Characterization of Rabaptin-5 γ Isoform
E. V. Korobko1, S. L. Kiselev1,2, and I. V. Korobko1*
1Institute of Gene Biology, Russian Academy of Sciences, ul. Vavilova 34/5, 119334 Moscow, Russia; fax: (499) 135-4105; E-mail: igorvk@igb.ac.ru; korobko1305@gmail.com2Present address: Vavilov Institute of General Genetics, ul. Gubkina 3, 119911 Moscow, Russia
* To whom correspondence should be addressed.
Received March 3, 2014; Revision received May 29, 2014
Rab GTPases are key regulators of intracellular membrane traffic acting through their effector molecules. Rabaptin-5 is a Rab5 effector in early endosome fusion and connects Rab5- and Rab4-positive membrane compartments owing to its ability to interact with Rab4 GTPase. Recent studies showed that Rabaptin-5 transcript is subjected to extensive alternative splicing, thus resulting in expression of Rabaptin-5 isoforms mostly bearing short deletions in the polypeptide chain. As interactions of a Rab GTPase with different effectors lead to different responses, functional characterization of Rabaptin-5 isoforms becomes an attractive issue. Indeed, it was shown that Rab GTPase effector properties of Rabaptin-5 and its α and δ isoforms are different. This work focused on another Rabaptin-5 isoform, Rabaptin-5γ. Despite its ability to interact with Rab5, endogenously produced Rabaptin-5γ was absent from early endosomes. Rather, it was found to be tightly associated with trans-Golgi network and partially localized to a Rab4-positive membrane compartment. The revealed intracellular localization of Rabaptin-5γ indicates that it is not involved in Rab5-driven events; rather, it functions in other membrane transport steps. Our study signifies the role of alternative splicing in determination of functional activities of Rab effector molecules.
KEY WORDS: intracellular membrane transport, early endosomes, trans-Golgi network, Rab5 small GTPase, Rab4 small GTPase, small GTPase effectors, Rabaptin-5γDOI: 10.1134/S000629791409003X
Abbreviations: 3AT, 3-amino-(1,2,4)-triazole; EGF, epidermal growth factor; EGFP, enhanced green fluorescent protein; GAL4AD, transcriptional activating domain of GAL4 transcription factor; GAL4BD, GAL4 transcription factor DNA binding domain; GST, glutathione-S-transferase; mAb, monoclonal antibody; PBS, phosphate buffered saline; PFA, paraformaldehyde; PNS, post-nuclear supernatant; TGN, trans-Golgi network.
The current concept of intracellular membrane transport implies Rab
GTPases as key regulators of specific transport steps. Rab GTPases act
as molecular switches cycling between “active” GTP-bound
and “inactive” GDP-bound forms, and their regulatory
functions in membrane transport events rely on specific interactions of
GTPase in “active” state with corresponding effector
molecules. Notably, activation of a single GTPase can initiate various
molecular events and pleiotropy of processes that relies on the ability
of GTPase to interact with different effector molecules. Rab5 is a key
regulator of early endocytic events, exerting its actions through
interaction with various effector molecules [1, 2].
Rabaptin-5 is a well-studied Rab5 effector being an essential component in early endosome fusion [3]. The role of Rabaptin-5 in early endosome fusion relies on its complexing with Rabex-5, a guanine nucleotide exchange factor for Rab5. Recruitment of Rabaptin-5/Rabex-5 by GTP-bound Rab5 to early endosomes is believed to result in local Rab5-GTP concentration increase, thus promoting Rab5-driven fusion [4, 5]. Yet another role of Rabaptin-5 in early endosome fusion has been proposed. Owing to a second Rab5 binding site, Rabaptin-5 has been suggested to act as a molecule tethering two Rab5-GTP-positive endosomes prior to their fusion [6].
The role of Rabaptin-5 as a Rab5 effector molecule in early endosome fusion is not the only function of Rabaptin-5. In addition to Rab5, Rabaptin-5 can interact with another GTPase, Rab4 [7], and contribute to Rab4-mediated regulation of rapid endosomal recycling [8]. Sequential interaction of Rabaptin-5 with Rab5 and Rab4 was suggested to be important in linking two sequential membrane domains in endosomal transport and maintaining its continuity [7, 9].
Rabaptin-5 functions turned out to be not restricted to early endosomal traffic. It was revealed that Rabaptin-5 can interact with clathrin adaptors, GGA proteins and the γ1-adaptin subunit of AP-1 clathrin adaptor complex, that are involved in transport from trans-Golgi network (TGN) to the endosomal compartment [10-12]. The interaction of Rabaptin-5 with GGA clathrin adaptors interferes with their ability to recruit clathrin, thus affecting their functions [10]. In addition, interaction of Rabaptin-5 with several other molecules, such as protein kinase MAK-V [13], Rab33b GTPase [14], Rabphilin-3 [15, 16], neuronal growth-associated protein GAP-43 [17], and tuberin [18] has been reported, although these interactions were not extensively validated and their functional consequences are mostly unknown.
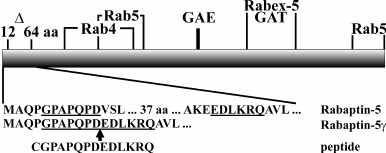
Rabaptin-5 is encoded by the RAB5EP gene, whose pre-mRNA is subjected to extensive alternative splicing. Besides Rabaptin-5, four different proteins encoded by the RAB5EP gene have been described so far – Rabaptin-5α, Rabaptin-5γ, Rabaptin-5δ (also called Rabaptin-5 isoforms) [19-21], and neurocrescin [22]. Rabaptin-5 isoforms arise from transcripts with skipped exons, while neurocrescin is translated from the transcript with an extra exon inserted. However, the repertoire of proteins expressed from the RAB5EP gene likely is much broader, as several fragments of RAB5EP transcripts characterized by removal of different exons were amplified [23]. In Rabaptin-5 isoform transcripts, exon skipping results in in-frame deletions, and the transcribed proteins bear short deletions compared to Rabaptin-5 [19]. Despite their small sizes (33-43 amino acids (a.a.)), these deletions can affect the functional properties of the proteins. In Rabaptin-5α, a C-terminally located 33 a.a. deletion leads to emphasized Rab4 effector properties of the protein [21]. In another isoform, Rabaptin-5δ, a 40 a.a. deletion in the N-terminal part of the protein results in disruption of the Rab4 binding site, thus rendering impossible Rab4 effector functions [24]. In this work we focused on properties of a previously unexplored Rabaptin-5 isoform, Rabaptin-5γ, which is characterized by 43 a.a. deleted close to the N-terminus of the protein (Fig. 1).
Fig. 1. Schematic representation of Rabaptin-5 protein. A deletion in Rabaptin-5γ covering amino acids 12-64 of Rabaptin-5 is shown (Δ). Regions critical for binding of Rabaptin-5 interacting proteins are shown according to following references: Rab4 binding site (Rab4) [24], N- and C-terminal Rab5 binding sites (Rab5) [3, 6], γ-adaptin and GGA protein GAE binding motif (GAE) and GGA1/GGA2 protein GAT binding region (GAT) [10], and Rabex-5 binding site (Rabex-5) [4]. Below, N-terminal amino acid sequences of Rabaptin-5 and Rabaptin-5γ proteins are shown along with the sequence of a peptide used as immunogen for anti-Rabaptin-5γ hybridoma production. Amino acids corresponding to the peptide sequence are underlined in Rabaptin-5 and Rabaptin-5γ sequence fragments. The site of deletion in Rabaptin-5γ is marked by an arrow.
MATERIALS AND METHODS
Plasmids. pPC97-Rab5, pPC97-Rab5Q79L, pPC97-Rab5S34N, pPC97-Rab4, pPC97-Rab4Q67L, and pPC97-Rab4S22N plasmids for expression of Rab5 and Rab4 and their GTPase-deficient (Q79L and Q67L) and GDP-bound (S34N and S22N) mutants fused to the DNA-binding domain of GAL4 transcription factor (GAL4BD) in yeast were described previously [24]. Plasmids pPC86-Rabaptin-5 [24] and pPC86-Rabaptin-5γ [25] for expression of Rabaptin-5 and Rabaptin-5γ fused to the transcription activating domain of the GAL4 transcription factor (GAL4AD) were described earlier. Plasmids for expression of myc-tagged Rab5Q79L [26], Rab4, Rab4Q67L, and enhanced green fluorescent protein (EGFP)-tagged Rabaptin-5 were described previously [24, 25]. The plasmid for expression of EGFP-tagged Rabaptin-5γ was constructed in a similar way. To construct plasmids for production of glutathione-S-transferase (GST) fused to amino acids 1-140 of mouse Rabaptin-5γ or amino acids 1-183 of mouse Rabaptin-5, the respective cDNA fragments restricted on their 3′-end by a HindIII site were cloned into pGEX-4T-2 vector (GE Healthcare, USA).
Antibodies. Anti-Rabex-5 polyclonal serum [4] was a gift from M. Zerial (Max-Plank Institute of Molecular Cell Biology and Genetics, Dresden, Germany). Mouse monoclonal 2G7 antibodies (mAb) against EGFP were kindly provided by A. Surovoy (Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia). Rabbit polyclonal antiserum against Rabaptin-5 was described before [24]. Secondary horseradish peroxidase-conjugated anti-rabbit IgG for immunoblotting were from GE Healthcare, UK. For immunocytochemical detection, secondary highly cross-absorbed anti-mouse and anti-rabbit IgG conjugated to AlexaFluor488 or AlexaFluor546 dyes (Molecular Probes, USA) were used. Other antibodies were: rabbit polyclonal anti-FLAG antibodies, mouse anti-myc mAb, clone 9E10, mouse anti-α-tubulin mAb, clone DM1α, mouse anti-γ-adaptin mAb, clone 100/3, mouse anti-clathrin light chain mAb, clone CON.1 (Sigma, USA), mouse anti-human golgin-97 mAb, clone CDF4 (Molecular Probes), and mouse anti-gm130 mAb, clone 35/GM130 (Transduction Laboratories, USA).
To generate anti-Rabaptin-5γ mAb, mice were immunized with peptide CGPAPQPDEDLKRQ corresponding to amino acids 5-17 of mouse Rabaptin-5γ (Fig. 1) conjugated through an additional N-terminal cysteine residue to human thyroglobulin (Sigma) with m-maleimidobenzoyl-N-hydroxysuccinimide ester (Sigma). Hybridomas were obtained according to a standard protocol [27]. Supernatants from hybridomas were screened for their interaction in enzyme-linked immunoassay with GST protein fused to amino acids 1-140 of Rabaptin-5γ and the lack of interaction with GST fused to a similar (amino acids 1-183) fragment of Rabaptin-5, which were expressed in BL21 E. coli strain and purified on glutathione-Sepharose 4B (GE Healthcare, Sweden) according to manufacturer’s recommendations. The clone secreting antibodies capable of recognizing Rabaptin-5γ fragment but not the fragment of Rabaptin-5 was selected for further application. Anti-Rabaptin-5γ mAb were affinity purified from hybridoma supernatant on thiol-activated Sepharose (Sigma) with immobilized peptide CGPAPQPDEDLKRQ as described previously [28]. To label anti-Rabaptin-5γ mAb with AlexaFluor546 dye, the AlexaFluor546 Protein Labeling Kit (Molecular Probes) was used.
Cell culture. BHK-21, HeLa B, and COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 µg/ml streptomycin (Invitrogen, USA). If required, cells were transfected with Unifectin-56 transfection reagent (Unifect Group, Russia).
Analysis of protein–protein interaction in yeast. For protein–protein interaction analysis, a yeast two-hybrid system based on Y153 reporter yeast strain and pPC97/pPC86 vectors [29, 30] was used. All manipulations were done as described previously [24]. Protein interaction was revealed by the ability of yeast to grow on appropriate minimal synthetic medium supplemented with 3-amino-(1,2,4)-triazole (3AT).
Cell fractioning. Membrane and cytosol fractions were prepared from COS-7 as described previously [3] with minor modifications. Cells from four 100-mm Petri dishes were scraped into phosphate-buffered saline (PBS), pelleted, and resuspended in 400 µl of 250 mM sucrose, 10 mM sodium phosphate, pH 7.2, by passing through a 27G needle. Cell breakage and nuclear integrity were monitored by phase-contrast microscopy. Nuclei and cell debris were pelleted by centrifugation for 10 min in a microcentrifuge at 4500 rpm yielding post-nuclear supernatant (PNS). The PNS was then fractioned into membrane (pellet) and cytosolic (supernatant) fractions by centrifugation at 4oC for 1 h at 60,000 rpm in a Beckman TLA-100 rotor (USA).
GST pull-down assay and co-immunoprecipitation analysis. GST pull down assay and co-immunoprecipitation analysis were done essentially as described previously [24].
Western blotting. Proteins after separation in SDS-polyacrylamide gel were transferred to Hybond-P membrane (GE Healthcare, USA) using the semi-dry transfer method. Western blot detection was done with ECL+ detection reagents (GE Healthcare) according to the manufacturer’s protocol.
Immunofluorescence microscopy. Cells were plated on coverslips and transfected if required. Twenty-four hours after transfection or next day after plating cells were washed in PBS and fixed with 3% (w/v) paraformaldehyde (PFA). Free aldehyde groups were quenched with 50 mM ammonium chloride, and the cells were permeabilized by incubation in PBS containing 0.1% Triton X-100. Alternatively, cells were fixed for 6 min in methanol chilled to –20°C. Coverslips were incubated with primary antibodies diluted in PBS containing 5% nonfat dry milk and 0.1% Tween 20 for 1 h. After washing with PBS, the coverslips were incubated with appropriate secondary antibodies as above, washed, and mounted in Mowiol (Calbiochem, USA). For simultaneous detection of two proteins with primary mouse antibodies, cells were stained with a single primary mouse antibody followed by secondary AlexaFluor488 conjugated anti-mouse IgG. After that, the coverslips were blocked in PBS containing 10% preimmune mouse serum for 1 h, and the cells were fixed with 3% PFA. After quenching of free aldehyde groups, the coverslips were incubated with anti-Rabaptin-5γ mAb conjugated with AlexaFluor546 dye as described above. The coverslips were examined with a Radiance 2100 confocal microscope (Bio-Rad, UK) and images were taken with ×60 magnification.
RESULTS
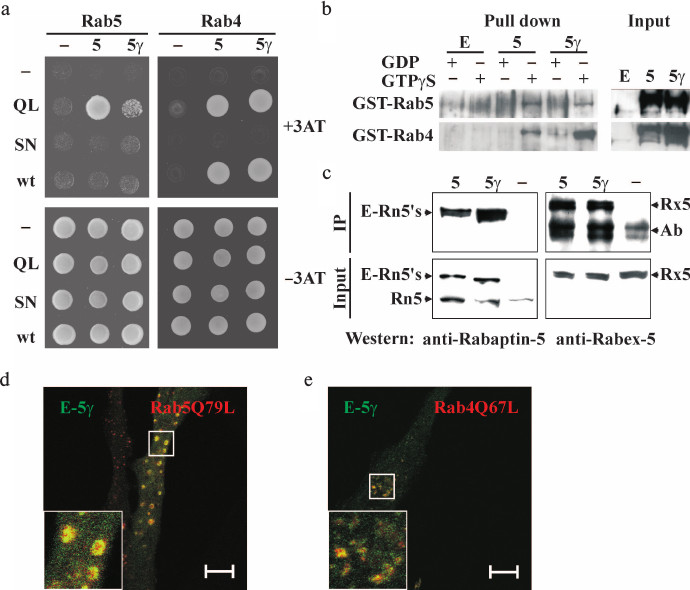
Interaction of Rabaptin-5γ with Rab4, Rab5, and Rabex-5. Rabaptin-5 specifically interacts with Rab5 GTPase in its active GTP-bound state [3]. This interaction along with complex formation with Rab5 guanine nucleotide exchange factor, Rabex-5, is crucial for the function of Rabaptin-5 in Rab5-dependent early endosome fusion [4, 5]. The additional Rab4 binding site in Rabaptin-5 makes it a linker molecule between sequential Rab5- and Rab4-positive membrane domains, and this interaction regulates Rab4-dependent rapid recycling of early endosomes [7, 9]. We analyzed whether Rabaptin-5γ could also interact with these proteins as it is a pre-requisite for its activities in early endosome fusion and Rab5/Rab4 membrane domain linking. In the yeast two-hybrid system, Rabaptin-5γ was able to interact with both GTPases in a GTP-dependent manner similar to Rabaptin-5 (Fig. 2a). To confirm these results, we carried out GST pull-down analysis and found that GTPγS- but not GDP-loaded Rab4 and Rab5 GTPases fused to GST pulled down Rabaptin-5γ from lysates of transfected cells (Fig. 2b). Together these results demonstrate that interactions of Rabaptin-5γ with Rab4 and Rab5 GTPases are similar to those previously observed for Rabaptin-5. Moreover co-immunoprecipitation analysis showed that exogenously produced in BHK-21 cells EGFP-tagged Rabaptin-5γ is complexed with Rabex-5 (Fig. 2c). The ability of Rabaptin-5γ to interact with both Rab5 and Rabex-5 suggests that Rabaptin-5γ should be competent in Rab5-dependent early endosome fusion. Indeed, co-expression of myc-tagged GTPase-deficient Rab5Q79L mutant with EGFP-tagged Rabaptin-5γ in HeLa B cells resulted in formation of typical enlarged Rab5Q79L-positive early endosomes, which were also positive for Rabaptin-5γ (Fig. 2d). Akin, similarly to Rabaptin-5, Rabaptin-5γ was also co-localized with Rab4 GTPase-deficient mutant, Rab4Q67L, on perinuclear membrane structures (Fig. 2e).
Fig. 2. Interaction of exogenously expressed Rabaptin-5γ with Rab5 and Rab4 GTPases and Rabex-5. a) Analysis of interaction between Rabaptin-5 (5) or Rabaptin-5γ (5γ) fused to GAL4AD domain or GAL4AD alone (–) and Rab5 or Rab4 GTPases (wt), their GTPase deficient (QL), or GDP-bound (SN) mutants fused to GAL4BD or GAL4BD alone (–). Y153 reporter yeast cells transformed with plasmids for expression of indicated protein combinations were spotted on synthetic minimal medium supplemented (top) or not supplemented (bottom) with 25 mM 3AT (3AT). b) Interaction of EGFP-tagged Rabaptin-5γ with Rab4 and Rab5 GTPases in GST pull-down assay. Proteins were pulled down from cytosols prepared from BHK-21 cells transiently expressing EGFP-tagged Rabaptin-5 (5) or Rabaptin-5γ (5γ) or EGFP alone (E) with GST-Rab4 or GST-Rab5 immobilized on glutathione-Sepharose 4B beads and loaded with either GDP or GTPγS as indicated. EGFP-tagged Rabaptin-5 and its γ isoform were detected in pull-downs (Pull down) and in cytosol (Input) by immunoblotting with polyclonal antiserum against Rabaptin-5. c) Cytosolic EGFP-tagged Rabaptin-5γ is complexed with Rabex-5. Cytosols were prepared from BHK-21 cells expressing EGFP-tagged Rabaptin-5 (5) or Rabaptin-5γ (5γ) proteins or EGFP alone (–). Cytosols (Input) or proteins immunoprecipitated from cytosols with anti-EGFP antibodies (IP) were immunoblotted with antiserum against Rabaptin-5 or Rabex-5 as indicated. Arrows indicate EGFP-tagged Rabaptin-5 and its γ isoform (E-Rn5′s), endogenous Rabaptin-5 (Rn5), Rabex-5 (Rx5), and antibody used for immunoprecipitation (Ab). d, e) EGFP-tagged Rabaptin-5γ co-localizes with Rab5 (d) and Rab4 (e) GTPase-deficient mutants in HeLa B cells. Cells co-transfected with plasmids for expression of EGFP-tagged Rabaptin-5γ (E-Rn5γ) and myc-tagged GTPase-deficient mutants of Rab5, Rab5Q79L (d), or Rab4, Rab4Q67L (e), were fixed and subjected to immunocytochemical analysis. EGFP-Rabaptin-5γ was imaged by green EGFP fluorescence, and myc-tagged GTPases were detected with anti-myc epitope mouse monoclonal 9E10 antibodies followed by anti-mouse IgG antibodies conjugated to red AlexaFluor546 dye. Merged images are shown with EGFP-Rabaptin-5γ colored green and myc-tagged GTPases colored red, with yellow color indicating their co-localization. Bars, 10 µm.
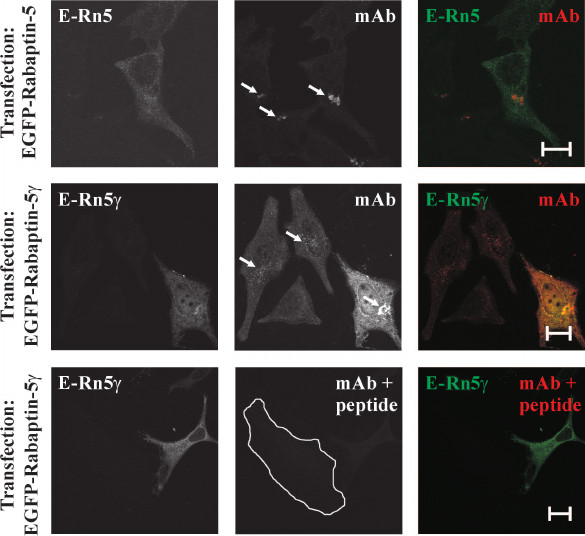
Generation of anti-Rabaptin-5γ monoclonal antibodies. Our results showed that Rabaptin-5 and its γ isoform are similar with respect to their interaction specificities with Rab4, Rab5, and Rabex-5. This observation suggests that Rabaptin-5γ, similarly to Rabaptin-5, acts in early endosome fusion and links Rab4 and Rab5 membrane domains. However, experimental evidences serving as a bias for this suggestion are based on observations made for exogenously produced Rabaptin-5γ protein. Exogenous expression of proteins with a heterogeneous tag in a cell might result in their mistargeting inside the cell and unphysiological interactions with other proteins. To avert this, we generated monoclonal antibodies against newly formed in Rabaptin-5γ epitope at the site of the exon skipping (Fig. 1). These antibodies were able to specifically recognize Rabaptin-5γ in immunocytochemical application without cross-reaction with Rabaptin-5. Indeed, anti-Rabaptin-5γ antibodies detected exogenously produced in cells EGFP-tagged Rabaptin-5γ but not EGFP-tagged Rabaptin-5 (Fig. 3). Importantly, while in non-transfected cells antibodies stained perinuclear membrane structures, the staining was blocked by preincubation of the antibodies with the peptide corresponding to the unique for Rabaptin-5γ epitope against which they were raised (Fig. 3), thus further confirming their specificity. With the use of these antibodies, tracing of endogenous Rabaptin-5γ protein becomes possible.
Fig. 3. Specificity of anti-Rabaptin-5γ monoclonal antibodies. HeLa B cells transiently expressing EGFP-Rabaptin-5 (E-Rn5) or EGFP-Rabaptin-5γ (E-Rn5γ) as indicated were fixed with 3% PFA and stained with anti-Rabaptin-5γ mAb followed by secondary anti-mouse IgG antibodies conjugated with AlexaFluor546 dye. EGFP-tagged proteins were detected by green fluorescence (left panels and green color in merged images) and anti-Rabaptin-5γ mAb immunoreactivity was detected by red fluorescence (middle column and red color in merged images). While anti-Rabaptin-5γ mAb readily detected EGFP-tagged Rabaptin-5γ as evident from co-localization of EGFP and AlexaFluor546 fluorescences (middle row), no staining of EGFP-tagged Rabaptin-5 by anti-Rabaptin-5γ mAb was observed (top row). Both detection of EGFP-tagged Rabaptin-5γ and endogenous Rabaptin-5γ on Golgi-like membrane structures (marked by arrows) by anti-Rabaptin-5γ mAb was abolished by preincubation of the antibodies with peptide CGPAPQPDEDLKRQ used as immunogen for hybridoma production (bottom row; non-transfected cell edges are shown by white line). Bars, 10 µm.
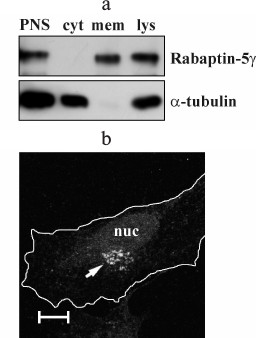
Rabaptin-5γ is a membrane-associated protein. Using anti-Rabaptin-5γ antibodies, we assayed intracellular partitioning of endogenous Rabaptin-5γ protein between cytosolic and membrane fractions. We found that in COS-7 cells Rabaptin-5γ, unlike Rabaptin-5 that is known to be a cytosolic protein part of which is recruited to membranes [3], is present only in membrane fraction (Fig. 4a). This observation indicates that in cells endogenous Rabaptin-5γ is localized exclusively to membranes. To confirm this result, we carried out an immunocytochemical analysis of endogenous Rabaptin-5γ in HeLa B cells. Indeed, anti-Rabaptin-5γ mAb showed intense staining of perinuclear membrane structures, thus further confirming that Rabaptin-5γ is predominantly associated with membranes in cells (Fig. 4b). Distinct intracellular distributions of Rabaptin-5 and its γ isoform suggest that these proteins are involved in different processes despite their similarity in ability to interact with Rab4, Rab5, and Rabex-5.
Fig. 4. Endogenous Rabaptin-5γ is a membrane-associated protein. a) Post-nuclear supernatant (PNS) and total cell lysate (lys) were prepared from COS-7 cells. PNS was used to prepare membrane (mem) and cytosolic (cyt) fractions, and proportional amounts of samples were subjected to immunoblotting. Endogenous Rabaptin-5γ protein was detected with anti-Rabaptin-5γ mAb (Rabaptin-5γ). Membrane was also probed with anti-α-tubulin DM1α monoclonal antibodies to demonstrate correct distribution of soluble α-tubulin protein (α-tubulin). b) Detection of Rabaptin-5γ in HeLa B cells. Cells were fixed with PFA, briefly permeabilized with 0.1% Triton X-100, and immunostained with anti-Rabaptin-5γ mAb followed by secondary anti-mouse IgG antibodies conjugated to AlexaFluor546 dye. The arrow indicates a perinuclear Golgi-like structure visualized with anti-Rabaptin-5γ mAb. Cell edges are depicted by white line and nucleus is marked (nuc). Bar, 10 µm.
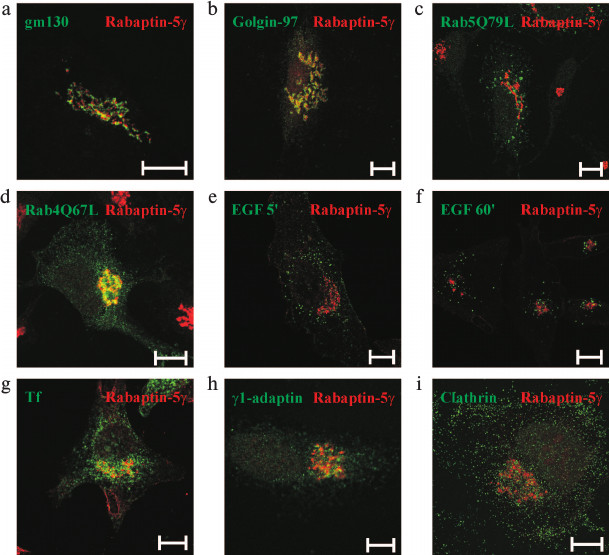
Rabaptin-5γ is localized to trans-Golgi network (TGN)/endosomal interface. Rabaptin-5γ was found to be concentrated on perinuclear membrane structures (Fig. 4b). To reveal the nature of the Rabaptin-5γ-positive compartment, co-localization studies with markers of various intracellular membrane compartments were done. Immunostaining for the cis-Golgi resident protein gm130 [31] showed that Rabaptin-5γ has spatially a close but yet distinct from gm130 pattern of staining (Fig. 5a). At the same time, significant co-localization was observed for Rabaptin-5γ and golgin-97, the protein known to function in transport between endosomal compartment and TGN [32] (Fig. 5b). This observation suggests that Rabaptin-5γ associates with membranes of TGN and endosomes. Rabaptin-5 was found on Rab5- and Rab4-positive endosomes [3, 7]. Therefore, co-localization of endogenous Rabaptin-5γ with these proteins was assessed. Unlike Rabaptin-5, Rabaptin-5γ was absent from Rab5Q79L-positive enlarged early endosomes (Fig. 5c). However when GTPase-deficient mutant, Rab4Q67L (Fig. 5d), or wild-type Rab4 (data not shown) were expressed in cells, they were found in part on Rabaptin-5γ-positive membranes. Together these results show that Rabaptin-5γ resides on the interface of TGN and Rab4-positive endosomal compartment but is absent from Rab5-positive early endosomes. In turn, this fact supposes that γ isoform of Rabaptin-5 does not function in Rab5-dependent early endosome fusion. This suggestion is further confirmed by the lack of co-localization between Rabaptin-5γ and transferrin or epidermal growth factor (EGF), two cargoes known to enter the cell through a Rab5-positive membrane domain (Fig. 5, e-g).
Fig. 5. Intracellular localization of Rabaptin-5γ protein. Untransfected HeLa B cells (a, b, h, i) or cells transfected with plasmids for expression of myc-tagged Rab5Q79L (c) or Rab4Q67L (d) were fixed with PFA, briefly permeabilized with 0.1% Triton X-100, and stained with mouse monoclonal antibodies against gm130 (a), golgin-97 (b), myc epitope (c, d), γ-adaptin (h), or clathrin light chain (i) followed by secondary anti-mouse IgG conjugated with AlexaFluor488 dye. After blocking with pre-immune mouse serum, cells were fixed with PFA and stained with anti-Rabaptin-5γ mAb conjugated with AlexaFluor546 dye. HeLa B cells were also labeled with EGF conjugated to AlexaFlour555 fluorophore for 5 (e) or 60 min (f), or with transferrin (Tf) conjugated to AlexaFluor555 dye (g), then fixed with PFA followed by brief permeabilization with 0.1% Triton X-100 (e, f) or fixed with methanol (g), and stained with mouse anti-Rabaptin-5γ mAb followed by secondary anti-mouse IgG conjugated with AlexaFluor488 dye. Rabaptin-5γ is pseudo-colored red and simultaneously visualized proteins are pseudo-colored green. Bars, 10 µm.
DISCUSSION
In this work we studied properties of the γ isoform of the Rab5 effector Rabaptin-5, which is generated by alternative splicing resulting in a 43-a.a. deletion in its N-terminus (Fig. 1). In particular, we elucidated whether Rabaptin-5γ participates in the same membrane transport events as Rabaptin-5, and how a deletion present in Rabaptin-5γ protein might affect its functions. The results of our study show that biochemically Rabaptin-5γ is identical to Rabaptin-5 in terms of its ability to interact with Rab5, Rabex-5, and Rab4, molecules whose binding to Rabaptin-5 is crucial for Rabaptin-5 functions in early endocytosis. These observations straightforwardly suggested that similarly to Rabaptin-5, Rabaptin-5γ is also a Rab5 effector in early endosome fusion and links Rab5- and Rab4-positive membrane domains, while the deletion in Rabaptin-5γ might affect some other unknown functions of Rabaptin-5. This suggestion was initially supported by co-localization of exogenously expressed Rabaptin-5γ with membranes positive for GTPase-deficient mutants of Rab4 and Rab5. However, the intracellular distribution of endogenous untagged Rabaptin-5γ protein argued against this hypothesis. Our data indicate that Rabaptin-5γ could not act as a Rab5 effector because no co-localization of Rabaptin-5γ with Rab5 was observed. In agreement with this suggestion, Rabaptin-5γ was absent from membrane structures positive for transferrin and EGF, whose internalization depend on Rab5. These results indicate that Rabaptin-5γ does not function in EGF (degradation) and transferrin (recycling) membrane transport pathways and further confirm irrelevance of Rabaptin-5γ to Rab5 function as both cargoes are internalized in a Rab5-dependent manner. Instead, we found that Rabaptin-5γ associates with membranes of TGN and perinuclear endosomal compartment as concluded from co-localization with golgin-97 and Rab4.
Yet there is obvious discrepancy between intracellular distribution of endogenous and exogenously produced Rabaptin-5γ proteins. On one hand, the difference could be reasoned by unphysiologically high expression level of exogenously produced Rabaptin-5γ protein. On the other hand, to detect exogenously produced Rabaptin-5γ, a tag needs to be introduced into the protein on either N- or C-terminus of the polypeptide chain, which might affect interactions with other proteins. Indeed, introduction of C-terminal FLAG tag into Rabaptin-5 polypeptide chain interfered with Rabaptin-5 interaction with Rab5 (unpublished observation) most likely due to spatial constriction of interaction with Rab5, whose binding site is on the extreme C-terminus of Rabaptin-5 [3, 33]. This observation prompted us to link a tag for detection on the N-terminus of Rabaptin-5 and its γ isoform. However, the N-terminal tag resides in close proximity (11 a.a.) to a site of deletion in Rabaptin-5γ, the only structurally different from Rabaptin-5 determinant present in the γ isoform and thus being most likely responsible for altered intracellular location of Rabaptin-5γ. Possibly the presence of the foreign sequence affects the fold of this determinant or sterically hinders its interaction with other moieties, thus precluding N-terminally tagged Rabaptin-5γ protein from mimicking the localization of the endogenous protein.
Along with association with a distinct membrane domain, Rabaptin-5γ differed from Rabaptin-5 in distribution between soluble and membrane associated states. While Rabaptin-5 dynamically shuttles between cytosol and membranes due to Rab5-GTP-dependent interaction [3], its γ isoforms is almost completely depleted from cytoplasm. As the only difference of Rabaptin-5γ from Rabaptin-5 is a small deletion in the polypeptide chain, the newly formed interaction interface in the site of deletion is likely to determine stable association of Rabaptin-5γ with membranes. Indeed, to detect endogenous Rabaptin-5γ by immunocytostaining with mAb against a newly formed epitope, a treatment with Triton X-100 or protein denaturation with methanol is required to unmask the antigen, while Tween 20 and saponin had no effect (data not shown) indicating that the epitope in Rabaptin-5γ is buried, possibly because of sticking to a membrane-associated moiety. Identification of a moiety anchoring Rabaptin-5γ on its specific membrane compartment would facilitate understanding of its functional role in membrane traffic and placing it into a particular route of membrane transport, although this is a challenge for a future research.
In this work we found that Rabaptin-5γ occupies a specific membrane domain characterized by the presence of Rab4 and golgin-97, thus placing Rabaptin-5γ on the interface of endosomal compartment and TGN and prompting the suggestion that it could function in a transport between these two membrane domains. Golgin-97, which is characterized by the most prominent co-localization with Rabaptin-5γ, is also known to function in transport between TGN and endosomes [32]. Interestingly, Rabaptin-5 was implicated in anterograde transport to endosomal compartment through interaction with γ1-adaptin [12], a subunit of AP-1 clathrin adaptor complex involved in transport between TGN and endosomes [34]. However, despite the presence in Rabaptin-5γ structural determinants responsible for association with γ1-adaptin (Fig. 1), no co-localization between these proteins was observed (Fig. 5h), thus indicating that Rabaptin-5γ has distinct (if any) from that of Rabaptin-5 function in TGN-to-endosome communication. In part, communication between TGN and endosomes is realized through clathrin-coated vesicles [35]. However, clathrin was absent from the Rabaptin-5γ-positive compartment (Fig. 5i), suggesting that if Rabaptin-5γ is involved in TGN-to-endosome communication, it acts in other than clathrin-dependent transport pathways.
In conclusion, results of our study clearly demonstrate that Rabaptin-5 and its γ isoform are enrolled in different molecular processes despite their minute structural differences consisting in 43 a.a. residues over an 862-a.a.-long protein. Our findings emphasize the role of minor isoforms of Rab effector proteins and exemplify the importance of alternative splicing in defining their functions.
We thank Dr. Marino Zerial for antiserum against Rabex-5 and Dr. Andrey Surovoy for anti-EGFP monoclonal antibodies. We are also grateful to Dr. Elena Sukhacheva (Moscow, Russia) for hybridoma production.
This work was supported by the Russian Foundation for Basic Research (grant Nos. 01-04-49852 and 04-04-49558).
REFERENCES
1.Zerial, M., and McBride, H. (2001) Rab proteins as
membrane organizers, Nat. Rev. Mol. Cell Biol., 2,
107-117.
2.Stenmark, H., and Olkkonen, V. M. (2001) The Rab
GTPase family, Genome Biol., 2, REVIEWS3007.1-3007.7.
3.Stenmark, H., Vitale, G., Ullrich, O., and Zerial,
M. (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in
endocytic membrane fusion, Cell, 83, 423-432.
4.Horiuchi, H., Lippe, R., McBride, H. M., Rubino,
M., Woodman, P., Stenmark, H., Rybin, V., Wilm, M., Ashman, K., Mann,
M., and Zerial, M. (1997) A novel Rab5 GDP/GTP exchange factor
complexed to Rabaptin-5 links nucleotide exchange to effector
recruitment and function, Cell, 90, 1149-1159.
5.Lippe, R., Miaczynska, M., Rybin, V., Runge, A.,
and Zerial, M. (2001) Functional synergy between Rab5 effector
Rabaptin-5 and exchange factor Rabex-5 when physically associated in a
complex, Mol. Biol. Cell, 12, 2219-2228.
6.Korobko, E. V., Palgova, I. V., Kiselev, S. L., and
Korobko, I. V. (2006) Apoptotic cleavage of rabaptin-5-like proteins
and a model for rabaptin-5 inactivation in apoptosis, Cell
Cycle, 5, 1854-1858.
7.Vitale, G., Rybin, V., Christoforidis, S.,
Thornqvist, P., McCaffrey, M., Stenmark, H., and Zerial, M. (1998)
Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with
GTP-bound Rab4 and Rab5, EMBO J., 17, 1941-1951.
8.Pagano, A., Crottet, P., Prescianotto-Baschong, C.,
and Spiess, M. (2004) In vitro formation of recycling vesicles
from endosomes requires adaptor protein-1/clathrin and is regulated by
rab4 and the connector rabaptin-5, Mol. Biol. Cell, 15,
4990-5000.
9.De Renzis, S., Sonnichsen, B., and Zerial, M.
(2002) Divalent Rab effectors regulate the sub-compartmental
organization and sorting of early endosomes, Nat. Cell Biol.,
4, 124-133.
10.Mattera, R., Arighi, C. N., Lodge, R., Zerial,
M., and Bonifacino, J. S. (2003) Divalent interaction of the GGAs with
the Rabaptin-5-Rabex-5 complex, EMBO J., 22, 78-88.
11.Deneka, M., Neeft, M., Popa, I., van Oort, M.,
Sprong, H., Oorschot, V., Klumperman, J., Schu, P., and van der Sluijs,
P. (2003) Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4
and gamma(1)-adaptin in membrane recycling from endosomes, EMBO
J., 22, 2645-2657.
12.Shiba, Y., Takatsu, H., Shin, H. W., and
Nakayama, K. (2002) Gamma-adaptin interacts directly with Rabaptin-5
through its ear domain, J. Biochem., 131, 327-336.
13.Korobko, I. V., Korobko, E. V., and Kiselev, S.
L. (2000) The MAK-V protein kinase regulates endocytosis in mouse,
Mol. Gen. Genet., 264, 411-418.
14.Valsdottir, R., Hashimoto, H., Ashman, K., Koda,
T., Storrie, B., and Nilsson, T. (2001) Identification of rabaptin-5,
rabex-5, and GM130 as putative effectors of rab33b, a regulator of
retrograde traffic between the Golgi apparatus and ER, FEBS
Lett., 508, 201-209.
15.Coppola, T., Hirling, H., Perret-Menoud, V.,
Gattesco, S., Catsicas, S., Joberty, G., Macara, I. G., and Regazzi, R.
(2001) Rabphilin dissociated from Rab3 promotes endocytosis through
interaction with Rabaptin-5, J. Cell. Sci., 114,
1757-1764.
16.Ohya, T., Sasaki, T., Kato, M., and Takai, Y.
(1998) Involvement of Rabphilin3 in endocytosis through interaction
with Rabaptin5, J. Biol. Chem., 273, 613-617.
17.Neve, R. L., Coopersmith, R., McPhie, D. L.,
Santeufemio, C., Pratt, K. G., Murphy, C. J., and Lynn, S. D. (1998)
The neuronal growth-associated protein GAP-43 interacts with rabaptin-5
and participates in endocytosis, J. Neurosci., 18,
7757-7767.
18.Xiao, G. H., Shoarinejad, F., Jin, F., Golemis,
E. A., and Yeung, R. S. (1997) The tuberous sclerosis 2 gene product,
tuberin, functions as a Rab5 GTPase activating protein (GAP) in
modulating endocytosis, J. Biol. Chem., 272,
6097-6100.
19.Korobko, E. V., Kiselev, S. L., and Korobko, I.
V. (2002) Multiple Rabaptin-5-like transcripts, Gene,
292, 191-197.
20.Korobko, E. V., Smirnova, E. V., Kiselev, S. L.,
Georgiev, G. P., and Korobko, I. V. (2000) Identification of a new
alternative-splicing transcript of Rabaptin-5 interacting with protein
kinase MAK-V, Dokl. Biokhim., 370, 1-3.
21.Nagelkerken, B., Van Anken, E., Van Raak, M.,
Gerez, L., Mohrmann, K., Van Uden, N., Holthuizen, J., Pelkmans, L.,
and Van Der Sluijs, P. (2000) Rabaptin-4, a novel effector of the small
GTPase rab4a, is recruited to perinuclear recycling vesicles,
Biochem. J., 346, 593-601.
22.Nishimune, H., Uyeda, A., Nogawa, M., Fujimori,
K., and Taguchi, T. (1997) Neurocrescin: a novel neurite-outgrowth
factor secreted by muscle after denervation, Neuroreport,
8, 3649-3654.
23.Kawasaki, T., Kunisato, A., Hazama, K., Uyeda,
A., and Taguchi, T. (2001) Identification of active regions for neurite
outgrowth activity of neurocrescin, Biochem. Biophys. Res.
Commun., 281, 761-765.
24.Korobko, E., Kiselev, S., Olsnes, S., Stenmark,
H., and Korobko, I. (2005) The Rab5 effector Rabaptin-5 and its isoform
Rabaptin-5delta differ in their ability to interact with the small
GTPase Rab4, FEBS J., 272, 37-46.
25.Korobko, E. V., Kiselev, S. L., and Korobko, I.
V. (2006) Dimerization properties of Rabaptin-5 and its isoforms,
Biochemistry (Moscow), 71, 1307-1311.
26.Stenmark, H., Parton, R. G., Steele-Mortimer, O.,
Lutcke, A., Gruenberg, J., and Zerial, M. (1994) Inhibition of rab5
GTPase activity stimulates membrane fusion in endocytosis, EMBO
J., 13, 1287-1296.
27.Dean, C. J. (1992) Preparation and testing of
monoclonal antibodies to recombinant proteins, in Methods for
Molecular Biology, Vol. 10. Immunochemical Protocols
(Manson, M. M., ed.) Humana Press, Totowa, NJ, pp. 43-63.
28.Korobko, I. V., Zavalishina, L. E., Kiselev, S.
L., Raikhlin, N. T., and Frank, G. A. (2004) Protein kinase MAK-V/Hunk
as a possible diagnostic and prognostic marker of human breast
carcinoma, Arkh. Patol., 66, 6-9.
29.Durfee, T., Becherer, K., Chen, P. L., Yeh, S.
H., Yang, Y., Kilburn, A. E., Lee, W. H., and Elledge, S. J. (1993) The
retinoblastoma protein associates with the protein phosphatase type 1
catalytic subunit, Genes Dev., 7, 555-569.
30.Chevray, P. M., and Nathans, D. (1992) Protein
interaction cloning in yeast: identification of mammalian proteins that
react with the leucine zipper of Jun, Proc. Natl. Acad. Sci.
USA, 89, 5789-5793.
31.Nakamura, N., Rabouille, C., Watson, R., Nilsson,
T., Hui, N., Slusarewicz, P., Kreis, T. E., and Warren, G. (1995)
Characterization of a cis-Golgi matrix protein, GM130, J.
Cell Biol., 131, 1715-1726.
32.Lu, L., Tai, G., and Hong, W. (2004) Autoantigen
Golgin-97, an effector of Arl1 GTPase, participates in traffic from the
endosome to the trans-Golgi network, Mol. Biol. Cell,
15, 4426-4443.
33.Zhu, G., Zhai, P., Liu, J., Terzyan, S., Li, G.,
and Zhang, X. C. (2004) Structural basis of Rab5-Rabaptin5 interaction
in endocytosis, Nat. Struct. Mol. Biol., 11, 975-983.
34.Kirchhausen, T. (1999) Adaptors for
clathrin-mediated traffic, Annu. Rev. Cell. Dev. Biol.,
15, 705-732.
35.Kreis, T. E., and Pepperkok, R. (1994) Coat
proteins in intracellular membrane transport, Curr. Opin. Cell
Biol., 6, 533-537.