REVIEW: Innate Immunity Underlies Symbiotic Relationships
E. P. Kisseleva
Institute of Experimental Medicine, Russian Academy of Medical Sciences, ul. Akademika Pavlova 12, 197376 St. Petersburg, Russia; fax: (812) 234-9489; E-mail: ekissele@yandex.ru
Received July 15, 2014
Here, the modern data regarding interactions between normal microbiota and barrier tissues in plants, humans and animals are reviewed. The main homeostatic mechanisms responsible for interactions between epithelium and innate immune cells with symbiotic bacteria are described. A key step in this process is recognition of soluble microbial products by ligation to pattern-recognition receptors expressed on the host cells. As a result, epithelial cells secrete mucus, antibacterial peptides and immunoregulatory molecules. The main outcomes from immunological reactions towards symbiotic bacteria involve development of conditions for formation and maintenance of microbial biocenosis as well as providing safety for the host. Also, it is considered important to preserve and transfer beneficial bacteria to progeny.
KEY WORDS: normal microbiota, pattern-recognition receptors, epithelium, innate immunity, symbiotic relationshipsDOI: 10.1134/S0006297914120013
According to the modern views, human body represents the biggest chimera containing 1013 own cells and 1014 different symbiotic microorganisms residing on the skin and mucous layers [1]. Thus, human cells comprise only 10% out of total number, whereas 90% of them belong to bacteria.
Symbiotic interactions evolved very early, and are described in plants and invertebrates. It is believed that symbiosis is a powerful factor in eukaryotic evolution, as animals that let beneficial bacteria to dwell in their bodies acquired considerable advantages for survival associated with defense from predators and pathogens, more advanced nutrition as well as biosynthesis of hormones, vitamins etc. With time, these relationships got more complicated, and vertebrates progressively developed more numerous and diverse symbiosis with more than 500 different microbiotic species. Immune system experiences strong evolutionary pressure and it must provide an opportunity to co-exist with foreign bacteria.
In invertebrates, all issues of interactions with symbionts were managed by innate immunity. Later, adaptive immunity emerged in evolution that also became involved in maintaining co-existence with beneficial bacteria. Historic discussion about an idea for development of adaptive immunity was enriched by Margaret McFall-Ngai and colleagues who hypothesized that it evolved not merely to fight against pathogens, but rather to establish some special relationships with normal microbiota [2].
Irrespective of whether or not the followers of this hypothesis are right or wrong, there are no doubts that a special type of immune response determining relationships between co-evolving macroorganism and microbes has emerged. Currently, there is no special term describing such interactions. For instance, a term “oral tolerance” is mainly related to oral immunization with different antigens and is used in context of vaccination or treatment of some diseases. Modern publications most commonly use term “mucosal immunity” covering a wide range of issues considering not only relationships with commensal microbes, but also infection-induced inflammatory changes in mucosal layers as well as reactions developed in response to autoimmune and allergic diseases.
By contrast, interaction of the body with normal microbiota undoubtedly represents a physiological process, as the main condition underlying symbiotic relationships is impermissibility of causing any harm to each other. In 2002 V. B. Klimovich proposed a term “acceptive immunity” and put forward an idea that interactions between immune system and symbiotic microbes represent a distinct type of immune response unrelated to anti-infectious defense [3]. Unfortunately, this term is not widely accepted yet, and immunological interaction with commensal microbes is still commonly viewed in the same way as the fight against pathogens.
Like in case of protection from pathogens the main strategy for immune system regarding symbiotic microbes is discrimination of “self” and “non-self”, however, it does not result in immune response followed by elimination of the foreign microbes, but rather a peaceful co-habitation with them.
Such interactions are aimed at: demarcation of beneficial bacteria and creation of conditions for their habitat, generation of specialized organs and body systems; restricting penetration of bacteria into internal environment of the body; surveillance and control for inhabitant microbes; creation and continuous maintenance of conditions hampering development of inflammation; preservation and transfer of beneficial microbes to progeny.
The abovementioned tasks are implemented both at the innate and adaptive immune response levels. A great deal of studies is dedicated to investigation of mucosal adaptive immunity mainly considering different types of CD4+ T-cell differentiation [4, 5]. The state of specific tolerance to antigens derived from normal microbiota is believed to develop in vivo. Herein, IgA production represents the dominant type of immune response being surveyed by regulatory T cells [6]. Due to the fact that lymphocytes cannot function on their own, without the participation of mobile innate immune cells, it is important to determine the contribution of these cells to the creation of symbiotic interactions. A key role in this process is played by the barrier epithelium, which may be considered as sessile cells of innate immunity. Current review discusses the role for epithelium and mobile cells of innate immunity that they play during interaction with symbiotic microorganisms.
Investigation of commensal host-immune crosstalk in humans and other mammals is difficult because of the high species diversity of normal microbiota. Thus, it is relevant to apply a phylogenetic approach.
PRINCIPLES OF SYMBIOSIS ORGANIZATION IN INVERTEBRATES
Immunological studies in invertebrates attract our interest because, on one hand, microbiota is represented by only a few species in these animals, and, on the other hand, they have only innate immunity, whereas adaptive immunity is absent.
The key innate immune cells are phagocytes that protect the internal milieu of the body against invasion by microorganisms. For recognition of foreign molecules, the phagocytes use pattern-recognition receptors (PRRs) that interact with pathogen-associated molecular patterns (PAMPs) of microorganisms. Phagocytes are unable to discriminate pathogenic bacteria from beneficial symbionts and are always ready to destroy any foreign material; therefore, one of the first tasks given by Nature for the host organism was to isolate and protect bacteria that were useful for the host.
Some principle approaches have been described for protection of symbiotic bacteria against innate immunity reactions in invertebrates [2, 7]. In some cases, the microorganisms can be separated by a barrier consisting of a chitin layer as in termites or, in other cases, they can be placed into specialized cells – bacteriocytes. This can be exemplified by the bacterium Buchnera aphidicola (in the pea aphid Acyrthosiphon pisum) that is obligate intracellular endosymbiont. This mechanism is widespread among invertebrates but has not been described in vertebrates. However, it is possible that something like this can be used in mammals during lactation for the intracellular transfer of microorganisms from the intestine into the mammary gland. Although it seems rather unlikely, nevertheless this hypothesis is discussed in a paper by Fernandez et al. [8].
In other invertebrates, specialized organs are generated for isolation of extracellular symbiotic bacteria and providing conditions for their survival. This is exemplified by formation of a light organ in the bobtail Hawaiian squid Euprymna scolopes [9]. The symbiosis of the squid with the bioluminescent bacteria Vibrio fischeri is reasonable because the squid uses light produced by the symbiont to mask itself from predators when it is foraging at night (an effect known as counter-illumination).
Formation of the light organ in squids has been described in detail [9-11]. A mollusk and a Gram-negative bacterium free-living in marine water realize a complicated exchange of various molecules, and the key step is an interaction of the bacterial PAMPs with the pattern-recognition receptors of the squid’s epithelium. In squid Toll-like receptors (TLR), five peptidoglycan-recognizing proteins (PGRP) and three lipopolysaccharide-binding proteins (LPS-binding protein, LBP) have been described [11].
The initial stage of this interaction is the influence of bacterial peptidoglycan dissolved in water on the epithelium of the juvenile light organ, which results in production of mucus by the epithelium. This mucus shedding on the surface of the organ facilitates aggregation of bacteria from the surrounding water.
During some few days, two main bacterial molecules which are PAMPs, namely LPS and peptidoglycan, act synergistically to induce apoptosis and proliferation of the squid’s epithelium, which results in morphogenetic changes and formation of the light organ.
The presence of symbiotic V. fischeri in the light organ induces synthesis of many molecules, in particular, the production of cytotoxic nitroxide and superoxide anions that are known to cause the major oxygen-dependent mechanism of intracellular killing of pathogens. However, in the situation with symbionts these molecules play another role and do not damage the bacteria due to an effective detoxification system acting against them. Moreover, both LPS and peptidoglycan produced by the bacteria also can attenuate the activity of NOS/NO [10].
It should be noted that a fragment of peptidoglycan synthesized by V. fischeri is similar to the tracheal cytotoxin (TCT) of pathogenic bacteria that causes a pronounced lesion of tissues in whooping cough and gonorrhea infections [9]. However, the organism responds differently to the same stimuli: in the case of pathogens, LPS and peptidoglycan act as inducers of NOS/NO, whereas in the case of symbiosis the same microbial products have an opposite effect and decrease the production of NO in the host organism [10]. Wang and Ruby believe that this exemplifies the difference between protective immunity and symbiotic relationships.
Thus, the main principles of the organization of symbiotic relationships in invertebrates are as follows: phagocytes and microbiota are separated by a barrier; the key stage is the recognition of the symbiotic bacteria PAMPs by the PRRs of the host organism; this interaction results not in the destruction of the bacteria but in production of mucus, various mediators, antibacterial peptides, and formation of special organs for the residence of the bacteria.
In order to describe symbiont recognition, Margaret McFall-Ngai and colleagues [9] proposed in 2004 more general term – microbe-associated molecular patterns (MAMPs), which supplemented the fundamental concept of pathogen-associated molecular patterns (PAMPs) introduced by Janeway [12].
SYMBIOSIS IN PLANTS
The symbiosis of leguminous plants with root nodule bacteria of the genus Rhizobium (from Greek rhizo – root and bio – life, i.e. life on roots) is another example of the mutually advantageous commensalism. This symbiosis plays an important role in the nitrogen cycle in Nature because soil bacteria settling on plant roots have the ability to fix atmospheric nitrogen and convert it into ammonium, which can be easily assimilated by plants. The molecular mechanisms of this symbiosis are well-studied [13].
The formation of symbiosis starts from production by plant roots of flavonoids, which act as chemoattractants for bacteria from the surrounding soil. Moreover, the roots also produce a mucous substance (mucus) for attracting bacteria and growth factors to stimulate the reproduction of the settled bacteria and then induce in them the production of a nodule factor (NOD-factor). Then the bacteria synthesizing the NOD-factor are included into the program of molecular interactions.
The NOD-factor is a lipo-chito-oligosaccharide, which is a MAMP, recognized by the pattern-recognition receptor of the root epidermis. This receptor is a leucine-rich repeat tyrosine kinase that initiates a cascade of intracellular signals in the epidermal cells. Then, under the influence of the phytohormone cytokinin, the activating signal is transmitted to other cells of the cortex, initiates their reproduction, and promotes further penetration of the bacteria.
As a result, a significant transformation of both partners occurs: on the roots, new organs appear as nodules into which the bacteria settle, and the soil nitrosobacteria upon settling lose their cellular membrane and differentiate into Y-shaped bacteroids. Now they can fix atmospheric nitrogen with the help of a nitrogenase enzymatic complex.
It has been shown that NO, which is a universal molecule recognizable by both bacteria and plants, also acts as a mediator in these interactions [10]. During formation of the symbiosis between leguminous plants and rhizobia, NO activates 100 genes in plants and 57 genes in bacteria.
THE BASIS OF SYMBIOTIC RELATIONSHIPS IN MAMMALS
Unfortunately, in contrast to leguminous plants, mechanisms of establishment of the microbiocenosis and its dialogue with the immune system in humans and other mammals are insufficiently studied. These relationships are usually investigated on a model of germ-free animals, or gnotobionts, grown under sterile conditions. These animals are infected with one or more species of the normal microbiota, and then the interaction of these bacteria with the immune system is studied. In other cases, dysbiosis is induced by antibiotics, and changes in the microbiota are compared with immunological disorders.
Gnotobionts are known to have a poorly developed lymphoid system of the intestine: they have a significantly decreased number of the lamina propria lymphocytes, the maturation of Peyer’s patches and mesenterial nodes is disturbed, generation of germinal centers is lowered, as well as the synthesis of immunoglobulins. It seems to be proved that products of normal microbiota influence immune system maturation in human and animals, mainly the formation of gut-associated lymphoid tissue (GALT) [14].
Also, it has been shown in experiments that commensal bacteria participate in the development and surface differentiation of the epithelium, and these changes create a niche for to be settled with microorganisms [15]. Microbial products contribute to the normal formation of the vascular network of intestinal villi due to induction of synthesis of angiogenic factors [16, 17]. In germ-free animals, mucus degradation is damaged because of the absence of bacteria which are responsible for this process [5]; the production of some antibacterial peptides is also affected (see below). However, these functions are restored upon settling of microorganisms, and this suggests that they are formed in the organism on demand.
These data are fragmentary and do not give an idea how symbiotic relationships develop in children. It is unknown whether factors acting as chemoattractants and stimulators of bacterial growth work in the above-described systems, what patterns interact with receptors of the epithelium, how signals are transmitted, etc. We can only ascertain some final events of this interaction.
The role of epithelium in relationships with the normal microbiota. Obviously, the most important role in the organization of symbiotic relationships on the level of the innate immune reactions in humans and other animals belongs to barrier tissues, especially to the intestinal epithelium. Just this tissue consisting of one layer of epithelium with thickness of only about 20 µm plays the main discriminating role in the separation of symbiotic bacteria from the internal body environment.
Due to mosaic organization, epithelial cells are adapted to perform very different functions, some of which form a basis for creation of a specialized milieu for the maintenance of bacteria. These functions include, first, the ability to recognize by PRRs the products secreted by the normal microbiota (MAMPs) and to react to them by synthesis of diverse factors that can be ascribed to three main types as follows: synthesis of mucus, synthesis of antibacterial peptides, synthesis of cytokines and mediators.
For recognition of MAMPs, intestinal cells express PRRs among which Toll-like receptors (TLR2-5, TLR9) and nuclear oligomerization domain-like receptors (NLRs) are thought to be most important. The genetic elimination of these receptors in experimental animals is associated with loss of the interconnection between the host cells and bacteria that results in changes in the microbiotal composition [4]. Receptors bound with G-proteins (GPCR) that are constantly expressed on the apical surface of the intestine epithelial cells and interact with short fatty acids, adenosine, and other microbial metabolites are also of great importance [5].
The production of mucus and antibacterial peptides by barrier tissues of the host is the most ancient mechanism for the development of symbiotic relationships and is the main innate immune response. Moreover, it should be noted that experimental data on the dependence of the synthesis of mucus and antibacterial peptides on bacterial stimuli are very contradictory (see below).
Production of mucus. The production of mucus has been conserved during evolution because it supports biofilm formation and colonization of bacteria in plants and in animals – from corals to humans [18]. Mucus is produced by goblet cells of the intestine and consists of different proteins of the extracellular matrix, such as mucins, collagens, elastin, fibronectin, fibrinogen, and laminin, and also proteoglycans. A constant component of mucus is secretory immunoglobulin A (sIgA), which is synthesized by plasma cells localized in the submucosa. The major structural component of mucus in the large intestine is the extracellular matrix protein mucin Muc2. Mucin and IgA are two of the most abundantly produced macromolecules in the body, which obviously indicates their important physiological role [19].
Studies on mucus production in the intestine were started not so long ago. This is partially associated with collapse of the mucus layer in histological preparations after conventional fixation with formaldehyde. However, upon fixation in Carnoy fixative, the mucous layer does not shrink, and then its thickness and structure were described for the first time in different parts of the gastrointestinal tract of rats [20].
It was unexpectedly found that there are two layers of mucus in the large intestine. The inner layer is a very firm stratified polymer tightly attached to the epithelium and free of bacteria; moreover, it prevents the penetration of bacteria across the epithelium. The outer layer is loose; bacteria live in it and create a biofilm. This layer is constantly degraded under the influence of microorganisms residing in it. The mucus is replenished from the inside, the inner layer transforms to the loose outer layer, which is degraded by bacteria and eliminated with feces [21].
In the small intestine there is only one layer of mucus, which is discontinuous and less well defined. Mucin is secreted in crypts and moves upward between the villi. The villi are not always covered with mucus; therefore, for protection against the penetration of bacteria the production of antibacterial peptides is very important here.
Mucin is a hyperglycosylated protein, and carbohydrates contribute up to 80% of the Muc2 mucin mass. Human mucin Muc2 contains a PTS domain consisting of frequently repeated amino acids: proline, threonine, and serine. This region of the molecule is glycosylated by O-glycans in the Golgi apparatus so densely that the mucin molecule resembles a bottle brush, with the central protein core decorated with O-glycan “bristles” extending in all directions. This conformation of the molecule allows it to actively bind water and contributes to its gel-forming properties. Upon secretion into the extracellular medium, mucins increase 100-1000-fold in volume due to water adsorbed by glycans.
Under the influence of bacterial proteases, the carbohydrate residues are degraded and become sources of energy for the symbiotic bacteria. Thus, it seems that Nature has cared about the provision of metabolic needs of the bacteria in the case of long-term interruptions in the entrance of food into the host organism. Moreover, the mucus components can be a good substrate for adhesion of many intestinal bacteria. Thus, mucus is a highly specialized microenvironment for the habitation of bacteria [22].
Not long ago another important feature of Muc2 was described – it can have an immunoregulatory influence. During phagocytosis by dendritic cells of Muc2-covered bacteria, mucin was shown to transmit to the dendritic cells tolerogenic signals, this being manifested by suppression of the NF-κB activity and induction of synthesis of IL-10 and TGFβ. Only glycosylated Muc2 caused such effect, which was mediated through an interaction of the carbohydrate residues with receptors of dendritic cells [23].
Formation of the biofilm in humans is the most prominent in the proximal part of the large intestine and gradually decreases to the distal part. This suggests that the vermiform appendix, with its most prominent biofilm and narrow lumen, should play a unique role as a “safe house” for the microbiota in the case of diarrhea caused by an intestinal infection [19].
The synthesis of Muc2 in the intestinal epithelium depends on microbial stimuli received from the normal microbiota. Experiments on mice showed that deletion of the myd88 gene in intestinal epithelium cells, accompanied by the switching out of the MyD88-dependent pathway of signal transduction from TLRs, led to a decrease in the expression of Muc2 protein [24]. The expression of the muc2 gene is also influenced by short fatty acids, propionate and butyrate, produced by microbial fermentation [25].
The production of mucus by epithelial cells is controlled by the innate and adaptive immunity and regulated by IL-9 and IL-13 [26, 27]. In transgenic mice with overexpression of IL-9, the goblet cells were activated in the intestinal mucosa, which was characterized by an increase in the expression of the gene responsible for the synthesis of Muc2 and of some other genes associated with these cells [26]. These cytokines are mainly produced by Th2 and Th9 lymphocytes. Thus, epithelial cells of the intestine represent an efferent arm of the adaptive immune response. In addition to Th2 and Th9 lymphocytes, IL-9 can also be synthesized by Th17 and T-regulatory cells, as well as by many innate immune cells including mast cells, innate immune lymphocytes (ILC), and NKT-cells [27].
Synthesis of antimicrobial factors. Another very important function of the intestinal epithelium retained during the evolution from the very ancient times is the production of antibacterial peptides and proteins that form a biochemical barrier for protection against adhesion and translocation of intestinal microbiota. Enterocytes synthesize α- and β-defensins, cathelicidins, and lysozyme, whereas Paneth cells localized in the depth of crypts in the human and mouse small intestine secrete α-defensins (cryptidins), C-type lectins, angiogenins, lysozyme, and phospholipase A2 [28]. Perhaps not all of these factors are associated with host-commensal interactions, but they may have other functions in the organism. Their antimicrobial activity is well studied, and their role in protective immunity is of no doubt [29], whereas the participation of each of these factors in symbiotic relationships needs further elucidation.
The participation of various factors in the interaction with normal microbiota, in particular, can be ascertained by inducibility of its synthesis by the body cells in response to a contact with beneficial bacteria or their products.
The literature on the dependence of α-defensin synthesis in Paneth cells on bacterial stimuli are contradictory. This problem was studied using different models: α-defensins were studied in germ-free mice, in mice with dysbiosis upon treatment with antibiotics, and in mice with deficiencies of different genes encoding pattern-recognition receptors or transcriptional factors involved in the regulation of immune response genes.
In the work of Putsep et al. [30], five defensin isoforms were isolated from the small intestine of germ-free mice and studied. The secretion of α-defensins by Paneth cells in the gnotobionts did not differ from the secretion in mice colonized with microorganisms, and thus was constitutive. However, later in the same laboratory it was found that the production of another peptide, cryptidin-related sequences 4C (CRS4C), was threefold lower in the germ-free mice than in the normal animals [31], i.e. some isoforms of α-defensins could be synthesized constitutively, whereas production of the others depended on the presence of symbiotic microorganisms.
This issue was further studied on the level of expression of the Defa genes responsible for synthesis of α-defensins in the small intestine of mice, and it was found to be significantly decreased when the microbiotal composition was changed by antibiotics [32]. These contradictions might be explained by the higher number of defensin-encoding genes known in mice (17 genes in mice) than the number of isolated peptides. In animals deficient in genes tlr, nod2, or myd88, expression of the Defa genes and expression of cryptidins (α-defensins) were also decreased, which confirms the inducibility of their synthesis [24, 32, 33].
There is no disagreement concerning production by Paneth cells of some other antibacterial factors, in particular of C-type lectins. It is established that synthesis of the Reg3γ protein (a C-type lectin regenerating islet-derived protein 3γ) depends on the presence of bacteria [34, 35] and is mediated through the MyD88-dependent pathway of signal transduction in epithelial cells [24, 36].
The cationic protein angiogenin 4 with antimicrobial and angiogenic properties, which is produced by the Paneth cells, also depends on stimuli from the normal microbiota [17].
Thus, for normal functioning of the mucous barrier and production of at least some antimicrobial factors, TLRs and NLRs are important, as well as the MyD88-dependent pathway of signal transduction and the presence of microbiota.
There is no doubt that antibacterial proteins and peptides play an important role in the relationships with symbiotic bacteria. Their antimicrobial action creates a biochemical barrier, i.e. a sterile zone near the epithelium that prevents the translocation of bacteria into the internal environment of the body, this barrier playing an especially important role in the small intestine [4]. Moreover, it is known that the antibacterial effect of peptides is manifested only over short distances and is lacking within the limits of the biofilm [37]; therefore, it does not create any danger for commensals inhabiting it. The microbicidal effect of peptides can be prevented by bacterial exopolysaccharides, nucleic acids, proteoglycans, and other molecules.
Another important function of antimicrobial peptides is their ability to serve as a kind of a biological filter limiting the penetration of many microbial products into the body. Thus, defensins and cathelicidins bind LPS and lipoteichoic acid and neutralize their activity, and thus play an important role in preventing inflammation in the mucosa [38]. There are also data on the involvement of antibacterial peptides in the inhibition of inflammatory reactions in phagocytes. Thus, cathelicidin LL-37 can suppress synthesis of proinflammatory cytokines in macrophages in response to LPS in vitro and increase the production of anti-inflammatory cytokines [39].
Changes in the synthesis of defensins in the organism, like increase (in transgenic mice with the human defensin gene HD5) or decrease (in animals with disorders in processing of cryptidins), both significantly influence the composition of the normal microbiota and homeostasis of the intestine [28].
Functions of antimicrobial proteins and peptides depend on their concentrations: in micromolar concentration, they display antimicrobial activity, but in nano- and picomolar concentrations they display angiogenic, wound-healing, and immunoregulatory effects. They can be chemoattractants for T-lymphocytes and dendritic cells and influence the synthesis of cytokines [38]. The angiogenic and immunoregulatory effects of antimicrobial factors can be significant not only during the development of inflammation (as it is usually thought), but also under normal physiological conditions: for organization of the mucosa-associated lymphoid tissue of the intestine, attraction of lymphocytes, and also for formation of the capillary network of villi and normal functioning of the intestinal epithelium [16, 17].
Thus, at present the activity of antimicrobial proteins and peptides in the intestinal mucosa is considered as a protection of the host organism, which suggests their important barrier role. However, some data have begun to appear about a possible promicrobial activity of antibacterial peptides [40]. Nevertheless, it seems that whether antibacterial peptides can perform promicrobial functions we shall know only in the future.
The production of antibacterial peptides by epithelial cells is controlled by the innate and adaptive immunity. The most important cytokines involved in the regulation of synthesis of antibacterial peptides in epithelial cells are IL-17 and IL-22 [41]. Moreover, these cytokines induce the proliferation and differentiation of the intestinal epithelium. These molecules are mainly produced by Th17 cells, whereas the epithelial cells act as an efferent arm of this type of adaptive response. Together with Th17 cells, other cells of the intestinal mucosa are involved in the control of processes on the level of the epithelium. IL-17 is produced by NK, NKT, γδT-, and CD8+ T-cells, while IL-22 is produced by Th22 and ILC. Additionally, hyperplasia of Paneth cells and increase in the synthesis of cryptidins, angiogenin 4, and phospholipase A2 can also be caused by other cytokines such as IL-9 and IL-13 [26].
Synthesis of cytokines and mediators. Production of cytokines and immune mediators is the third most important function of the epithelium. The follicle-associated epithelium localized above Peyer’s patches has a unique ability to recognize microbial stimuli and respond to them by production of cytokines. Moreover, these cells can also discriminate signals from normal microbiota and pathogens.
Normally, direct contact of microorganisms with receptors is limited to prevent inflammation. Thus, expression of TLR2 and TLR4 is suppressed on epithelial cells. TLR5 is localized only on the basolateral side of the cells, and it is not expressed into the intestinal lumen. TLR3, TLR7, TLR8, and TLR9 are localized in intracellular endosomal structures, and NLR – in the cytoplasm [5].
Under the integrated influence of various soluble microbial products and metabolites, normally functioning epithelium produces a number of anti-inflammatory cytokines and mediators, including such important products as transforming growth factor β (TGFβ), interleukin 10 (IL-10), thymic stromal lymphopoietin (TSLP), and retinoic acid (RA), which is a metabolite of nutritional vitamin A [4, 5]. These factors act as immunoregulators of innate immune cells localized in the mucosa and also influence the development of the adaptive immune response. These anti-inflammatory mediators also promote the immunoglobulin synthesis switching for class IgA. Factors produced by the epithelial cells together with cytokines released by the innate immune cells create a microenvironment favorable for differentiation of T-regulatory cells.
On invasion of pathogens, the reaction of epithelial cells is different, associated first of all with different “behavior” of the microorganisms. The normal microbiota colonizing the surface of the mucosa is an ecosystem whose members have been carefully selected by Nature in many parameters including the ability to live on the surface of the epithelium without damaging it. On the contrary, in pathogenic bacteria virulence genes are activated on contact with the epithelium. The pathogenic factors cause damage to the epithelium that is necessary for their penetration into the internal body environment. The damage to epithelial cells has two important consequences for the development of immune response.
First, normally latent pattern-recognition receptors of epithelial cells are activated and “alarm signalization” is switched on, which includes synthesis of proinflammatory cytokines and chemokines. They attract phagocytes and initiate the development of inflammation.
Second, the damaged epithelial cell, similarly to any other cell, produces molecules signalizing about its damage – damage-associated molecular patterns (DAMPs) – which are perceived by phagocytes as additional activating factors [42].
As a result, in the case of a pathogen entrance, the immune system receives from the epithelium other information – dendritic cells are activated under the influence of PAMPs, DAMPs, and proinflammatory cytokines, and the protective immune response develops, which is directed to killing the particular infective agent. This reaction is significantly different from the immune response to microbial products of commensals in the absence of pathogens.
Permeability of the intestinal epithelium and antigen sampling. Permeability of the intestinal epithelium is the most important regulator of homeostasis of the intestinal mucosa. Metabolites as well as nutritional and microbial antigens are absorbed paracellularly across tight junctions [43]. Various protein molecules are transported here, but microbial products such as LPS can also be transferred. Proinflammatory cytokines TNFα and IFNγ increase paracellular permeability, whereas the anti-inflammatory cytokine TGFβ and glucocorticoid hormones display a stabilizing effect. LPS increases the intestinal permeability. Recently, it was shown that in addition to the paracellular pathway, low molecular weight soluble antigens could enter the body through goblet cells [4].
Thus, the intestinal epithelium covered with mucus and antibacterial peptides forms a complex biological filter that controls the entrance into the body of microbial products, which influence the immune system. Innate and adaptive immune cells localized in submucosa have pattern-recognition receptors and can sense MAMPs penetrating across the epithelial barrier.
To control the composition and amount of normal microbiota, bacteria can be delivered through M-cells [44]. M-cells, or microfold cells, localized above Peyer’s patches, are epithelial cells specialized for the vesicular transport of various molecules and bacteria. Bacteria transferred through M-cells are phagocytized by dendritic cells, and then the inductive phase of the immune response to antigens of the normal microbiota starts. Moreover, a limited number of the bacteria can be sampled directly in the intestinal lumen by the dendritic cells that extend their processes through the epithelium.
Commensals and microbial products interact with the immune system mainly in the small intestine via specialized lymphoid tissue – Peyer’s patches. Moreover, the porous layer of mucus allows the body cells to have nearer contact with the bacteria. Therefore, the protection by antibacterial peptides is especially important for the small intestine epithelium. An important role is also played by IgA, which promotes the transport of bacteria through M-cells [45]. The number of bacteria inhabiting the small intestine is not high in comparison with their number in the large intestine and is strictly controlled by the organism (from 103 bacteria per ml of the intestinal contents in the proximal part and to 108 in the distal part). An increase in the number of bacteria in the small intestine is associated with development of small bowel bacterial overgrowth syndrome that is usually accompanied by inflammation.
The situation in the large intestine is quite different. Here food digestion under the influence of human enzymes is virtually finished, and undigested fibers are available for the commensals. In the large intestine the most numerous microbiocenosis of the human organism is present (1012 bacteria per g feces). The epithelium of the large intestine is covered with a very dense layer of mucus that prevents contact of the microorganisms with the cells of the body. Therefore, there are no zones of immune response induction here, and the immunoregulatory influence of the microbiota is mediated through different metabolites, among which short fatty acids are best-studied [46].
The role of mobile innate immune cells in symbiotic interactions. The innate immune cells are an essential part of the mucosa-associated lymphoid tissue of the intestine. These cells include macrophages, dendritic cells, mast cells, natural killer cells (NK), NKT-cells, γδT-cells, B1 lymphocytes, and innate immune lymphocytes (ILC). These cells settle mainly the lamina propria region and are a part of the unorganized mucosa-associated lymphoid tissue.
The innate immune cells can recognize signals entering from the commensals using PRRs and react to these signals, producing various cytokines. These cells are involved in very different processes – from regulation of epithelial functions to influence on the adaptive immunity (Table 1). Thus, RORγt+ ILC through the produced lymphotoxin influence organization of the intestinal lymphoid tissue and the entrance of T- and B-lymphocytes. Macrophages, dendritic and mast cells, γδT-cells, and ILC synthesize cytokines and retinoid acid that are favorable for synthesis of IgA. The innate immune cells receive signals from the epithelial cells and respond by synthesis of factors maintaining the normal functioning of the epithelium according to the feedback principle. Moreover, as mentioned above, they control production of mucus and antibacterial peptides in the epithelial cells.
Table 1. The role of innate immune cells in
host–symbiont crosstalk in intestinal mucosa
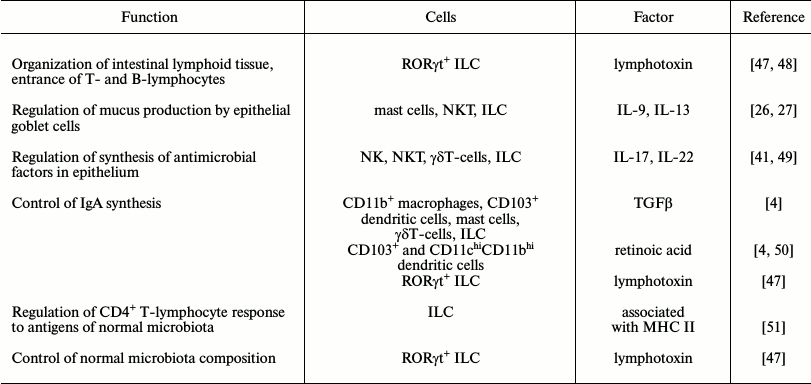
Creation of a specific microenvironment for differentiation of T-regulatory cells is an important function of innate immune cells of the intestinal mucosa [6]. It occurs with involvement of all mobile cells of the innate immunity and of the intestinal epithelial cells that produce anti-inflammatory factors such as TGFβ, IL-10, and RA.
Antigen-presenting cells – dendritic cells and macrophages – play a special role in this cell community. In addition to their major function of phagocytosis of microorganisms and presentation of antigens to naive T-lymphocytes, some of the antigen-presenting cells localized in the lamina propria acquire unusual features of tolerogenic dendritic cells and anergic macrophages. Many researchers believe that just these cells promote the formation of T-regulatory cells and induction of tolerance [5, 52]. Macrophages with the CD11bhighCX3CR1+ phenotype synthesize for this anti-inflammatory cytokines IL-10 and TGFβ, whereas CD103+ myeloid dendritic cells can store and produce large amounts of RA [53].
Mechanisms of induction and activation of such cells are insufficiently studied. Their stimulation can be associated with products of intestinal bacteria. Some species of intestinal bacteria are shown to suppress production of proinflammatory cytokines by macrophages [54]. The primary conditioning of dendritic cells is performed by the intestinal epithelial cells that produce anti-inflammatory factors – TGFβ and RA [55]. Tolerogenic signals can also be transmitted to dendritic cells during phagocytosis of bacteria through mucus fragments containing Muc2 [23] and through IgA the covers the intestinal bacteria [56] and induce in dendritic cells synthesis of anti-inflammatory cytokines.
Intracellular mechanisms of activity of tolerogenic dendritic cells are thought to include a down-regulation of TLR responses, suppression of the signal transduction pathway through NF-κB mediated by NOD2 [5], and the necessary involvement of transcriptional factor TRAF6 [57].
Nevertheless, tolerogenic dendritic cells are only a part of the total pool of antigen-presenting cells. The normal intestinal mucosa contains different dendritic cells that produce IL-12, IL-23, and other cytokines promoting the differentiation of CD4+ T-lymphocytes into the Th1-, Th2-, and Th17-types of the immune response [5].
It should be noted that the intestinal dendritic cells fully retain their ability to participate in the protective response to possible pathogens, notwithstanding their involvement in relationships with the commensals [5].
TGFβ is a dominating cytokine in the intestinal mucosa because it is important for induction of T-regulatory cells and synthesis of IgA. Moreover, this cytokine stabilizes the permeability of the intestinal epithelium and also suppresses the expression of TLR in the epithelial cells. TGFβ is synthesized not only by dendritic cells and macrophages but also by mast cells, intraepithelial γδT-cells, ILC, and the intestinal epithelial cells. This cytokine is a universal mediator of homeostasis in the intestinal mucosa and acts as a connecting link between innate and adaptive immunity.
Recently, the involvement of B1-lymphocytes in IgA synthesis in the intestinal mucosa has been actively discussed in the literature [4, 58]. B1-lymphocytes are cells of the innate immunity and can T-independently synthesize polyreactive antibodies of the M-class. In the peritoneal cavity of mice there are many B1-lymphocytes that migrate from there into the lamina propria region and proliferate under the influence of IL-5 and IL-15. It is thought that B1-lymphocytes localized in the organized gut-associated mucosal tissue (Peyer’s patches, isolated lymphoid follicles, and mesenterial lymph nodes) can switch to IgA synthesis in T-independent mode. However, this requires activation of their TLRs and production of TGFβ, BAFF (B cell-activating factor) and APRIL (a proliferation-inducing ligand) by dendritic cells. It is supposed that in the case of T-independent synthesis of IgA in the lamina propria region the molecules (BAFF, APRIL, TSLP) switching the immunoglobulin class are produced directly by the epithelium itself. However, the exact localization of this process is still unknown.
However, the question about the involvement of B1-lymphocytes in the synthesis of IgA in humans and contribution of these molecules to the total pool of secretory IgA in the gastrointestinal tract is even more disputed [58]. As differentiated from mouse, an adult human has a small number of B1-lymphocytes because they manifest themselves mainly during embryogenesis. Until recently, the T-independent switching to IgA class synthesis was believed to be impossible [59]. In 2007, it was shown that in humans the synthesis could be switched from IgM to IgA2 and from IgA1 to IgA2 in T-independent mode with participation of APRIL cytokine produced by intestinal epithelial cells [60]. However, the problem of phenotyping of B1-lymphocytes in humans is not solved finally because there is no consensus on what B1-lymphocytes are and what markers are characteristic for them [58, 61].
THE ROLE OF IMMUNOLOGICAL FACTORS IN TRANSMISSION OF SYMBIOTIC
BACTERIA TO PROGENY
It is quite reasonably to suppose that as animals have found for themselves some beneficial bacteria and a mutually advantageous symbiosis occurs, a mechanism should be provided for retention and transmission of these bacteria to progeny. In fact, insects possess numerous pathways for vertical transmission of microbiota [7]. Thus, in beewolf females the antenna-located symbionts are secreted into the brood chamber where they are captured by the larvae; in carpenter ants bacteria are located in specialized bacteriocytes in the reproductive tissue and invade the oocytes; in the plataspid stinkbugs special capsules with symbiotic bacteria are ingested by newborn nymphs; in other species of these stinkbugs bacteria are transferred onto the surface of the eggs by a special “lubricating organ”.
However, the transmission of the endosymbiotic bacteria Wigglesworthia in the tsetse fly seems to be the most surprising. These flies are viviparous and have a “milk gland” to feed larvae in their uterus. Moreover, the Wigglesworthia bacteria are harbored within the lumen of the “milk gland” and are probably transmitted into the developing larvae via milk secretions [62].
The human baby is sterile until birth. It was thought that the baby’s organism is introduced to maternal microbiota mainly during the passing through the birth canal, and after that bacteria settle from the environment. Milk was considered a source of nutrition and protection against infections, and only recently we started to understand that milk is an important factor for transmission of symbiotic bacteria and formation of the biocenosis [8].
In 2012, the microbiome of human milk was studied for the first time, and it was found to be a separate microbial community not similar in composition to other microbiocenoses of human body [63]. The same bacterial species were isolated from the baby’s feces. In 1 ml of mature milk, there are 103-104 bacteria (in colostrum the number of bacteria is much higher). A baby drinks about 800 ml of milk per day and thus gets 105-107 bacteria daily. How bacteria enter the breast milk is unknown. There is a hypothesis that they can be transferred from the intestine to milk gland endogenously by macrophages or dendritic cells [8].
Breast-feeding plays the most important role in formation of the intestinal microbiota of a child. There is an opinion that milk can program creation of the intestinal microbiocenosis and development of the immune system of the child, and that immunological factors contribute to this [64, 65]. In insects, only the innate immunity is involved in transmission of microbiota to progeny, whereas in humans both mechanisms of innate and adaptive immune responses participate in this, and immune cells play an important role in this process.
Human milk contains up to 1 million of the mother’s cells per ml (colostrum contains up to 10 million). These cells are mainly macrophages, but neutrophils and lymphocytes (5 and 10%, respectively) are also present. Lymphocytes are represented by both T- and B-cells, the majority of which are memory cells. CD8+, CD4+, CD4+CD25+ lymphocytes, γδT cells, and plasma cells are also present. Data obtained on domestic and laboratory animals (cited in the article by Arvola et al. [66]) indicate that lymphocytes and other cells of milk and colostrum can penetrate across the intestine of newborns, enter mesenterial lymph nodes, and further come into the circulation, retaining viability and functional activity.
Secretory IgA, which is a factor of adaptive immunity, is one of the major immunological components of milk. Milk contains up to 1 g/liter of sIgA, and colostrum contains 5 g/liter. It is supposed that the interaction with maternal IgA promotes adhesion of bacteria and is very important during their settling in the child’s intestine and formation of the microbial biocenosis [45]. Other researchers also think that IgA and mucin play an important role in formation of the biofilm [67]. Mucin is also present in milk.
The next innate immune factor, lactoferrin, is another major component of milk. The lactoferrin content in milk and colostrum is 2 and 7 g/liter, respectively. Lactoferrin is an iron-binding glycoprotein that is traditionally considered as a bacteriostatic factor preventing the reproduction of microorganisms because it binds iron, which is necessary for their growth [29]. However, the role of this factor is now being reconsidered, and it is believed to be a bifidogenic factor [68].
It occurs that, depending on the iron concentration in vitro, lactoferrin is able not to suppress but even to stimulate growth of particular species of bifidobacteria. Some mechanisms are being considered to explain this effect: the use by the bacteria of lactoferrin-bound iron, of polysaccharide chains of lactoferrin as a source of energy, and the growth-stimulating capacity of cationic lactoferrin peptides. Oligosaccharides and lysozyme that are present in milk also have bifidogenic activity [68]. Bifidobacteria are major component of the intestinal microbiocenosis in infants, and therefore, to restore their number, formula containing bovine lactoferrin is successfully used.
There are data indicating that lactoferrin can be also used by Helicobacter pylori for their reproduction, and they utilize iron selectively from human and not from bovine lactoferrin. Note that transferrin, which is also a component of breast milk, is not used by these bacteria as a source of iron [69]. At present, H. pylori are considered dualistically: on one hand, they are believed to be a risk factor for development of stomach ulcer and cancer, but on the other hand, they seem to be useful for the body, otherwise they would not be retained as a component of the normal microbiota [70]. In fact, these bacteria are necessary for stimulation of production of two important hormones of the stomach, ghrelin and leptin, whose absence can provoke a disease of the esophagus. There are data indicating that children not possessing H. pylori are more sensitive to development of asthma and allergies [71].
Such milk components as growth factors (epidermal growth factor (EGF)), hormones, cytokines, polyunsaturated fatty acids, and oligosaccharides can promote maturation and functioning of the epithelial cells of the infant’s intestine [72]. Human milk has a complex composition, and studies on the roles of its components in formation of the intestinal microbiota are only at their beginning.
CONCLUDING REMARKS
Symbiotic relationships with bacteria have existed on Earth from very ancient times, are distributed among many plants and animals, and are an important achievement in the evolution of living beings. An important role in these relationships belongs to the immune system, which is responsible for maintaining integrity of the macroorganism and protecting it against entrance of unnecessary or deleterious foreign material. As relating to symbionts, the immune response strategy is designed for creating conditions for production and retention of the microbial biocenosis, on one hand, and for providing the safety of the host’s organism, on the other hand. The retention and delivery of beneficial microorganisms to the progeny is also an important task.
Recognition of soluble microbial products by PRRs is a key molecular mechanism of the immune system interaction with the normal microbiota. As differentiated from interaction with pathogens, in this case there is no direct contact with bacteria because they are separated from the immune system cells by the epithelial barrier. It can be supposed that highly sensitive and non-mediating phagocytosis TLRs have been created by Nature just for the distant interaction with microorganisms. It is usually called “microbial sensing”, i.e. the cells localized under the epithelium can “sense” the microbiota with their PRRs. Only a small part of bacteria in the small intestine is phagocytized by dendritic cells or passes across M-cells to trigger an adaptive immune response. However, all the main signals are presented not by living bacterial cells, but by their products or fragments of the dead microbes that reach the immune system cells penetrating across the epithelium. This can be a reason for the presence of TLRs in all cells of the immune system, including T- and B-lymphocytes.
Just the barrier tissue is the primary link for contact with bacteria. The interaction of PRRs with MAMPs triggers in epithelial cells a cascade of the intracellular signal events leading to activation of the cell. It is interesting that this process is similar in plants, invertebrates, and mammals (Table 2). PRRs of plants are somewhat like NOD-like receptors of mammals.
Table 2. Comparative characteristics of
interaction of surface epithelium with products of symbiotic bacteria
in plants, invertebrates, and mammals
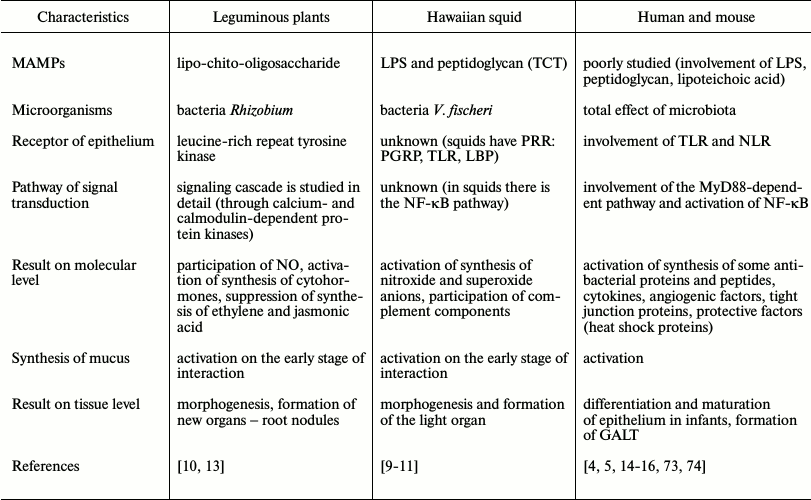
All stages of the interaction with symbiotic bacteria are best-studied in leguminous plants: a microbial pattern, receptor, signal transduction pathways, and the result of this interaction. Humans have not a single symbiont as plants, but hundreds of species, thus, the effect of integrated microbial signals is usually considered. In the literature there are only few indications that intestinal epithelial cells are influenced by LPS and peptidoglycans via TLRs and NLRs, and that MyD88-dependent pathway and the NF-κB activation are involved.
Production of mucus is an initial reaction of the host to the presence of bacteria. In addition to mucus, in the epithelium of invertebrates and mammals microbial signals induce synthesis of antibacterial peptides, and in mammals of cytokines whose totality presents major effector molecules of the innate immunity. The universal molecule NO also plays an important role in interaction of the epithelium with bacteria. Mucus is a habitat and energy source for microorganisms, antibacterial peptides protect the host’s body, and cytokines form a microenvironment for triggering adaptive immune responses.
It should be noted that, as differentiated from the interaction with pathogens, the activation of PRRs on epithelial cells in the case of symbiotic relationships leads to morphogenetic changes. Thus, in leguminous plants it is formation of root nodules, in squids – of the light organ, in mammals – of the GALT.
Production of secretory IgA, partially contributed by the innate B1 cells, is the major response of the adaptive immune system to microbial stimulation. sIgA is released into the intestinal lumen where it interacts with bacteria and participates in formation of the biofilm.
Thus, homeostasis of mucosae is maintained by a complex of the innate and adaptive immune responses that finally results in host–bacterial mutualism.
REFERENCES
1.Whitman, W. B., Coleman, D. C., and Wiebe, W. J.
(1998) Prokaryotes: the unseen majority, Proc. Natl. Acad. Sci.
USA, 95, 6578-6583.
2.McFall-Ngai, M. (2007) Care for the community,
Nature, 445, 153.
3.Klimovich, V. B. (2002) Actual problems of
evolutionary immunology, J. Evol. Biochem. Physiol., 38,
562-574.
4.Brown, E. M., Sadarangani, M., and Finlay, B. B.
(2013) The role of the immune system in governing host-microbe
interactions in the intestine, Nature Immunol., 14,
660-667.
5.Hill, D. A., and Artis, D. (2010) Intestinal
bacteria and the regulation of immune cell homeostasis, Annu. Rev.
Immunol., 28, 623-667.
6.Josefowicz, S. Z., Lu, L.-F., and Rudensky, A. Y.
(2012) Regulatory T cells: mechanism of differentiation and function,
Annu. Rev. Immunol., 30, 531-564.
7.Feldhaar, H., and Gross, R. (2009) Genome
degeneration affects both extracellular and intracellular bacterial
endosymbionts, J. Biol., 8, 31.1-31.5.
8.Fernandez, L., Langa, S., Martin, V., Maldonaldo,
A., Jimenez, E., Martin, R., and Rodriguez, J. M. (2013) The human milk
microbiota: origin and potential roles in health and disease,
Pharmacol. Res., 69, 1-10.
9.Koropatnick, T. A., Engle, J. T., Apicella, M. A.,
Stabb, E. V., Goldman, W. E., and McFall-Ngai, M. J. (2004) Microbial
factor-mediated development in a host-bacterial mutualism,
Science, 306, 1186-1188.
10.Wang, Y., and Ruby, E. G. (2011) The roles of NO
in microbial symbioses, Cell. Microbiol., doi:
10.1111/j.1462-5822.2011.01576.x.
11.Nyholm, S. V., and Graf, J. (2012) Knowing your
friends: invertebrate innate immunity fosters beneficial bacterial
symbioses, Nature Rev. Microbiol., 10, 815-827.
12.Medzhitov, R., and Janeway, C. A. (2002) Decoding
the patterns of self and nonself by innate immune system,
Science, 296, 298-300.
13.Ferguson, B. J., Indrasumunar, A., Hayashi, S.,
Lin, M.-H., Lin, Y.-H., Reld, D. E., and Gresshoff, P. M. (2010)
Molecular analysis of legume nodule development and autoregulation,
J. Integr. Plant Biol., 52, 61-76.
14.Renz, H., Brandtzaeg, P., and Hornef, M. (2012)
The impact of perinatal immune development of mucosal homeostasis and
chronic inflammation, Nature Rev. Immunol., 12, 9-23.
15.Hooper, J. V., and Gordon, J. L. (2001) Commensal
host–bacterial relationships in the gut, Science,
292, 1115-1118.
16.Stappenbeck, S., Hooper, L. V., and Gordon, J. I.
(2002) Developmental regulation of intestinal angiogenesis by
indigenous microbes via Paneth cells, Proc. Natl. Acad. Sci.
USA, 99, 15451-15455.
17.Hooper, L. V., Stappenbeck, T. S., Hong, C. V.,
and Gordon, J. I. (2003) Angiogenins: a new class of microbicidal
proteins involved in innate immunity, Nat. Immunol., 4,
269-273.
18.Bollinger, R. R., Barbas, A. S., Bush, E. L.,
Lin, S. S., and Parker, W. (2007) Biofilms in the large bowel suggest
an apparent function of the human vermiform appendix, J. Theor.
Biol., 249, 826-831.
19.Laurin, M., Everett, M. L., and Parker, W. (2011)
The cecal appendix: one more immune component with a function disturbed
by post-industrial culture, Anat. Record., 294,
567-579.
20.Atuma, C., Strugala, V., Allen, A., and Holm, L.
(2001) The adherent gastrointestinal mucus gel layer: thickness and
physical state in vivo, Am. J. Physiol. Gastrointest. Liver
Physiol., 280, G922-G929.
21.Johansson, M. E. V., Phillipson, M., Petersson,
J., Velcich, A., Holm, L., and Hansson, G. C. (2008) The inner of the
two Muc2 mucin-dependent mucus layers in colon devoid of bacteria,
Proc. Natl. Acad. Sci. USA, 105, 15064-15069.
22.Johansson, M. E. V., Holmen Larsson, J. M., and
Hansson, G. C. (2011) The two mucus layers of colon are organized by
the MUC2 mucin, whereas the outer layer is a legislator of
host–microbial interactions, Proc. Natl. Acad. Sci. USA,
108, Suppl. 1, 4659-4665.
23.Shan, M., Gentile, M., Yeiser, J. R., Walland, A.
C., Bornstein, V. U., Chen, K., He, B., Cassis, L., Bigas, A., Cols,
M., Comerma, L., Huang, B., Blander, J. M., Xiong, H., Mayer, L.,
Berin, C., Augenlicht, L. H., Velcich, A., and Cerutti, A. (2013) Mucus
enhances gut homeostasis and oral tolerance by delivering
immunoregulatory signals, Science, 342, 447-453.
24.Frantz, A. L., Rogier, E. W., Weber, C. R., Shen,
L., Cohen, D. A., Fenton, L. F., Bruno, M. E. C., and Kaetzel, C. S.
(2012) Targeted deletion of MyD88 in intestinal epithelial cells
results in compromised antibacterial immunity associated with
down-regulation of polymeric immunoglobulin receptor, mucin-2, and
antibacterial peptides, Mucosal Immunol., 5, 501-512.
25.Burger-van Paassen, N., Vincent, A., Puiman, P.
J., van der Sluis, M., Bouma, J., Boehm, G., van Goudoever, J. B., van
Seuningen, I., and Renes, I. B. (2009) The regulation of intestinal
mucin MUC2 expression by short-chain fatty acids: implications for
epithelial protection, Biochem. J., 420, 211-219.
26.Steenwinckel, V., Louahed, J., Lemaire, M. M.,
Sommereyns, C., Warnier, G., McKenzie, A., Brombacher, F., Van Snick,
J., and Renauld, J.-C. (2009) IL-9 promotes IL-13-dependent Paneth cell
hyperplasia and up-regulation of innate immunity mediators in
intestinal mucosa, J. Immunol., 182, 4737-4743.
27.Zhao, P., Xiao, X., Ghobrial, R. M., and Li, X.
C. (2013) IL-9 and Th9 cells: progress and challenges, Intern.
Immunol., 25, 547-551.
28.Salzman, N. H. (2011) Microbiota–immune
system interaction: an uneasy alliance, Curr. Opin. Microbiol.,
14, 99-105.
29.Kokryakov, V. N. (2006) Essays about the
Innate Immunity [in Russian], Nauka, St. Petersburg.
30.Putsep, K., Axelsson, L. G., Boman, A., Midtvedt,
T., Normark, S., Boman, H. G., and Andersson, M. (2000) Germ-free and
colonized mice generate the same products from enteric prodefensins,
J. Biol. Chem., 275, 40478-40482.
31.Karlsson, J., Putsep, K., Chu, H., Kays, R. J.,
Bevins, C. L., and Andersson, M. (2008) Regional variations in Paneth
cell antimicrobial peptide expression along mouse intestinal tract,
BMC Immunol., 9, 37.
32.Menendez, A., Willing, B. P., Montero, M.,
Wlodarska, M., So, C. C., Bhinder, G., Vallance, B. A., and Finlay, B.
B. (2013) Bacterial stimulation of the TLR-MyD88 pathway modulates the
homeostatic expression of ileal Paneth cell α-defensins, J.
Innate Immun., 5, 39-49.
33.Kobayashi, K. S., Chamaillard, M., Ogura, Y.,
Henegariu, O., Inohara, N., Nunez, G., and Flavell, R. A. (2005)
Nod2-dependent regulation of innate and adaptive immunity in the
intestinal tract, Science, 307, 731-734.
34.Cash, H. L., Whitham, C. V., Behrendt, C. L., and
Hooper, L. V. (2006) Symbiotic bacteria direct expression of an
intestinal bacterial lectin, Science, 313, 1052-1054.
35.Sanos, S. L., Bui, V. L., Mortha, A., Oberle, K.,
Heners, C., Johner, C., and Diefenbach, A. (2009)
ΡΟΡγt and commensal microflora are required for
the differentiation of mucosal interleukin-22 producing NKp46+ cells,
Nat. Immunol., 10, 11-12.
36.Brandl, K., Pitas, G., Schnabl, B., DeMatteo, R.
P., and Pamer, E. G. (2007) MyD88-mediated signals induce the bacterial
lectin RegIIIγ and protect mice against intestinal Listeria
monocytogenes infection, J. Exp. Med., 204,
1891-1900.
37.Otto, M. (2006) Bacterial evasion of
antimicrobial peptides by biofilm formation, Curr. Top. Microbiol.
Immunol., 306, 251-258.
38.Bevins, C. L., and Ganz, T. (2004) Antimicrobial
peptides of the alimentary tract of animals, in Mammalian Host
Defense Peptides (Diamond, G., Laute, D., and Klein-Paul, M., eds.)
Cambridge University, UK, pp. 161-188.
39.Scott, M. G., Davidson, D. J., Gold, M. R.,
Bowdish, D., and Hancock, R. E. W. (2002) The human antimicrobial
peptide LL-37 is a multifunctional modulator of innate immune
responses, J. Immunol., 169, 3883-3891.
40.Masuda, K., Nakamura, K., Yoshioka, S., Fukaya,
R., Sakai, N., and Ayabe, T. (2011) Regulation of microbiota by
antimicrobial peptides in the gut, Adv. Otorhinolaryngol.,
72, 97-99.
41.Weaver, C. T., and Hatton, R. D. (2009) Interplay
between the Th17 and Treg cell lineages: a (co)evolutionary
perspective, Nat. Rev. Immunol., 9, 883-889.
42.Matzinger, P. (2007) Friendly and dangerous
signals: is the tissue in control? Nat. Immunol., 8,
11-13.
43.Turner, J. R. (2009) Intestinal mucosal barrier
function in health and disease, Nat. Rev. Immunol., 9,
799-809.
44.Pickard, J. M., and Chervonsky, A. V. (2010)
Sampling of the intestinal microbiota by epithelial M cells,
Curr. Gastroenterol. Rep., 12, 331-339.
45.Everett, M. L., Palestrant, D., Miller, S. E.,
Bollinger, R. R., and Parker, W. (2004) Immune exclusion and immune
inclusion: a new model of host–bacterial interactions in the gut,
Clin. Appl. Immunol. Rev., 4, 321-332.
46.Macia, L., Thorburn, A. N., Binge, L. C., Marino,
E., Rogers, K. E., Maslowski, K. M., Vieira, A. T., Kranich, J., and
Mackay, C. R. (2012) Microbial influences on epithelial integrity and
immune function as a basis for inflammatory diseases, Immunol.
Rev., 245, 164-176.
47.Kruglov, A. A., Grivennikov, S. I., Kuprash, D.
V., Winsauer, C., Prepens, S., Seleznik, G. M., Ebert, G., Littman, D.
R., Heikenwalder, M., Tumanov, A. V., and Nedospasov, S. A. (2013)
Nonredundant function of soluble LTα3 produced by innate lymphoid
cells in intestinal homeostasis, Science, 342,
1243-1246.
48.Pearson, C., Uhlig, H. H., and Powrie, F. (2012)
Lymphoid microenvironments and innate lymphoid cells in the gut,
Trends Immunol., 33, 289-296.
49.Sonnenberg, G. F., Fouser, L. A., and Artis, D.
(2011) Border patrol: regulation of immunity, inflammation and tissue
homeostasis at barrier surfaces by IL-22, Nat. Immunol.,
12, 383-390.
50.Macpherson, A. J., Geuking, M. B., and McCoy, K.
D. (2012) Homeland security: IgA immunity at the frontiers of the body,
Trends Immunol., 33, 160-167.
51.Hepworth, M. R., Montichelli, L. A., Fung, T. C.,
Ziegler, C. G. K., Grunberg, S., Sinha, R., Mantegazza, A. R., Ma,
H.-L., Crawford, A., Angelosanto, J. M., Wherry, E. J., Koni, P. A.,
Bushman, F. D., Elson, C. O., Eberl, G., Artis, D., and Sonnenberg, G.
F. (2013) Innate lymphoid cells regulate CD4+ T-cell responses to
intestinal bacteria, Nature, 498, 113-117.
52.Smith, P. D., Smythies, L. E., Shen, R.,
Greenwell-Wild, T., Gliozzi, M., and Wahl, S. M. (2011) Intestinal
macrophages and response to microbial encroachment, Immunology,
4, 31-42.
53.Honda, K., and Takeda, K. (2009) Regulatory
mechanisms of immune responses to intestinal bacteria, Mucosal
Immunol., 2, 187-196.
54.Pena, J. A., and Versalovic, J. (2003)
Lactobacillus rhamnosus GG decreases TNF-α production in
lipopolysaccharide-activated murine macrophages by contact-independent
mechanism, Cell. Microbiol., 5, 277-285.
55.Iliev, I. D., Mileti, E., Matteoli, G., Chieppa,
M., and Rescigno, M. (2009) Intestinal epithelial cells promote
colitis-protective regulatory T-cell differentiation through dendritic
cell conditioning, Mucosal Immunol., 2, 340-350.
56.Mantis, N. J., Rol, N., and Corthesy, B. (2011)
Secretory IgA’s complex roles in immunity and mucosal homeostasis
in the gut, Mucosal Immunol., 4, 603-611.
57.Han, D., Walsh, M. C., Cejas, P. J., Dang, N. N.,
Kim, Y. F., Kim, J., Charrier-Hisamuddin, L., Chau, L., Zhang, Q.,
Bittinger, K., Bushman, F. D., Turka, L. A., Shen, H., Reizis, B.,
DeFranco, A. L., Wu, G. D., and Choi, Y. (2013) Dendritic cell
expression of the signaling molecule TRAF6 is critical for gut
microbiota-dependent immune tolerance, Immunity, 28,
1-12.
58.Pabst, O. (2012) New concepts in the generation
and functions of IgA, Nat. Rev. Immunol., 25,
139-143.
59.Snoeck, V., Peters, I. E., and Cox, E. (2006) The
IgA system: a comparison of structure and function in different
species, Vet. Res., 37, 455-467.
60.He, B., Xu, W., Santini, P. A., Polydorides, A.
D., Chiu, A., Estrella, J., Shan, M., Shadbun, A., Villanacci, V.,
Plebani, A., Knowles, D. M., Rescigno, M., and Cerutti, A. (2007)
Intestinal bacteria trigger T-cell-independent IgA2 class switching by
inducing epithelial cell secretion of the cytokine APRIL,
Immunity, 26, 812-826.
61.Brandtzaeg, P. (2013) Secretory IgA: designed for
anti-microbial defense, Front. Immunol., 4, 1-17.
62.Attardo, G. M., Lohs, C., Heddi, A., Alam, U. H.,
Yildirim, S., and Aksoy, S. (2008) Analysis of milk gland structure and
function in Glossina morsitans: milk protein production,
symbiont populations and fecundity, Insect. Physiol., 54,
1236-1242.
63.Cabrera-Rubio, R., Collado, M. C., Laitinen, K.,
Salminen, S., Isolauri, E., and Mira, A. (2012) The human milk
microbiome changes over lactation and is shaped by maternal weight and
mode of delivery, Am. J. Clin. Nutr., 96, 544-551.
64.Field, C. J. (2005) The immunological components
of human milk and their effect on immune development in infants, J.
Nutr., 135, 1-4.
65.Chirico, G., Marzollo, R., Cortinovis, S., Fonte,
C., and Gasparoni, A. (2008) Antiinfective properties of human milk,
J. Nutr., 138, 1801S-1806S.
66.Arvola, M., Gustafsson, E., Svensson, L.,
Jansson, L., Holmdahl, R., Heyman, B., Okabe, M., and Mattsson, R.
(2000) Immunoglobulin-secreting cells of maternal origin can be
detected in B cell-deficient mice, Biol. Reprod., 63,
1817-1824.
67.Mathias, A., and Corthesy, B. (2011) N-glycans on
secretory component. Mediators of the interaction between secretory IgA
and Gram-positive commensals sustaining intestinal homeostasis, Gut
Microbes, 2, 287-293.
68.Oda, H., Wakabayashi, H., Yamauchi, K., and Abe,
F. (2014) Lactoferrin and bifidobacteria, Biometals, 27,
915-922.
69.Husson, M. O., Legrand, D., Spik, G., and
Leclerc, H. (1993) Iron acquisition by Helicobacter pylori:
importance of human lactoferrin, Infect. Immun., 61,
2694-2697.
70.Blaser, M. (2011) Stop the killing of beneficial
bacteria, Nature, 476, 393-394.
71.Chen, Y., and Blaser, M. J. (2007) Inverse
associations of Helicobacter pylori with asthma and allergy,
Arch. Intern. Med., 167, 821-827.
72.He, Y., Liu, S., Leone, S., and Newburg, D. S.
(2014) Human colostrum oligosaccharides modulate major immunologic
pathways of immature human intestine, Mucosal Immunol., doi:
10.1038/mi.2014.20 (Epub ahead of print).
73.Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaheh,
F., Edberg, S., and Medzhitov, R. (2004) Recognition of commensal
microflora by Toll-like receptors is required for intestinal
homeostasis, Cell, 118, 229-241.
74.Goto, Y., and Ivanov, I. I. (2013) Intestinal
epithelial cells as mediators of the commensal–host immune
crosstalk, Immunol. Cell Biol., 91, 204-214.