, E., Burch-Smith, T. M., and Zambryski,
P. C. (2012) Proc. Natl. Acad. Sci. USA, 109,
5098-5103.
10.Micheli, F. (2001) Trends Plant. Sci.,
6, 414-419.
11.Pelloux, J., Rusterucci, C., and Mellerowicz, E.
J. (2007) Trends Plant. Sci., 12, 267-277.
12.Markovic, O., and Janecek, S. (2004)
Carbohydrate Res., 339, 2281-2295.
13.Nemecek-Marshall, M., MacDonald, R. C., Franzen,
J. J., Wojciechowski, C. L., and Fall, R. (1995) Plant Physiol.,
108, 1359-1368.
14.Dorokhov, Y. L., Komarova, T. V., Petrunia, I.
V., Frolova, O. Y., Pozdyshev, D. V., and Gleba, Y. Y. (2012) PLoS
Pathog., 8, e1002640.
15.Huve, K., Christ, M. M., Kleist, E., Uerlings,
R., Niinemets, U., Walter, A., and Wildt, J. (2007) J. Exp.
Bot., 58, 1783-1793.
16.Diachenko, L., Lau, Y. F., Campbell, A. P.,
Chenchik, A., Mogadam, F., Huang, B., Lukyanov, S., Lukyanov, K.,
Gurskaya, N., Sverdlov, E. D., and Siebert, D. (1996) Proc. Natl.
Acad. Sci. USA, 93, 6025-6030.
17.Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D.,
Osorio, S., Fernie, A. R., and Frommer, W. B. (2012) Science,
335, 207-211.
18.Bihmidine, S., Hunter, C. T., 3rd, Johns, C. E.,
Koch, K. E., and Braun, D. M. (2013) Front. Plant Sci.,
4, 177.
19.Li, Y., Wu, M. Y., Song, H. H., Hu, X., and Qiu,
B. S. (2005) Arch. Virol., 150, 1993-2008.
20.Chen, Z., Silva, H., and Klessig, D. F. (1993)
Science, 262, 1883-1886.
21.Sanchez-Casas, P., and Klessig, D. F. (1994)
Plant Physiol., 106, 1675-1679.
22.Ernst, A. M. (2012) Proc. Natl. Acad.
Sci. USA, 109, E1980-1989.
23.Lee, J., Han, C. T., and Hur, Y. (2013) Mol.
Biol. Rep., 40, 197-209.
24.Oikawa, P. Y., Giebel, B. M., Sternberg, L., Li,
L., Timko, M. P., Swart, P. K., Riemer, D. D., Mak, J. E., and Lerdau,
M. T. (2011) New Phytol., 191, 1031-1040.
25.Wolf, S., and Greiner, S. (2012)
Protoplasma, 249, Suppl. 2, S169-175.
26.Hasunuma, T., Fukusaki, E., and Kobayashi, A.
(2004) J. Biotechnol., 111, 241-251.