Polyphosphates as an Energy Source for Growth of Saccharomyces cerevisiae
L. V. Trilisenko and T. V. Kulakovskaya*
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, pr. Nauki 5, 142290 Pushchino, Moscow Region, Russia; fax: (495) 956-3370; E-mail: alla@ibpm.pushchino.ru* To whom correspondence should be addressed.
Received January 24, 2014; Revision received February 4, 2014
Cells of the yeast Saccharomyces cerevisiae with a low content of polyphosphates (polyP) are characterized by disturbance of growth in medium with 0.5% glucose. The parent strain with polyP level reduced by phosphate starvation had a longer lag phase. The growth rate of strains with genetically determined low content of polyP due to their enhanced hydrolysis (CRN/pMB1_PPN1 Sc is a superproducer of exopolyphosphatase PPN1) or reduced synthesis (the BY4741 vma2Δ mutant with impaired vacuolar membrane energization) was lower in the exponential phase. The growth of cells with high content of polyP was accompanied by polyP consumption. In cells of strains with low content of polyP, CRN/pMB1_PPN1 Sc and BY4741 vma2Δ, their consumption was insignificant. These findings provide more evidence indicating the use of polyP as an extra energy source for maintaining high growth rate.
KEY WORDS: polyphosphates, growth rate, mutant, exopolyphosphatase, Saccharomyces cerevisiaeDOI: 10.1134/S0006297914050125
Inorganic polyphosphates (polyP) are linear polymers consisting of orthophosphoric acid residues linked by macroergic phosphoanhydride bonds. Along with the function of phosphorus and energy accumulation, polyP are involved in formation of transport channels in membranes, regulation of the pool of metal cations, overcoming stresses, providing for the survival of microorganisms under unfavorable conditions, regulation of enzyme activity, gene expression [1-3], and cell wall structure and function [4].
The energy of the phosphoanhydride bond in polyP is approximately the same as in the ATP molecule; therefore, it is believed that energy reserve is one of the most important functions of these polymers [2]. However, there is little evidence to support the hypothesis of polyP being used as an energy source in microorganisms.
It has been shown that the content of polyP in the bacteria Corynebacterium xerosis [5] and Escherichia coli [6] decreases in the period of active growth and increases in the stationary phase. This effect is observed under conditions of phosphate excess in the medium. In cells of Saccharomyces cerevisiae, the degree of polymerization of high molecular mass polyP fractions decreases during active growth [7]. It has been suggested that the energy released as a result of hydrolysis of phosphoanhydride bonds in polyP is used to maintain the high rate of yeast growth in the exponential phase [7].
One approach to solving this problem is comparative study of the growth of cells with low and high content of polyP. Cells with low polyP content are obtained as a result of phosphate starvation [8]. At present, there are yeast strains with genetically determined low content of polyP. The strain with mutation in the vma2 gene encoding one of the subunits of vacuolar H+-ATPase [9] has a nearly 5-fold lower content of polyP [10, 11]. However, this strain demonstrates no changes in polyphosphatase activity [10]. The mutation in the vma2 gene disturbs proton translocation activity of the vacuolar membrane ATPase (V-ATPase) [9]. We suppose that polyP synthesis by the polyphosphate synthase Vtc4p localized in the yeast vacuolar membrane [12] needs this membrane to be energized [11], and the lower level of polyP in strain BY4741 vma2Δ is accounted for by reduction in their synthesis. The recently obtained transformant strain CRN/pMB1_PPN1 Sc contains a plasmid with the gene of S. cerevisiae exopolyphosphatase PPN1. The level of polyP in the transformant decreases due to their enhanced cleavage by the PPN1 enzyme [13].
The goal of this work was to study the effect of alterations in polyP content on growth characteristics of the yeast S. cerevisiae in the active growth stage.
MATERIALS AND METHODS
The strains of the yeast S. cerevisiae presented in Table 1 were the objects of research. The strains CRY and CRN were kindly provided by researchers of A. Kornberg’s laboratory (USA) [14] and the strains BY4741 and BY4741 vma2Δ were kindly provided by Dr. P. Kane (SUNY Upstate Medical University, USA) [9]. The transformed S. cerevisiae strain CRN/pMB1_PPN1 Sc containing the PPN1 expression vector was obtained at the Center for Bioengineering of the Russian Academy of Sciences (Moscow) [13]. The expression vector also contained the URA3 gene.
Table 1. Strains used in the work
The CRY, CRN, BY4741, and BY4741 vma2Δ yeast cultures were maintained on agarized YPD medium; the strain CRN/pMB1_PPN1 Sc was maintained on agarized minimal synthetic medium without uracil [13]. The agarized YPD medium contained 1% yeast extract, 2% peptone, 2% glucose, and 2% agar. The selective liquid medium contained (per liter) 20 g of glucose and 6.7 g of dry YNB medium (Bacto-yeast nitrogen base; Difco, USA). The medium was supplemented with L-tryptophan, L-histidine, L-methionine (20 mg each per liter) and L-leucine (60 mg per liter). CRY and CRN were grown in medium containing also adenine and uracil (20 mg each per liter); the medium for CRN/pMB1_PPN1 Sc contained only 20 mg adenine per liter. For cultivation of strains BY4741 and BY4741 vma2Δ, 20 mg uracil was added per liter of the medium, and the pH was adjusted to 5.0 with 50 mM succinate-NaOH buffer [9].
The cultures were grown at 28-29ºC in 1-liter flasks with 200 ml of the minimal synthetic medium on shakers (145 rpm). The biomass of strain CRY was obtained by adding 5 ml of the inoculum into the flasks with phosphate-containing (7.1 mM) YNB medium and phosphate-deficient (0.08 mM) YNB medium with 2% glucose. The culture was grown for 24 h; then both variants were re-inoculated into flasks with the minimal synthetic medium containing phosphate and 0.5% glucose. The strains CRN, CRN/pMB1_PPN1 Sc, BY4741, and BY4741 vma2Δ were first grown in the minimal synthetic medium with phosphate and 2% glucose. After 24-h cultivation, they were re-inoculated into the same medium but with 0.5% glucose. The growth was analyzed by the culture optical density at 530 nm in a 3.07-mm cuvette. The biomass was harvested by centrifugation at 3000g for 10 min, twice washed with cold distilled water, and used for assaying the content of inorganic polyP in the cells.
PolyP was extracted from the cells as two fractions: cold and hot perchlorate extracts [13]. The cold perchlorate extract was the acid-soluble polyP fraction (polyPI). The hot perchlorate extract polyPII included polyP2, polyP3, polyP4, and polyP5 fractions [7]. Aliquots of the fractions were hydrolyzed in the presence of 1 N HCl in a boiling water bath for 10 min. The amount of orthophosphate formed was assayed according to Heinonen and Lahti [15]. The presented data are the mean values of three independent experiments.
RESULTS AND DISCUSSION
We compared the growth characteristics and changes in the content of polyP during active growth of S. cerevisiae cells with high and low content of these polymers. The effect of polyP on cell growth was assessed under carbon deficiency: the medium contained 0.5% glucose, while the initial culture density was increased to A530 ~ 0.25. The cells of the parent strain CRY with low content of polyP were obtained by phosphate starvation. The mutant strains with reduced polyP synthesis (strain BY4741 vma2Δ) [10, 11] or enhanced polyP hydrolysis (overproducer of exopolyphosphatase PPN1) were also used [13].
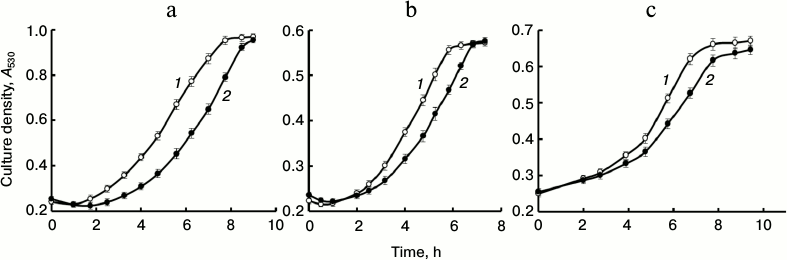
Figure 1 shows culture growth on a glucose-deficient (0.5%) and Pi-excessive (7 mM) medium. Pi-starved cells of the parent strain CRY, as well as the mutant strains, were characterized by growth inhibition.
Fig. 1. Growth curves for S. cerevisiae strains in medium with 0.5% glucose and 7 mM Pi. a) Parent strain CRY: 1) cells pre-grown in medium with 7 mM Pi; 2) cells pre-grown in medium with 0.08 mM Pi. b: 1) strain CRN; 2) transformant CRN/pMB1_PPN1 Sc. c: 1) parent strain BY4741; 2) mutant BY4741 vma2Δ.
The values of lag phase duration, specific growth rate (µ), and generation time (td) calculated by the known formulas [16], as well as total polyP content in the cells used for inoculation in the 0.5% glucose-containing medium, are given in Table 2. Indeed, the cells with reduced polyP content demonstrated growth inhibition irrespective of how this reduction was achieved. However, such inhibition was different in the cases of physiologically lower initial content of polyP in strain CRY and reduced content of polyP as a result of mutations. The CRY cells with low polyP content were characterized by delayed growth (Fig. 1a, curve 2) compared to the cells of the same strain with high polyP content (curve 1). However, growth rate and generation time in the exponential phase were the same for both variants of the CRY cells (Table 2).
Table 2. Growth parameters of S.
cerevisiae strains with different initial content of polyP
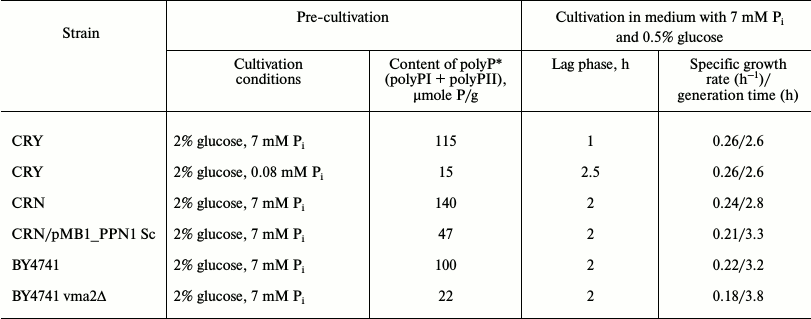
*Content of polyP was assayed in cells that reached stationary
growth stage and were used for re-inoculation in medium with 7 mM
Pi and 0.5% glucose.
The cells of strains CRN/pMB1_PPN1 Sc and BY4741 vma2Δ with genetically determined low level of polyP showed no difference in lag phase duration but demonstrated a decrease in growth rate in the exponential phase and an increase in generation time (Fig. 1 (b and c) and Table 2).
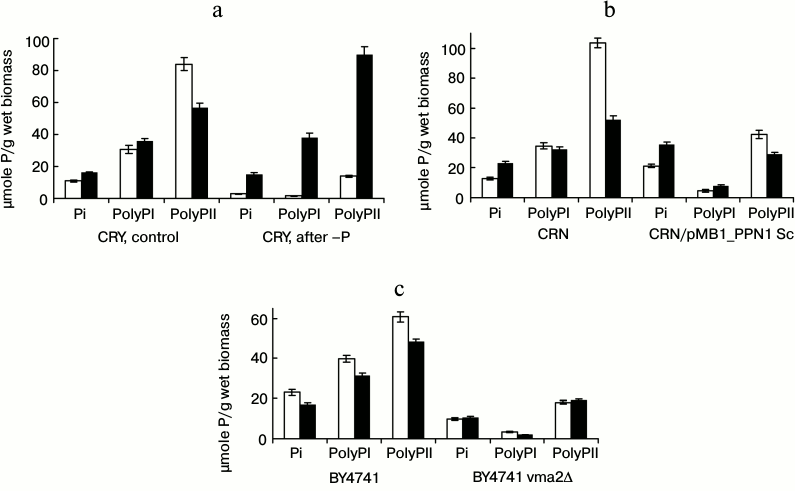
We also compared the content of Pi and polyP in the initial cells and the cells grown to the stationary phase in medium with 0.5% glucose (Fig. 2). All variants of the cells with high initial total content of polyP demonstrated its decrease by the stationary stage. This decrease was observed mostly in the long-chain polyPII. In these experiments, cultivation was performed in the Pi-excessive medium; hence, we suppose that polyP is used not as a phosphate source but as an energy source. The cells of the parent strain CRY, where the content of polyP is much lower as a result of preliminary cultivation under Pi starvation, did not consume, but, on the contrary, accumulated polyP, first of all, the long-chain polyPII (Fig. 2a). The effect of rapid over accumulation of polyP after the transfer of Pi-starved S. cerevisiae cells to a phosphate-containing medium is well known [8]. The system of polyP over synthesis is induced already during phosphate starvation [17]. Hence, intensive energy-consuming accumulation of polyP occurs in the first hours after re-inoculation of starved cells to Pi-containing medium, which may be the cause of the extension of the lag phase compared to the control cells (Table 2).
Fig. 2. Content of Pi, polyPI, and polyPII in cells of S. cerevisiae strains: in the initial cells grown till the stationary phase in medium with 2% glucose (white columns); in stationary cells during growth in medium with 0.5% glucose and 7 mM Pi (black columns). a) Strain CRY: pre-cultivation in medium with 7 mM Pi (CRY, control); pre-cultivation in medium with 0.08 mM Pi (CRY, after –P). b) Strain CRN and transformant CRN/pMB1_PPN1 Sc. c) Strain BY4741 and mutant BY4741 vma2Δ; b, c) pre-cultivation till stationary phase in medium with 2% glucose and 7 mM Pi.
The strain CRN and the transformant CRN/pMB1_PPN1 Sc were pre-grown in medium with Pi excess. The low content of polyP in the transformant is associated with the high level of exopolyphosphatase PPN1, which intensively hydrolyzes these polymers [13]. At the same time, the content of Pi in the cells increases (Fig. 2b). The transformant seems to preserve only the polyP inaccessible for the recombinant exopolyphosphatase PPN1. However, their level seems to be insufficient to maintain the same growth rate as in the parent strain CRN. The total content of polyP decreased in the stationary phase by 54 µmoles P/g wet biomass in strain CRN but only by 11 µmoles P/g wet biomass in the transformant CRN/pMB1_PPN1 Sc.
The strain BY4741 and the mutant BY4741 vma2Δ were also pre-grown in the medium with Pi excess. The content of polyP in the mutant BY4741 vma2Δ was even lower than in the transformant CRN/pMB1_PPN1 Sc (21 and 47 µmoles/g wet biomass, respectively). This is associated with the reduced ability of BY4741 vma2Δ cells to synthesize polyP [10, 11]. PolyP was not consumed by this mutant during the growth (Fig. 2c), and its growth rate was lower (Table 2).
Thus, the genetically determined low content of polyP in the cells reduced the growth rate of the yeast irrespective of what it had been caused by: disturbed synthesis of these compounds (mutant BY4741 vma2Δ) or their enhanced hydrolysis (transformant CRN/pMB1_PPN1 Sc). The physiologically determined decrease in the content of polyP (strain CRY after preliminary phosphate starvation) resulted in lag phase elongation but had no effect on growth rate. These results suggest that polyPs are additional energy sources for maintaining high growth rate in the period of active budding.
This work was supported by the Russian Foundation for Basic Research (project No. 14-04-00515).
REFERENCES
1.Kornberg, A., Rao, N. N., and Ault-Riche, D. (1999)
Annu. Rev. Biochem., 68, 89-125.
2.Kulaev, I. S., Vagabov, V. M., and Kulakovskaya, T.
V. (2004) The Biochemistry of Inorganic Polyphosphate,
Wiley.
3.Rao, N. N., Gomez-Garcia, M. R., and Kornberg, A.
(2009) Ann. Rev. Biochem., 78, 605-647.
4.Kalebina, T. S., Egorov, S. N., Arbatsky, N. P.,
Bezsonov, E. E., Gorkovsky, A. A., and Kulaev, I. S. (2008) Dokl.
Akad. Nauk, 420, 695-698.
5.Hughes, D. E., and Muhammed, A. (1962) in Proc.
Acids Ribonucleiques et Polyphosphates. Structure, Synthese et
Fonction, CNRS International Colloquion, Paris, pp. 591-602.
6.Nesmeyanova, M. A. (2000) Biochemistry
(Moscow), 65, 309-314.
7.Vagabov, V. M., Trilisenko, L. V., Shchipanova, I.
N., Sibel’dina, L. A., and Kulaev, I. S. (1998)
Mikrobiologiya, 67, 193-198.
8.Vagabov, V. M., Trilisenko, L. V., and Kulaev, I.
S. (2000) Biochemistry (Moscow), 65, 349-354.
9.Milgrom, E., Diab, H., Middleton, F., and Kane, P.
M. (2007) J. Biol. Chem., 282, 7125-7136.
10.Tomashevsky, A. A., Ryazanova, L. P.,
Kulakovskaya, T. V., and Kulaev, I. S. (2010) Biochemistry
(Moscow), 75, 1052-1054.
11.Trilisenko, L., Tomashevsky, A., Kulakovskaya,
T., and Kulaev, I. (2013) Folia Microbiol., 58,
437-441.
12.Hothorn, M., Neumann, H., Lenherr, E. D., Wehner,
M., Rybin, V., Hassa, P. O., Uttenweiler, A., Reinhardt, M., Schmidt,
A., Seiler, J., Ladumer, A. G., Hermann, C., Scheffzek, K., and Mayer,
A. (2009) Science, 324, 513-516.
13.El’darov, M. A., Baranov, M. V., Dumina, M.
V., Zhgun, A. A., Andreeva, N. A., Trilisenko, L. V., Kulakovskaya, T.
V., Ryazanova, L. P., and Kulaev, I. S. (2013) Biochemistry
(Moscow), 78, 946-953.
14.Sethuraman, A., Rao, N. N., and Kornberg, A.
(2001) Proc. Natl. Acad. Sci. USA, 98, 8542-8547.
15.Heinonen, Y. K., and Lahti, R. Y. (1981) Anal.
Biochem., 113, 313-317.
16.Hall, B. G., Acar, H., Nandipati, A., and Barlow,
M. (2013) Mol. Biol. Evol., 31, 232-238.
17.Trilisenko, L. V., Andreeva, N. A., Kulakovskaya,
T. V., Vagabov, V. M., and Kulaev, I. S. (2003) Biochemistry
(Moscow), 68, 577-581.