REVIEW: Molecular Barriers to Processes of Genetic Reprogramming and Cell Transformation
I. V. Chestkov1*, E. A. Khomyakova1, E. A. Vasilieva1, M. A. Lagarkova1,2*, and S. L. Kiselev1,3
1Vavilov Institute of General Genetics, Russian Academy of Sciences, ul. Gubkina 3, 119991 Moscow, Russia; fax: +7 (499) 135-4326; E-mail: ichestkov@vigg.ru2Research Institute of Physical-Chemical Medicine, Federal Medical-Biological Agency, ul. Malaya Pirogovskaya 1a, 119435 Moscow, Russia; fax: +7 (499) 246-4409; E-mail: maryalag@yahoo.com
3Kazan (Volga Region) Federal University, ul. Kremlevskaya 18, 420008 Kazan, Russia; fax: +7 (843) 292-4448; E-mail: public.mail@kpfu.ru
* To whom correspondence should be addressed.
Received June 25, 2014
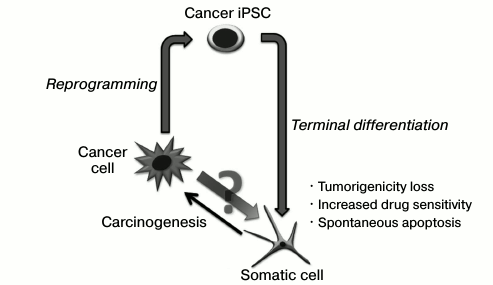
Genetic reprogramming by ectopic expression of transcription factor genes induces the pluripotent state in somatic cells. This technology provides an opportunity to establish pluripotent stem cells for each person, as well as to get better understanding of epigenetic mechanisms controlling cell state. Interestingly, some of the molecular processes that accompany somatic cell reprogramming in vitro are also characteristic for tumor manifestation. Thus, similar “molecular barriers” that control the stability of epigenetic state exist for both processes of pluripotency induction and malignant transformation. The reprogramming of tumor cells is interesting in two aspects: first, it will determine the contribution of epigenetic changes in carcinogenesis; second, it gives an approach to evaluate tumor stem cells that are supposed to form the entire cell mass of the tumor. This review discusses the key stages of genetic reprogramming, the similarity and difference between the reprogramming process and malignant transformation.
KEY WORDS: genetic reprogramming, transformation, tumor, epigenetics, induced pluripotent stem cellsDOI: 10.1134/S0006297914120037
DIFFERENT COMBINATIONS OF TRANSCRIPTION FACTORS CAN ALTER FATE OF SOMATIC CELLS
The Nobel Prize in physiology and medicine in 2012 was awarded to J. Gurdon and S. Yamanaka for the development of technologies for reprogramming of adult organism somatic cells into their pluripotent state. The technology of Gurdon is based on the fact that an oocyte contains all factors necessary for reprogramming of the somatic cell genome, while placed into the oocyte as an embryonic cell genome [1]. The technology developed by Yamanaka is based on the discovery of increased expression of four transcription factors combination – Oct3/4, Sox2, Klf4, and c-Myc – in the somatic cell transforming the latter into the embryonic state [2]. No doubt, both technologies opened a new era in study of the mechanisms of pluripotent stem cell self-maintenance and properties and also advanced the possibility of the use of pluripotent cells in regenerative medicine. It is necessary to note that the discovery of a transcription factor combination, so-called “Yamanaka cocktail”, in great degree was predetermined by the study showing that a terminally differentiated cell can be converted (trans-differentiated) into a somatic cell of another tissue type under ectopic expression of transcription factors. For example, expression of the MyoD gene is sufficient for development of mouse embryonic fibroblasts into myoblasts [3]. Thus, in the presence of the corresponding expression profile in a somatic cell, alteration of its epigenetic state can occur with participation of a minimum number of transcription factors. Some examples: as mentioned above, the MyoD gene is a master gene in myoblasts; exocrine cells of mouse pancreatic gland can be converted into β-cells under the action of three genes (Ngn3, Pdx1 and Mafa) [4]; ectopic expression of Brn2 (also known as Pou3f2), Ascl1, and Myt1l transcription factors leads to trans-differentiation of mouse fibroblasts into matured neurons [5]; elevated expression of only a single tissue-specific gene is sufficient for conversion of liver cells into insulin-producing cells (Pdx1 gene) [6], and callosal neurons into corticospinal motor neurons (Fezf2 gene) [7]. It is necessary to note that somatic cell trans-differentiation can be observed in vitro as well as in vivo [4, 6, 7].
The principal difference of the above-listed trans-differentiations from reprogramming is a definite type of cell as a substrate is necessary for trans-differentiation, i.e. a certain combination of the expression profile and the cell epigenetic state, while the four factors of the reprogramming process are universal, and they allow induction of the pluripotent state in practically any type of somatic cell [2, 8-12].
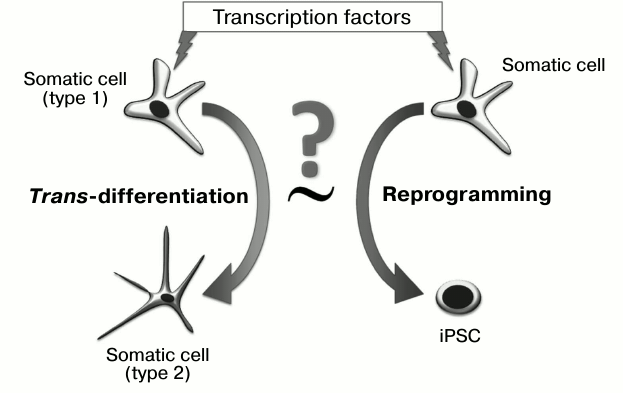
The use of a combination of transcription factors has allowed “time trans-differentiation” and obtaining of the unique resource of all human cell types – induced pluripotent stem cells (iPSC). We will consider below whether the process of pluripotent state formation represents an example of directed trans-differentiation (Fig. 1), and if certain stages of reprogramming can take place in the development of tumors.
Fig. 1. Are the processes of trans-differentiating and reprogramming equivalent to each other? To realize both processes, transcription factors are introduced into a cell, which change gene expression profiles and induce the epigenetic state.
Genetic reprogramming, in contrast to tumor transformation, is a directed process. Only one component of the Yamanaka cocktail – Oct3/4 – is a master gene of the pluripotent state [13]; however, to self-maintain and control the epigenetic state of embryonic stem cells (ESC), cooperation of this transcription factor with Sox2 and their joint participation in the heterodimer complex is necessary [14]. Moreover, during regulation of a number of pluripotent state genes the Oct3/4-Sox2 complex also interacts with Klf4 transcription factor [15] that, as shown earlier, participates in cell cycle regulation and proliferation processes. Klf4 inhibits transcription of p53 that causes the antiapoptotic effect [16]; however, simultaneously, interaction of Klf4 and p53 positively regulates expression of p21CIP1 transcription factor [17].
The c-Myc transcription factor is of special interest, whose role has recently been considered to be the amplifier of the reprogramming process. Indeed, this factor is not obligatory for pluripotent state induction, and reprogramming can occur with participation of only Oct3/4, Sox2, and Klf4, but the efficacy of the process decreases dramatically [18, 19]. It is well known that c-Myc participates in the regulation of many genes and its increased expression causes cell proliferation and increased transformation. This protein participates in cell cycle regulation through inhibiting the earlier-mentioned p21CIP1 [20]. Thus, the common role of Klf4 and c-Myc transcription factors in reprogramming is p53-p21 signal pathway inhibition, which leads to antiapoptotic effect and blocking of cell arrest in G1/S phase. This was partially confirmed in experiments on reprogramming dynamics. Analysis of different stages of somatic cell reprogramming in mice led to a conclusion regarding the existence of two phases in gene expression changing under pluripotent state induction. During the first phase – activation of genes involved in cell proliferation, metabolism, and cytoskeleton formation – reduction in development-associated gene expression takes place. During the second phase, the genes of early embryonic development and iPSC pluripotent state maintenance were activated. These conclusions have been confirmed by three independent full-genome characterization of cells: coding gene expression profile, non-coding microRNAs, and alteration of histone modification characteristic for active/inactive chromatin. Interestingly, the activation of the “first wave” genes (day 3-6 of reprogramming) was determined by ectopic expression of Klf4 and c-Myc transcription factors, while alteration of gene expression during the “second wave” was directed by Oct3/4, Sox2, and Klf4 factors (day 9-12). It was suggested that Klf4 plays dual role during transition of a somatic cell to the pluripotent state. Surprisingly, at the first stages of reprogramming (day 3) a cell population unable to induce pluripotency during further cultivation has already been defined. A few possible reasons for the existence of such cells have been suggested: 1) inability to pass the mesenchymal–epithelial transition; 2) aberrant activation of differentiation genes and genes associated with immune response; 3) inability to maintain directed global alteration of gene expression [21]. So, the transition of cells into the pluripotent state took place only during the second phase of the reprogramming process, under activation of early embryonic development genes. It becomes clear that the reprogramming process is not the reverse of the differentiation process in the course of ontogenesis.
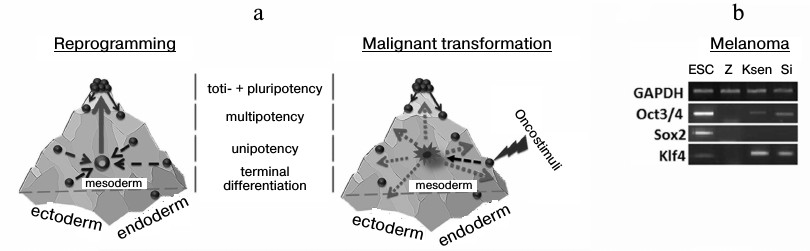
Conrad Waddington, when offering the term “epigenetics”, suggested the concept of an epigenetic surface (epigenetic landscape) along which cells roll, performing an ontogenetic program [22]. Developing his approach, it is possible to suggest a spatial model of ontogenesis in the form of a combination of three surfaces, each of which reflects the differentiation of cells inside one germ layer (Fig. 2a). Under the action of the ectopic expression of Klf4 and c-Myc, the cell transits into an epigenetic state equally distance from each side of such a pyramid or closer to the state of the initial cell. New epigenetic states are extremely unstable, and it is not possible to compare them with any phenotype during the development of the organism. To abandon such a state, the cell needs a stimulus, and expression of Oct3/4, Sox2, and Klf4 transcription factors represents such a stimulus in genetic reprogramming.
Fig. 2. Three-dimensional models of an epigenetic state of cells under reprogramming and transformation. Development of an organism is accompanied by reduction of the ability of cells for differentiation like sliding down from a mountain slope. At the same time, cell specialization is performed in the frame of one of the germ layers – ectoderm, endoderm, or mesoderm. Two phases exist in the change of a gene expression profile for a somatic cell on its way toward the pluripotent state: phase I (a: depicted in dashed arrows) induced by Klf4 and c-Myc transcription factors, and phase II that is characterized by activation of early embryonic development under the influence of Oct3/4, Sox2, and Klf4 transcription factors (a: depicted as a solid arrow). During malignant transformation in the absence of a determined phase II, there is stochastic alteration of gene expression (depicted as dotted arrows), and partial activation of early embryonic development genes, for example, in human melanoma lines (b).
A similar model can also be considered in the case of malignant cell transformation. A cell leaves its epigenetic surface under transforming events and acquires a new destabilized phenotype (first phase). It was thought earlier that not less than ten changes must simultaneously occur to transform a cell, but, taking into account that elevated expression of only 1-three transcription factors is enough for trans-differentiation [3-7], and the physical environmental conditions promote the reprogramming process [23], then the number of destabilizing factors per cell may be less than ten. In this situation, in contrast to the reprogramming process, there are no determined stimuli for the cell to leave the state (absence of a second phase), this resulting in stochastic activation or silencing of genes. So, we and others reveal Oct3/4 and Klf4 transcription during analysis of primary lines of tumor cells obtained from patients (Fig. 2b) [24]. However, in this case it is incorrect to suggest the similarity of tumor cells to the pluripotent state. Moreover, the presence of a transcript in a tumor cell is not always evidence for correct functioning of a studied gene because: 1) a gene may contain genetic disruptions, and 2) splicing variants and pseudogenes may be expressed. The latter situation was shown in case of the Nanog gene, which has three paralogs in the human genome that code a full protein sequence. At the same time, in the case of human ESC, mainly Nanog1 (whose role is to maintain the pluripotent state [25]) is transcribed and, partly, Nanog2. In most analyzed tumor lines (14 of 17) expression of NanogP8 was observed, although the total content of Nanog gene transcripts was lower than in pluripotent cells [26].
Thus, in accordance with the model depicted in Fig. 2, it is suggested that the tumor cell expression profile arises randomly as the result of searching for the epigenetic niche and the most stable state. Similar “epigenetic errors” arise not only as the result of malignant transformation, but also in the presence of a determining stimulus, for example, during genetic reprogramming [27]. The overall efficiency of the process, which does not exceed 1% for skin fibroblasts during the use of the classic “Yamanaka cocktail”, may indicate this. Importantly, in the case of malignant transformation, high proliferative potential of cancer cells leads to instability of the genetic material, which expands the epigenetic landscape of the tumor. Thus, the stochastic nature of gene activation/silencing in cancer cells takes place not only in the initial period, but also during every cell division (as evidenced by constant subclone formation in tumor cell lines and in some tumors).
TUMOR-SUPPRESSOR GENES AND INHIBITORS OF APOPTOSIS
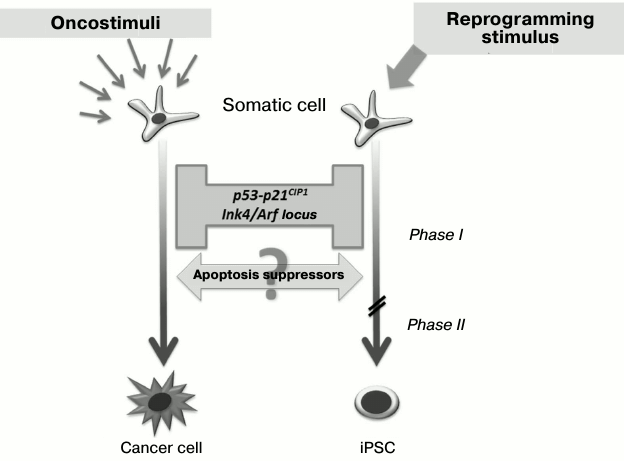
As discussed earlier, the role of Klf4 and c-Myc transcription factors as members of the “Yamanaka cocktail” in the first stages of reprogramming may be linked with double inhibition of the p53-p21 signaling pathway. Interestingly, this signaling pathway is one of the main barriers on the way to the pluripotent state [28-32]. Thus, in the case of reduction of p53 or p21CIP1 expression with the use of shRNA, there is three-fold increase in the reprogramming efficiency, and in case of p53–/–cells it is 10-fold increase. Moreover, in such p53 inhibition somatic cells can be reprogrammed by only two factors – Oct3/4 and Sox2 [28]. During the reprogramming process, there is activation of one of the key p53-associated locus – Ink4/Arf, whose inhibition leads to significant increase in the reprogramming efficiency [30].
Little is known about the influence of protein inhibitors of apoptosis (IAPs) on the efficiency of somatic cell transition into the pluripotent state. Nevertheless, it was found under increased expression of antiapoptotic Bcl-2 protein that reduced cell death during reprogramming also increased the efficiency of the process four-fold [28]. Interestingly, similar tumor-suppressor genes’ silencing and apoptosis suppressors’ activation accompany also the malignant transformation of cells during carcinogenesis (Fig. 3), and regulation of such processes may occur at the genetic as well as the epigenetic level [33-37]. Thus, a somatic cell overcomes similar “molecular barriers” in reprogramming as well as developing cancer, but the first process is directed and leads to an epigenetically stable state without genetic and malignant transformation.
Fig. 3. Barriers and drivers of genetic reprogramming and malignant transformation of cells. Inhibition of the p53-p21 signaling pathway and associated tumor suppressors increases efficiency of somatic cell reprogramming. At the same time, activation of apoptosis suppressors promotes genetic reprogramming as well as malignant transformation.
CANCER CELL REPROGRAMMING
There is still no answer to the question of what makes the key contribution to cell direction to malignant transformation. No doubt, differing from the differentiation processes in ontogenesis, genetic changes play key roles in carcinogenesis. Nevertheless, recent studies demonstrate the importance of epigenetic changes in cell acquisition of tumor phenotype [24, 33, 38], giving rise to hopes for identifying a real impact on such epimutations in cancer cells. There are drugs that influence the cancer cell epigenome, for example, CHR-3996 [39], a compound belonging to the class of inhibitors of histone deacetylases. Nevertheless, the question about the possibility of converting a tumor cell into a normal cell through global impact on its epigenetic status is still open.
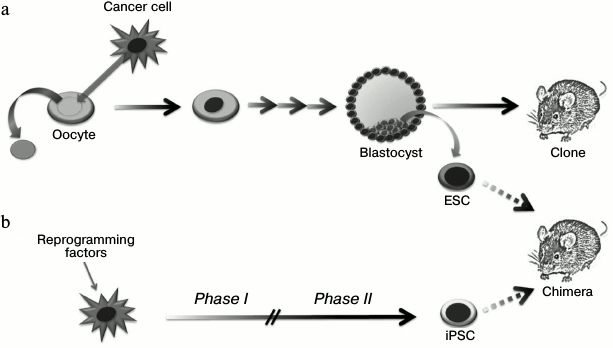
A set of experiments on tumor genome reprogramming have been performed using the somatic cell nuclear transfer technology (Fig. 4 and Table 1). In reprogramming of a nucleus of a normal somatic cell, there is a cascade of epigenetic alterations including changes in DNA methylation and histone modification levels, which allow the donor nucleus to transit into the embryonic state. Blastocysts obtained from such modified oocyte can be implanted into a surrogate female. Then an organism identical to the initial nucleus donor cell is developed (“cloning”) [40]. At the same time, internal blastocyst cells can be replaced in vitro to prepare stable ESC lines, introduction of which into recipient blastocysts leads to formation of a chimeric organism [41]. Thus, the technology of somatic cell nuclear transfer allows altering the epigenetic state of a cell without disruption of the nuclear genetic material. It is necessary to note that efficiency of blastocyst preparation using this technology depends strictly on the methylation status and the donor nucleus differentiation potential: thus, the efficiency of neuronal progenitor cell reprogramming reaches 50% [42], while reprogramming efficiency in terminally differentiated cells such as lymphocytes [43], NK cells [44], and neurons [45] does not exceed 10%.
Fig. 4. Application of nuclei transfer technology (a) and ectopic expression of transcription factors (b) for making changes in the epigenetic status of tumor cells and studying their epigenomic plasticity.
Table 1. Tumor cell reprogramming with the
use of nuclei transfer technology into an anucleated oocyte

It was first shown in experiments on frogs that kidney carcinoma cell nuclei can be reprogrammed through their transfer into an anucleated oocyte, and embryonic development is continued until the tadpole stage [46]. Then similar results were obtained in mice on transplantation of medulloblastoma cell nuclei. However, here there was incomplete formation of embryos due to development arrest [47]. Hochedlinger et al. demonstrated in 2004 that anucleated oocytes are capable of development into pre-implantation blastocyst after introduction of genetic material from cells of different cancer models, but genetic material from primary tumor cells did not possess such a property. Efficiency of blastocyst development from genetic material of different cancer model cells was not more than 13% of oocytes surviving after introduction of nuclei. It was shown that only in case of transplantation of R545 melanoma cell nuclei, the ESC line was obtained (from formed blastocysts) that not only formed in immunodeficient mice teratomas containing cells of three germ layers, but also colonized chimeric animal tissues after injection into recipient blastocysts [48]. However, it was impossible to develop a whole organism from R545-ESC only, although key stages of organogenesis at embryonic development stage E9.5 were revealed. It is necessary to note that the melanoma R545 cell line studied in the work was obtained with the use of the inducible system of H-RasV12G gene expression in murine cells (genotype: Tyr-rtTA+, Tet-RAS+, ink4a/Arf−/−) [49]. In addition to ink4a/Arf locus deletion, a trisomy of chromosomes 8 and 11 was revealed in the studied cell line. According to the authors, such genetic disruptions may represent the barrier for formation of the entire organism from the R545-ESC only [48]. Later it was demonstrated that reprogramming by cell nuclear transfer of teratocarcinoma cells does not lead to alteration of the epigenetic state of cells, and this was shown while comparing cell ability for differentiation and formation of a chimeric organism [50].
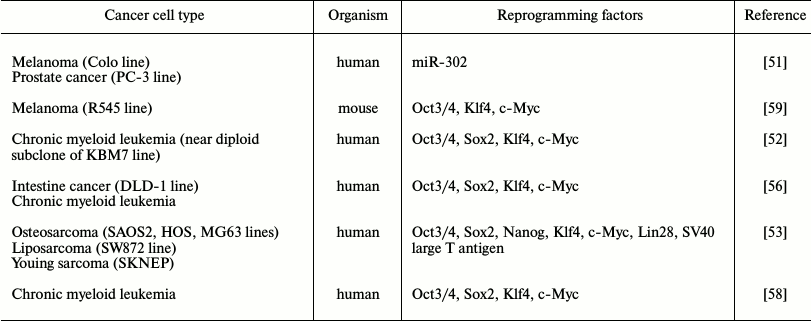
The famous work of Takahashi and Yamanaka published in 2006 highlighted a new era in the field of cell reprogramming, and it attracted researchers with a perspective of this new approach for studying carcinogenesis (Table 2). It was shown in 2008 that after introduction of miR-302 microRNA into melanoma cells (Colo line), pancreatic gland cancer cells (PC3 line), it is possible to induce expression of ESC-associated genes and conduct global demethylation of the tumor genome. A cell obtained as the result of reprogramming formed teratoma-like cysts with origins of three germ layers in immunodeficient mice [51]. It is necessary to note that results of this work have not yet been reproduced in other laboratories. Further, a few groups of researchers have demonstrated the possibility of tumor genome reprogramming using transcription factors (Table 2). It is important to note that it is necessary to account for heterogeneity of the initial cancer cell culture during characterization of reprogrammed transformed cells due to the ability of cells for subcloning. Thus, it is likely to compare an obtained iPSC with a subclone of the cell that was reprogrammed and demonstrate the loss of tumorigenic potential.
Table 2. Tumor cell reprogramming examples
using ectopic expression of transcription factors and microRNA
Is it possible to reprogram cancer cells to induced pluripotent state like somatic cells? How “labile” is the epigenetic state of cancer cells? Answers for these questions are determined by a number of genetic changes leading to cell transformation. Thus, it was demonstrated in 2010 that only cells of chronic myeloid leukemia KBM7 line subclone, possessing a near diploid set of chromosomes, are able to induce the pluripotent state on introduction of all four components of the “Yamanaka cocktail” [52]. Further, it was revealed that the tumor genome reprogramming (for a few lines of human sarcoma) proceeds without achieving the clearly pluripotent state, and the epigenetic status of cells is fixed near the multipotent state that was sufficient for cells to lose their malignant properties [53]. It is necessary to note that during the use of nucleus transfer technology, alteration of epigenetic status of the somatic cell genome is achieved by the action of multiple factors contained in oocyte cytoplasm, while the genetic reprogramming process can be performed with the use of only four transcription factors and this may be insufficient for removal of the tumor epigenetic markers and pluripotent state induction.
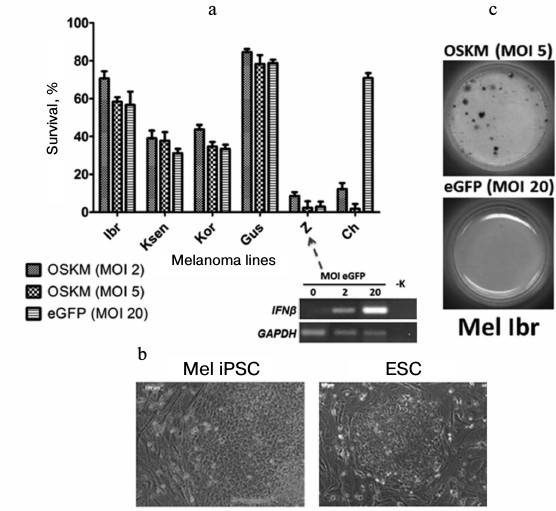
For example, when we performed genetic reprogramming of human melanoma primary lines [54], we revealed different behavior of cells on the fifth day after introduction of genes of four transcription factors (Oct3/4, Sox2, c-Myc, Klf4) with the use of recombinant lentivirus particles. Some tumor cells (Mel Ibr, Mel Kor, Mel Ksen, Mel Gus lines) overcame initial stages of the reprogramming process, but in the case of Mel Ibr and Mel Kor lines only formation of ESC-like compact colonies and increase in endogenous alkaline phosphatase activity took place, i.e. processes similar to somatic cell reprogramming were observed (Fig. 5). Interestingly, mass death of cells was observe in some cases as the result of: 1) inability to overcome the interferon response (Mel Z line) in lentivirus infection (such treatment now possibly can be used in case of leukemia [55]); 2) influence of reprogramming genes themselves (that may be linked with inability to overcome the reprogramming phase I; Mel Ch line). However, the epigenetic status of cells obtained from the reprogramming process was unstable, which was accompanied by characteristic morphology loss during 2-3 cultivation passages. At the same time, during the reprogramming of teratoma (formed from iPSC or ESC by injection of these cells into immune deficient mice) cells, there was highly effective formation of iPSC colonies that expressed the pluripotent state markers and were able to differentiate into derivatives of three germ layers. This means that these malformations formed by human pluripotent stem cells in immunodeficient mice in essence are not tumor cells because any somatic cells are readily reprogrammed.
Fig. 5. Primary lines of human melanoma demonstrate different survival on the fifth day after introduction of reprogramming genes (a). Nevertheless, compact colonies are formed in a few cases that are similar to human ESC (b), and activation of alkaline phosphatase takes place in some of them (c).
How can the reprogramming process help in study of carcinogenesis and cancer treatment? In 2010, while preparing differentiated derivatives from iPSC of intestine cancer line DLD-1, researchers revealed an increase in cell sensitivity to the chemotherapeutic agent 5-FU compared with the initial tumor cells [56]. Thus, on altering the epigenetic state of cancer cells it is possible to more effectively apply known medicinal preparations. On the other hand, some works demonstrate that the cells obtained as the result of reprogramming/differentiation of cancer cells are more resistant to currently used therapeutic agents [52, 57, 58]. In particular, differentiated derivatives of iPSC from chronic myeloid leukemia cells, such as neuronal and fibroblast-like cells, but not blood cells, were insensitive to imatinib – the inhibitor of tyrosine kinases used for treatment of BCR-ABL-associated myeloleucosis, proving that the medicinal agent influences cells that are in a definite epigenetic state only [52, 58]. Thus, changing the tumor cell epigenetic state remains a promising direction for choosing therapeutic means. Searching and identification of master genes specific for every type of somatic cells may convert cancer cells to the initial epigenetic state with loss of malignant phenotype (Fig. 6).
Fig. 6. Plausible technologies for correcting cancer cells epigenome with using reprogramming/differentiation or introduction of tissue-specific master genes of somatic cells.
Directed preparation of different types of somatic cells by trans-differentiation is one of the most perspective directions of modern cell biology. However, to trans-differentiate various types of somatic cells, it is necessary to identify key master genes inducing directed change in the epigenetic state of each cell type.
Pluripotent stem cells are a unique resource for preparation of specialized cells of an organism because they are able to proliferate unlimitedly and at the same time are stable in vitro. The technology of genetic reprogramming allows preparing pluripotent cells for each individual and from practically any available cells of the organism.
Acquisition of unlimited proliferative potential by cells proceeds as the result of malignant transformation of cells; however, it is accompanied by a loss of stability of their epigenetic state. This is evidenced, for example, by heterogeneity of cells inside a tumor. Such heterogeneity may be linked with the fact that a partial de-differentiation of somatic cells or transformation takes place in the progenitor cells of the organism may proceed during malignant transformation. Heterogeneity of a tumor may also be formed due to non-directed processes such as stochastic activation/silencing of genes during epigenomic destabilization, but a link with initial cell state is preserved. Such destabilization takes place also at initial stages of genetic reprogramming under the action of Klf4 and c-Myc transcription factors, although this process is directed.
It can be suggested that the malignant potential of cancer cells might be overcome by directed alteration and epigenetic state stabilization. Silencing of gene suppressors of tumor development and activation of apoptosis suppressors that accompany the cell transformation in carcinogenesis must promote the genetic programming of tumor cells. Nevertheless, only a small number of works that show the possibility to obtain induced pluripotent stem cancer cells has been published. Possibly, genetic disruptions may be a barrier for whole tumor genome remodeling and induction of the pluripotent state.
This work was supported by the Foundation for Promotion of Development of Small Enterprises in the Field of Research and Development within the Program “Umnik” (I. V. C.) and the Program of the Presidium of the Russian Academy of Sciences “Dynamics and Preservation of Genetic Reserves”.
REFERENCES
1.Gurdon, B. (1962) The developmental capacity of
nuclei taken from intestinal epithelium cells of feeding tadpoles,
J. Embryol. Exp. Morphol., 10, 622-640.
2.Takahashi, K., and Yamanaka, S. (2006) Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors, Cell, 126, 663-676.
3.Davis, R. L., Weintraub, H., and Lassar, A. B.
(1987) Expression of a single transfected cDNA converts fibroblasts to
myoblasts, Cell, 51, 987-1000.
4.Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J.,
and Melton, D. A. (2008) In vivo reprogramming of adult
pancreatic exocrine cells to beta-cells, Nature, 455,
627-632.
5.Pang, Z. P., Yang, N., Vierbuchen, T., Ostermeier,
A., Fuentes, D. R., Yang, T. Q., Citri, A., Sebastiano, V., Marro, S.,
Sudhof, T. C., and Wernig, M. (2011) Induction of human neuronal cells
by defined transcription factors, Nature, 476,
220-223.
6.Ferber, S., Halkin, A., Cohen, H., Ber, I., Einav,
Y., Goldberg, I., Barshack, I., Seijffers, R., Kopolovic, J., Kaiser,
N., and Karasik, A. (2000) Pancreatic and duodenal homeobox gene 1
induces expression of insulin genes in liver and ameliorates
streptozotocin-induced hyperglycemia, Nat. Med., 6,
568-572.
7.Rouaux, C., and Arlotta, P. (2013) Direct lineage
reprogramming of post-mitotic callosal neurons into corticofugal
neurons in vivo, Nat. Cell. Biol., 15,
214-221.
8.Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M.,
Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007) Induction of
pluripotent stem cells from adult human fibroblasts by defined factors,
Cell, 131, 861-872.
9.Aasen, T., Raya, A., Barrero, M. J., Garreta, E.,
Consiglio, A., Gonzalez, F., Vassena, R., Bilic, J., Pekarik, V.,
Tiscornia, G., Edel, M., Boue, S., and Izpisua Belmonte, J. C. (2008)
Efficient and rapid generation of induced pluripotent stem cells from
human keratinocytes, Nat. Biotechnol., 26, 1276-1284.
10.Loh, Y. H., Agarwal, S., Park, I. H., Urbach, A.,
Huo, H., Heffner, G. C., Kim, K., Miller, J. D., Ng, K., and Daley, G.
Q. (2009) Generation of induced pluripotent stem cells from human
blood, Blood, 113, 5476-5479.
11.Lagarkova, M. A., Shutova, M. V., Bogomazova, A.
N., Vassina, E. M., Glazov, E. A., Zhang, P., Rizvanov, A. A.,
Chestkov, I. V., and Kiselev, S. L. (2010) Induction of pluripotency in
human endothelial cells resets epigenetic profile on genome scale,
Cell Cycle, 9, 937-946.
12.Philonenko, E. S., Shutova, M. V., Chestkov, I.
V., Lagarkova, M. A., and Kiselev, S. L. (2011) Current progress and
potential practical application for human pluripotent stem cells,
Int. Rev. Cell Mol. Biol., 292, 153-196.
13.Niwa, H., Miyazaki, J., and Smith, A. G. (2000)
Quantitative expression of Oct-3/4 defines differentiation,
dedifferentiation or self-renewal of ES cells, Nat. Genet.,
24, 372-376.
14.Rodda, D. J., Chew, J. L., Lim, L. H., Loh, Y.
H., Wang, B., Ng, H. H., and Robson, P. (2005)Transcriptional
regulation of nanog by OCT4 and SOX2, J. Biol. Chem.,
280, 24731-24737.
15.Nakatake, Y., Fukui, N., Iwamatsu, Y., Masui, S.,
Takahashi, K., Yagi, R., Yagi, K., Miyazaki, J., Matoba, R., Ko, M. S.,
and Niwa, H. (2006) Klf4 cooperates with Oct3/4 and Sox2 to activate
the Lefty1 core promoter in embryonic stem cells, Mol. Cell
Biol., 26, 7772-7782.
16.Rowland, B. D., Bernards, R., and Peeper, D. S.
(2005) The KLF4 tumor suppressor is a transcriptional repressor of p53
that acts as a context-dependent oncogene, Nat. Cell Biol.,
7, 1074-1082.
17.Zhang, W., Geiman, D. E., Shields, J. M., Dang,
D. T., Mahatan, C. S., Kaestner, K. H., Biggs, J. R., Kraft, A. S., and
Yang, V. W. (2000) The gut-enriched Kruppel-like factor (Kruppel-like
factor 4) mediates the transactivating effect of p53 on the
p21WAF1/Cip1 promoter, J. Biol. Chem., 275,
18391-18398.
18.Nakagawa, M., Koyanagi, M., Tanabe, K.,
Takahashi, K., Ichisaka, T., Aoi, T., Okita, K., Mochiduki, Y.,
Takizawa, N., and Yamanaka, S. (2008) Generation of induced pluripotent
stem cells without Myc from mouse and human fibroblasts, Nat.
Biotechnol., 26, 101-106.
19.Wernig, M., Meissner, A., Cassady, J. P., and
Jaenisch, R. (2008) c-Myc is dispensable for direct reprogramming of
mouse fibroblasts, Cell Stem Cell, 2, 10-12.
20.Seoane, J., Le, H. V., and Massague, J. (2002)
Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome
of the p53 response to DNA damage, Nature, 419,
729-734.
21.Polo, J. M., Anderssen, E., Walsh, R. M.,
Schwarz, B. A., Nefzger, C. M., Lim, S. M., Borkent, M., Apostolou, E.,
Alaei, S., Cloutier, J., Bar-Nur, O., Cheloufi, S., Stadtfeld, M.,
Figueroa, M. E., Robinton, D., Natesan, S., Melnick, A., Zhu, J.,
Ramaswamy, S., and Hochedlinger, K. (2012) A molecular roadmap of
reprogramming somatic cells into iPS cells, Cell, 151,
1617-1632.
22.Waddington, C. H. (1957) The Strategy of the
Genes, Geo Allen and Unwin, London.
23.Yoshida, Y., Takahashi, K., Okita, K., Ichisaka,
T., and Yamanaka, S. (2009) Hypoxia enhances the generation of induced
pluripotent stem cells, Cell Stem Cell, 5, 237-241.
24.Suva, M. L., Riggi, N., and Bernstein, B. E.
(2013) Epigenetic reprogramming in cancer, Science, 339,
1567-1570.
25.Chambers, I., Colby, D., Robertson, M., Nichols,
J., Lee, S., Tweedie, S., and Smith, A. (2003) Functional expression
cloning of Nanog, a pluripotency sustaining factor in embryonic stem
cells, Cell, 113, 643-655.
26.Palla, A. R., Piazzolla, D., Abad, M., Li, H.,
Dominguez, O., Schonthaler, H. B., Wagner, E. F., and Serrano, M.
(2013) Reprogramming activity of NANOGP8, a NANOG family member widely
expressed in cancer, Oncogene, 33, 2513-2519.
27.Ohnishi, K., Semi, K., Yamamoto, T., Shimizu, M.,
Tanaka, A., Mitsunaga, K., Okita, K., Osafune, K., Arioka, Y., Maeda,
T., Soejima, H., Moriwaki, H., Yamanaka, S., Woltjen, K., and Yamada,
Y. (2014) Premature termination of reprogramming in vivo leads
to cancer development through altered epigenetic regulation,
Cell, 156, 663-677.
28.Kawamura, T., Suzuki, J., Wang, Y. V., Menendez,
S., Morera, L. B., Raya, A., Wahl, G. M., and Izpisua Belmonte, J. C.
(2009) Linking the p53 tumor suppressor pathway to somatic cell
reprogramming, Nature, 460, 1140-1144.
29.Hong, H., Takahashi, K., Ichisaka, T., Aoi, T.,
Kanagawa, O., Nakagawa, M., Okita, K., and Yamanaka, S. (2009)
Suppression of induced pluripotent stem cell generation by the p53-p21
pathway, Nature, 460, 1132-1135.
30.Li, H., Collado, M., Villasante, A., Strati, K.,
Ortega, S., Canamero, M., Blasco, M. A., and Serrano, M. (2009) The
Ink4/Arf locus is a barrier for iPS cell reprogramming, Nature,
460, 1136-1139.
31.Utikal, J., Polo, J. M., Stadtfeld, M., Maherali,
N., Kulalert, W., Walsh, R. M., Khalil, A., Rheinwald, J. G., and
Hochedlinger, K. (2009) Immortalization eliminates a roadblock during
cellular reprogramming into iPS cells, Nature, 460,
1145-1148.
32.Marion, R. M., Strati, K., Li, H., Murga, M.,
Blanco, R., Ortega, S., Fernandez-Capetillo, O., Serrano, M., and
Blasco, M. A. (2009) A p53-mediated DNA damage response limits
reprogramming to ensure iPS cell genomic integrity, Nature,
460, 1149-1153.
33.Herman, J. G., and Baylin, S. B. (2003) Gene
silencing in cancer in association with promoter hypermethylation,
N. Engl. J. Med., 349, 2042-2054.
34.Popov, N., and Gil, J. (2010) Epigenetic
regulation of the INK4b-ARF-INK4a locus: in sickness and in health,
Epigenetics, 5, 685-690.
35.Van den Hurk, K., Niessen, H. E., Veeck, J., van
den Oord, J. J., van Steensel, M. A., ZurHausen, A., van Engeland, M.,
and Winnepenninckx, V. J. (2012) Genetics and epigenetics of cutaneous
malignant melanoma: a concert out of tune, Biochim. Biophys.
Acta, 1826, 89-102.
36.Portt, L., Norman, G., Clapp, C., Greenwood, M.,
and Greenwood, M. T. (2011) Anti-apoptosis and cell survival: a review,
Biochim. Biophys. Acta, 1813, 238-259.
37.Howell, P. M., Jr., Liu, S., Ren, S., Behlen, C.,
Fodstad, O., and Riker, A. I. (2009) Epigenetics in human melanoma,
Cancer Control, 16, 200-218.
38.Jones, P. A., and Baylin, S. B. (2002) The
fundamental role of epigenetic events in cancer, Nat. Rev.
Genet., 3, 415-428.
39.Moffat, D., Patel, S., Day, F., Belfield, A.,
Donald, A., Rowlands, M., Wibawa, J., Brotherton, D., Stimson, L.,
Clark, V., Owen, J., Bawden, L., Box, G., Bone, E., Mortenson, P.,
Hardcastle, A., van Meurs, S., Eccles, S., Raynaud, F., and Aherne, W.
(2010) Discovery of
2-(6-{[(6-fluoroquinolin-2-yl)methyl]amino}bicyclo[3.1.0]hex-3-yl)-N-hydroxypyrimidine-5-carboxamide
(CHR-3996), a class I selective orally active histone deacetylase
inhibitor, J. Med. Chem., 53, 8663-8878.
40.Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A.
J., and Campbell, K. H. (1997) Viable offspring derived from fetal and
adult mammalian cells, Nature, 385, 810-813.
41.Tam, P. P., and Rossant, J. (2003) Mouse
embryonic chimeras: tools for studying mammalian development,
Development, 130, 6155-6163.
42.Blelloch, R., Wang, Z., Meissner, A., Pollard,
S., Smith, A., and Jaenisch, R. (2006) Reprogramming efficiency
following somatic cell nuclear transfer is influenced by the
differentiation and methylation state of the donor nucleus, Stem
Cells, 24, 2007-2013.
43.Hochedlinger, K., and Jaenisch, R. (2002)
Monoclonal mice generated by nuclear transfer from mature B and T donor
cells, Nature, 415, 1035-1038.
44.Inoue, K., Wakao, H., Ogonuki, N., Miki, H.,
Seino, K., Nambu-Wakao, R., Noda, S., Miyoshi, H., Koseki, H.,
Taniguchi, M., and Ogura, A. (2005) Generation of cloned mice by direct
nuclear transfer from natural killer T cells, Curr. Biol.,
15, 1114-1118.
45.Eggan, K., Baldwin, K., Tackett, M., Osborne, J.,
Gogos, J., Chess, A., Axel, R., and Jaenisch, R. (2004) Mice cloned
from olfactory sensory neurons, Nature, 428, 44-49.
46.McKinnell, R. G., Deggins, B. A., and Labat, D.
D. (1969) Transplantation of pluripotential nuclei from triploid frog
tumors, Science, 165, 394-396.
47.Li, L., Connelly, M. C., Wetmore, C., Curran, T.,
and Morgan, J. I. (2003) Mouse embryos cloned from brain tumors,
Cancer Res., 63, 2733-2736.
48.Hochedlinger, K., Blelloch, R., Brennan, C.,
Yamada, Y., Kim, M., Chin, L., and Jaenisch, R. (2004) Reprogramming of
a melanoma genome by nuclear transplantation, Genes Dev.,
18, 1875-1885.
49.Chin, L., Tam, A., Pomerantz, J., Wong, M.,
Holash, J., Bardeesy, N., Shen, Q., O’Hagan, R., Pantginis, J.,
Zhou, H., Horner, J. W., 2nd, Cordon-Cardo, C., Yancopoulos, G. D., and
DePinho, R. A. (1999) Essential role for oncogenic Ras in tumor
maintenance, Nature, 400, 468-472.
50.Blelloch, R. H., Hochedlinger, K., Yamada, Y.,
Brennan, C., Kim, M., Mintz, B., Chin, L., and Jaenisch, R. (2004)
Nuclear cloning of embryonal carcinoma cells, Proc. Natl. Acad. Sci.
USA, 101, 13985-13990.
51.Lin, S. L., Chang, D. C., Chang-Lin, S., Lin, C.
H., Wu, D. T., Chen, D. T., and Ying, S. Y. (2008) Mir-302 reprograms
human skin cancer cells into a pluripotent ES-cell-like state,
RNA, 14, 2115-2124.
52.Carette, J. E., Pruszak, J., Varadarajan, M.,
Blomen, V. A., Gokhale, S., Camargo, F. D., Wernig, M., Jaenisch, R.,
and Brummelkamp, T. R. (2010) Generation of iPSCs from cultured human
malignant cells, Blood, 115, 4039-4042.
53.Zhang, X., Cruz, F. D., Terry, M., Remotti, F.,
and Matushansky, I. (2013) Terminal differentiation and loss of
tumorigenicity of human cancers via pluripotency-based reprogramming,
Oncogene, 32, 2249-2260.
54.Mikhaylova, I. N., Kovalevsky, D. A., Morozova,
L. F., Golubeva, V. A., Cheremushkin, E. A., Lukashina, M. I.,
Voronina, E. S., Burova, O. S., Utyashev, I. A., Kiselev, S. L.,
Demidov, L. V., Beabealashvilli, R. Sh., and Baryshnikov, A. Y. (2008)
Cancer/testis genes expression in human melanoma cell lines,
Melanoma Res., 18, 303-313.
55.Batenchuk, C., Le Boeuf, F., Stubbert, L., Falls,
T., Atkins, H. L., Bell, J. C., and Conrad, D. P. (2013)
Non-replicating rhabdovirus-derived particles (NRRPs) eradicate acute
leukemia by direct cytolysis and induction of antitumor immunity,
Blood Cancer J., 3, e123.
56.Miyoshi, N., Ishii, H., Nagai, K., Hoshino, H.,
Mimori, K., Tanaka, F., Nagano, H., Sekimoto, M., Doki, Y., and Mori,
M. (2010) Defined factors induce reprogramming of gastrointestinal
cancer cells, Proc. Natl. Acad. Sci. USA, 107, 40-45.
57.Nagai, K., Ishii, H., Miyoshi, N., Hoshino, H.,
Saito, T., Sato, T., Tomimaru, Y., Kobayashi, S., Nagano, H., Sekimoto,
M., Doki, Y., and Mori, M. (2010) Long-term culture following ES-like
gene-induced reprogramming elicits an aggressive phenotype in mutated
cholangiocellular carcinoma cells, Biochem. Biophys. Res.
Commun., 395, 258-263.
58.Kumano, K., Arai, S., Hosoi, M., Taoka, K.,
Takayama, N., Otsu, M., Nagae, G., Ueda, K., Nakazaki, K., Kamikubo,
Y., Eto, K., Aburatani, H., Nakauchi, H., and Kurokawa, M. (2012)
Generation of induced pluripotent stem cells from primary chronic
myelogenous leukemia patient samples, Blood, 119,
6234-6242.
59.Utikal, J., Maherali, N., Kulalert, W., and
Hochedlinger, K. (2009) Sox2 is dispensable for the reprogramming of
melanocytes and melanoma cells into induced pluripotent stem cells,
J. Cell Sci., 122, 3502-3510.