Plant Cell Death Caused by Fungal, Bacterial, and Viral Elicitors: Protective Effect of Mitochondria-Targeted Quinones
D. B. Kiselevsky1, O. Yu. Frolova2, A. G. Solovyev3, Yu. L. Dorokhov3,4, S. Yu. Morozov3, and V. D. Samuilov1*
1Lomonosov Moscow State University, Faculty of Biology, Leninsky Gory 1-12, 119991 Moscow, Russia; fax: +7 (495) 939-4309; E-mail: vdsamuilov@mail.ru2Lomonosov Moscow State University, Institute of Mitoengineering, Leninsky Gory 1-75B, 119991 Moscow, Russia; fax: +7 (495) 939-5945
3Lomonosov Moscow State University, Belozersky Institute of Physico-Chemical Biology, Leninsky Gory 1-40, 119991 Moscow, Russia; fax: +7 (495) 939-0338
4Vavilov Institute of General Genetics, Russian Academy of Sciences, Gubkina st. 3, 119991 Moscow, Russia; fax: +7 (499) 132-8962
* To whom correspondence should be addressed.
Received April 27, 2014
Chitosan (partially deacetylated chitin), a component of fungal cell walls, caused epidermal cell (EC) death in the leaves of pea (Pisum sativum L.) and tobacco Nicotiana tabacum or Nicotiana benthamiana detected by destruction of cell nuclei. The mitochondria-targeted quinone SkQ1 prevented the destruction of EC nuclei induced by chitosan. Chitosan increased and SkQ1 suppressed the activity of protein kinases in N. benthamiana and P. sativum and eliminated the effect of chitosan. Chitosan induced the generation of reactive oxygen species (ROS) in the guard cells (GC) of pea plants. Treatment with chitosan or H2O2 did not cause destruction of GC nuclei; however, it resulted in disruption of the permeability barrier of the plasma membrane detected by propidium iodide fluorescence. Treatment with bacterial lipopolysaccharide but not peptidoglycan caused destruction of pea EC nuclei, which was prevented by SkQ1. Leaves of tobacco plants containing the N gene responsible for resistance to tobacco mosaic virus (TMV) were infiltrated with Agrobacterium tumefaciens cells. These cells contained a genetic construct with the gene of the helicase domain of TMV replicase (p50); its protein product p50 is a target for the N-gene product. As a result, the hypersensitive response (HR) was initiated. The HR manifested itself in the death of leaves and was suppressed by SkQ3. Treatment of tobacco epidermal peels with the A. tumefaciens cells for the p50 gene expression stimulated the destruction of EC nuclei, which was inhibited by SkQ1 or SkQ3. The p50-lacking A. tumefaciens cells did not induce the destruction of EC nuclei. The protective effect of mitochondria-targeted antioxidants SkQ1 and SkQ3 demonstrates the involvement of mitochondria and their ROS in programmed cell death caused by pathogen elicitors.
KEY WORDS: programmed cell death, mitochondria-targeted quinones, chitosan, lipopolysaccharide, peptidoglycan, tobacco mosaic virus, guard cells, epidermal cellsDOI: 10.1134/S0006297914120050
Abbreviations: DCF, 2′,7′-dichlorofluorescein; DCFH-DA, 2′,7′-dichlorofluorescin diacetate; EC, epidermal cells; ETI, effector-triggered immunity; GC, guard cells; HR, hypersensitive response; LPS, lipopolysaccharide; PAMP, pathogen-associated molecular patterns; PCD, programmed cell death; PG, peptidoglycan; PI, propidium iodide; PTI, PAMP-triggered immunity; ROS, reactive oxygen species; SkQ1, 10-(6′-plastoquinonyl)decyl triphenylphosphonium; SkQ3, 10-(6′-methylplastoquinonyl)decyl triphenylphosphonium; TMV, tobacco mosaic virus.
The strategy of innate immunity in humans and animals is as follows:
cell receptors recognize the conservative molecular structures of large
groups of pathogens, i.e. pathogen-associated molecular patterns
(PAMPs) [1, 2]. Plant innate
immunity has three branches: 1) basal (horizontal) resistance induced
by PAMPs (PAMP-triggered immunity, PTI) or DAMPs (damage-associated
molecular patterns); 2) vertical resistance induced by pathogen
effectors (effector-triggered immunity, ETI), and 3) RNA interference
or gene silencing, i.e. suppression of the expression of viral and
nonviral pathogen-related genes [3-7].
PAMPs include external molecular structures of fungi and bacteria located on the surface of their cells: fungal chitin, lipopolysaccharide (LPS) of the outer membrane of Gram-negative bacteria, peptidoglycan (PG) of bacterial cell walls, and the protein of bacterial flagella (flagellin). DAMPs are the compounds released from a plant under the influence of hydrolytic enzymes of the pathogen. Pathogen effectors are the agents that disturb the PTI-activating system of signal transduction from plant cell receptors interacting with PAMPs or realization of immune response in PTI. Their recognition in ETI is ensured by R (resistance)-genes [3, 5, 7, 8].
An example of ETI is the interaction between tobacco mosaic virus (TMV) and a tobacco plant. The resistance gene in TMV-resistant types of tobacco is the N-gene, which encodes the protein responsible for recognition of the effector – the helicase domain of TMV replicase (p50 protein) [7, 9].
PAMPs, DAMPs, and pathogen effectors are combined by the term “elicitors”. They are the agents inducing immune response in plants. An important element of the immune system is hypersensitive response (HR), when cell death is initiated in the place of pathogen penetration and the resultant barrier of dead cells prevents the spread of infection in plant tissues. Some features of HR-related cell death allow it to be considered as a specific form of programmed cell death (PCD) typical of plants, together with its main forms – vacuolar cell death and necrosis [10-12].
Some researchers believe that the PTI-activating PAMP recognition does not lead to the development of HR accompanied by cell death in an infected plant. They believe that PCD is initiated at the next stage of immune response as a result of interaction between the R-gene products and pathogen effectors (ETI), with a stronger immune response of a plant compared to PTI [10, 13, 14].
HR is accompanied by an “oxidative burst”, i.e. intensive reactive oxygen species (ROS) production by plant cells. ROS (O2-·, H2O2, OH·) are strong oxidants that can react with different cell components (lipids, DNA, proteins), disturbing their functions and causing cell death in both pathogen and plant. ROS production during HR occurs as a result of activation of NADPH oxidase of the plant plasma membrane, as well as with the involvement of the cell wall peroxidase [15-18].
Mitochondria play a key role in apoptosis, one of PCD forms typical of animal cells. On one hand, the following proteins initiating or promoting cell death are released from mitochondria – cytochrome c, flavoprotein AIF (apoptosis inducing factor), Smac/DIABLO (second mitochondrial activator of caspases/direct IAP binding protein with low pI), serine protease HtrA2/Omi, and endonuclease G [19]. On the other hand, the mitochondrial respiratory chain generates dangerous for the cell ROS, which cause apoptosis [20, 21]. ROS are also generated by plant mitochondria [22].
Can PCD during HR in plants be regulated by mitochondria and ROS produced in them? Plant cell death, including that during HR, depends on the redox state of ubiquinone in mitochondria and is suppressed by mitochondrial alternative oxidase, which catalyzes electron transfer from ubiquinol to O2 bypassing respiratory chain complexes III and IV (activation of the alternative oxidase inhibits PCD) [23-26].
Mitochondria-targeted quinones have been synthesized that contain a ubi- or plastoquinone and its derivatives covalently bound to membrane-penetrating cations via a decane or pentane linker. They include 10-(6′-plastoquinonyl)decyl triphenylphosphonium (SkQ1) and 10-(6′-methylplastoquinonyl)decyl triphenylphosphonium (SkQ3). Uptake along the gradient of the membrane potential (Δψ) allows the accumulation of these compounds in energized mitochondria [27, 28]. Mitochondria-targeted quinones at low (nanomolar) concentrations demonstrated antioxidant properties and protected animal cells from ROS-induced death [29]. They can be used as a tool for studying the role of mitochondrial ROS in PCD.
The goal of this work was to test the elicitors of various pathogens (fungi, bacteria, and viruses) as PCD inducers in plants. The involvement of mitochondrial ROS in plant cell death was assessed by studying the effects of mitochondria-targeted quinones SkQ1 and SkQ3 on PCD caused by pathogen elicitors.
MATERIALS AND METHODS
The experiments were performed with the Nicotiana tabacum cv. Samsun NN tobacco leaves carrying the N-gene and with the epidermal peels isolated from the lower leaf surface of N. tabacum and N. benthamiana or the pea (Pisum sativum L.). The plants were grown in daylight or under periodic illumination with fluorescent lamps or a metal-halide lamp of ~100 µE·m–2·s–1 (light, 16 h; darkness, 8 h) at 23-28ºC. Epidermal peels were separated from the leaves with forceps and put into distilled water; compositions of the reagents added to them are given in figure captions. Chitosan treatment was performed by mixing epidermal peels in chitosan suspension on a magnetic stirrer. Epidermal peels were incubated with the reagents in polystyrene plates at 22-25ºC.
After the incubation, the epidermis was treated with Battaglia fixative (chloroform, 96% ethanol, glacial acetic acid, and 40% formalin, 5 : 5 : 1 : 1 v/v) for 5 min, washed with ethanol for 10 min to remove the fixative, incubated in water for 5 min, and stained with Carazzi’s hematoxylin for 20 min. The stained epidermal peels were washed with tap water and examined under a light microscope. The portion of the cells with destroyed nuclei and without nuclei was determined per 300-500 of studied cells (in 3-4 epidermal peels). The experiments were repeated 2-3 times. Typical data obtained in one of the repeated experiments are presented.
Protein kinase activities were determined by the inclusion of [32P]phosphate from γ[32P]ATP in myelin basic protein. The epidermal peels in Tris buffer (pH 6.8) with glycerol, SDS, and mercaptoethanol were heated at 95ºC for 15 min and electrophoresed in 10% polyacrylamide gel with 0.25 mg/ml of myelin basic protein. Then the gel was washed in 25 mM Tris buffer (pH 7.5) with phosphatase inhibitors (5 mM NaF and 0.1 mM Na3VO4), 0.5 mM dithiothreitol (DTT), 0.1% Triton X-100, and 0.5 mg/ml of bovine serum albumin three times for 30 min, incubated in renaturing 25 mM Tris buffer (pH 7.5), 5 mM NaF, 0.1 mM Na3VO4, 1 mM DTT overnight, and preincubated in 20 mM Hepes (pH 7.4), 50 mM KCl, 5 mM MnCl2 for 30 min; then 50 µCi (~15 pM) of γ[32P]ATP was added, followed by 1-h incubation. Phosphorylation of myelin basic protein was stopped by a solution containing 0.5-1% sodium pyrophosphate and 5% trichloroacetic acid. The radioactive label of the myelin basic protein in the gel was detected by autoradiography.
ROS production in the cells was assessed using 2′,7′-dichlorofluorescin diacetate (DCFH-DA). Epidermal peels were stained with 20 µM DCFH-DA for 30 min. While penetrating the cells, DCFH-DA is deacetylated by intracellular esterases and oxidized by ROS, mainly by H2O2 with the involvement of peroxidases, transforming into intensively fluorescing 2′,7′-dichlorofluorescein (DCF). DCF formation may involve transition metal ions, e.g. Fe2+, and cytochrome c possessing an activity similar to that of peroxidase [30, 31]. DCF fluorescence was excited with a laser beam of 488 nm and recorded using a Carl Zeiss Axiovert 200M fluorescence microscope with an LSM 510 Meta confocal module (Carl Zeiss, Germany) at 500-530 nm.
The state of the plasma membrane of the cells was assessed using propidium iodide (PI), a fluorescent dye binding to DNA and not penetrating through undamaged cell membranes. Epidermis was stained with 2 µM PI for 20 min and examined under the Carl Zeiss Axiovert 200M fluorescence microscope with LSM 510 Meta confocal module. PI fluorescence was excited with a mercury lamp at 525-565 nm or with a laser at 543 nm and recorded at 575-640 or 565-615 nm in the fluorescent or confocal mode, respectively.
The following agents were tested as elicitors: low-viscosity chitosan from crab shells (Fluka, Switzerland), LPS from Escherichia coli 0127:B8 (Sigma, USA), PG from Bacillus subtilis (Sigma), and viral protein p50 (the helicase domain of TMV replicase) synthesized in tobacco cells as a result of their transformation by Agrobacterium tumefaciens carrying genetic constructions with the p50 gene [32]. The suspension of A. tumefaciens cells with plasmid pLH with or without (control) p50 gene was grown for 16 h on a shaker at 28°C in LB medium (10 g/liter tryptone, 5 g/liter NaCl, 5 g/liter yeast extract), with sterile antibiotics (rifampicin, streptomycin, and spectinomycin, 100 µg/ml of each; the plasmid pLH carries the genes of resistance to these antibiotics), 10 mM MES buffer, pH 5.5, and 20 µM acetosyringone (4′-hydroxy-3′,5′-dimethoxyacetophenone). Acetosyringone is a phenol compound released from damaged plant cells and recognized by the VirA protein. As a result the virulence genes (vir-genes) are activated in the Ti-plasmid of A. tumefaciens to ensure the process of agrobacterial transformation – the transfer of the region of the Ti-plasmid (T-DNA) into the cells and its integration into the plant genome [33, 34]. Agrobacterium tumefaciens cells were precipitated by centrifugation for 15 min at 3000 rpm and incubated for 3 h for transformation in 10 mM MES buffer (pH 5.5) with 10 mM MgCl2 and 150 µM acetosyringone.
RESULTS
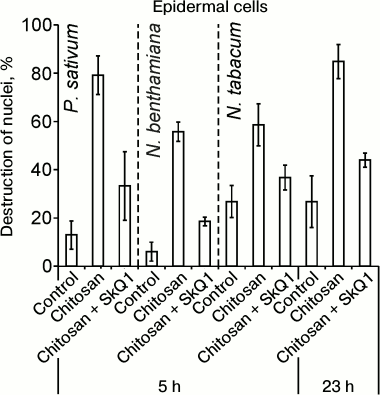
Chitosan (poly(β-1,4)-N-acetylglucosamine) formed as a result of incomplete deacetylation of chitin, a component of fungal cell walls, induced cell death detected by the destruction of cell nuclei. Chitosan caused destruction of EC (epidermal cell) nuclei in the epidermal peels from pea leaves, without affecting GC (guard cells) [35]. Anaerobiosis, antioxidants, and mitochondria-targeted quinones (SkQ) prevented chitosan-induced destruction of pea EC nuclei [35, 36]. Chitosan and mitochondria-targeted quinones had similar effects on the cells from leaves of N. benthamiana and N. tabacum: chitosan induced destruction of nuclei in ECs that was prevented by SkQ1 (Fig. 1).
Fig. 1. Effect of SkQ1 on chitosan-induced destruction of EC nuclei in epidermis from pea (P. sativum) and tobacco (N. benthamiana and N. tabacum) leaves. 100 nM SkQ1 was added to epidermal peels, incubated for 1 h, treated with 0.1 mg/ml of chitosan for 30 min on a magnetic stirrer, and incubated for 5 or 23 h in the dark.
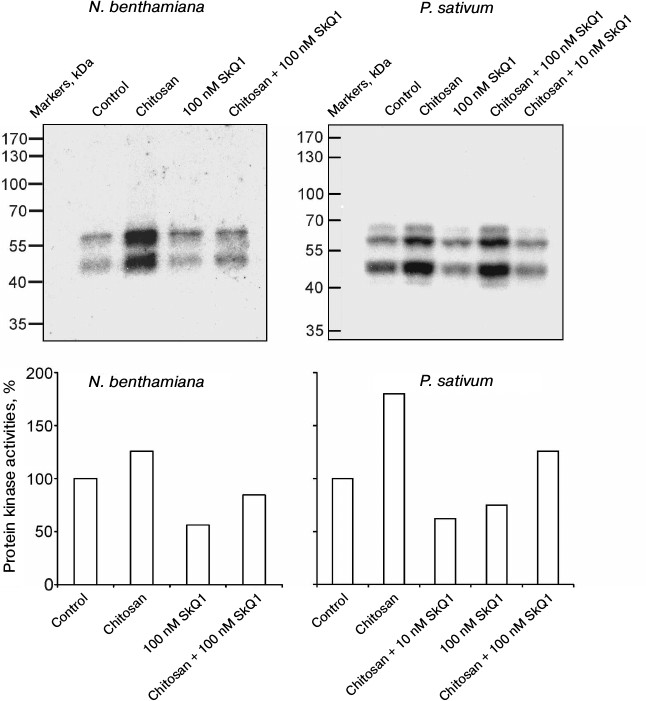
In N. benthamiana and P. sativum plants, it was demonstrated that chitosan stimulated and SkQ1 suppressed the activation of protein kinases detected by the incorporation of radioactive isotope 32P from γ[32P]ATP into myelin basic protein as a result of its phosphorylation in gel by the protein kinases from plant cells after electrophoresis. SkQ1 at concentrations of 10 and 100 nM inhibited chitosan-dependent activation of protein kinases (Fig. 2).
Fig. 2. Effects of chitosan and SkQ1 on protein kinase activities in epidermal peels from N. benthamiana and P. sativum leaves. SkQ1 was added to epidermal peels, incubated for 1 h, treated with 0.1 mg/ml of chitosan for 30 min on a magnetic stirrer, and incubated for 1 h in the dark. The figure shows the results of autoradiography of the gel with myelin basic protein after electrophoresis of the proteins from epidermal peels and its treatment with γ[32P]ATP (top) and the specific protein kinase activity (bottom) calculated with TotalLab by the ratio of the total optical densities of bands on the upper figures to the total optical densities of Coomassie-stained protein bands in the same region of the gel. The activity of protein kinases in the control (without the additives) was taken as 100%.
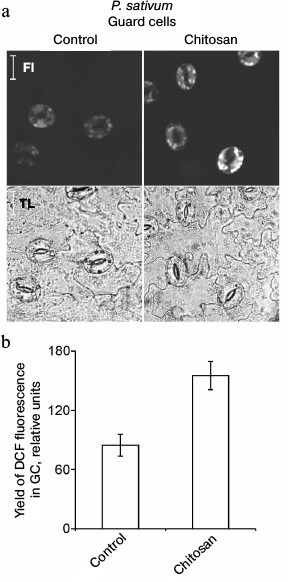
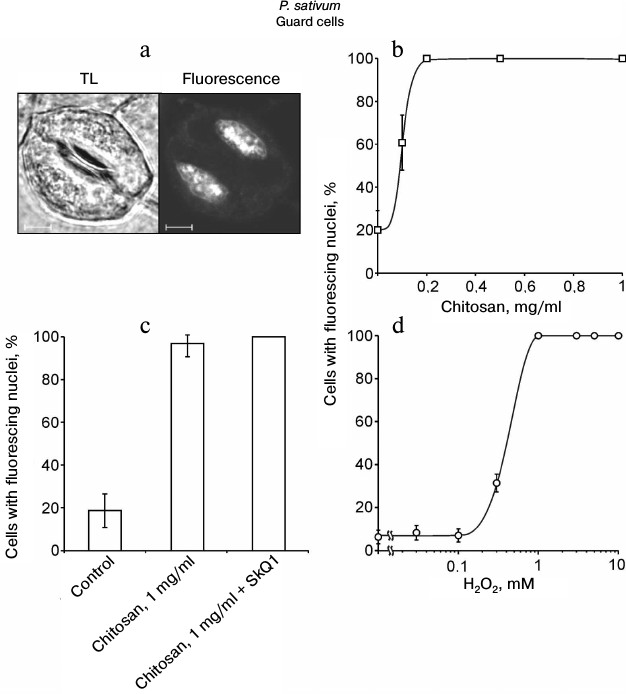
Chitosan did not induce destruction of GC nuclei [35], but it stimulated ROS generation in GC from pea leaves, which was detected by DCF fluorescence using confocal microscopy (Fig. 3). The treatment of epidermal peels from pea leaves with chitosan at a concentration of 0.1-1 mg/ml resulted in disruption of the permeability barrier of the GC plasma membrane for PI staining the DNA (Fig. 4, a and b). At the same time, GC nuclei remained intact. SkQ1 had no effect on chitosan-induced disruption of the permeability barrier of the GC plasma membrane for PI (Fig. 4c). The effect of H2O2 at a concentration of 0.3-10 mM on the pea GC was similar to that of chitosan (Fig. 4d).
Fig. 3. Effect of chitosan on the yield of DCF fluorescence in pea leaf epidermal cells. Epidermal peels were incubated for 3 h in distilled water in the light, stained with DCFH-DA, washed from excessive dye in distilled water, and treated with 1 mg/ml of chitosan for 20 min on a magnetic stirrer. Fl, DCF fluorescence; TL, transmitted light image; scale bar, 20 µm (a). b) Yield of DCF fluorescence in GC as the mean of measurements in 40-63 cells.
Fig. 4. Effects of chitosan (a-c) and H2O2 (d) on the GC plasma membrane permeability in pea leaf epidermis detected by the PI fluorescence in cell nuclei. a-c) Epidermal peels were put into chitosan solutions and, after the addition of 10 nM SkQ1, incubated for 22 h with mixing on a magnetic stirrer. TL, transmitted light image; scale bar, 5 µm (a). d) Epidermal peels were put into H2O2 solutions and incubated for 23-24 h. After the incubation, the peels were stained with PI. The portion of the cells (b-d) with PI-stained nuclei was determined as in (a).
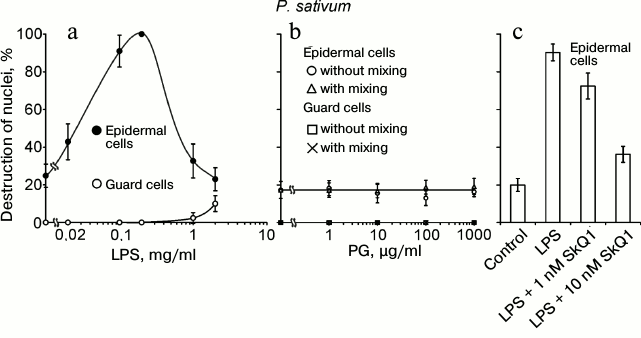
The following components of the outer membrane of Gram-negative bacteria and bacterial cell wall were tested as cell death inducers: LPS from E. coli and PG from B. subtilis. LPS, similar to chitosan, at a concentration of 20-200 µg/ml induced the destruction of EC nuclei in pea leaf epidermis (Fig. 5a). At higher concentrations (1-2 mg/ml), the effect of LPS vanished. GC with nuclei not destroyed by chitosan [35] were also resistant to LPS. PG did not induce the destruction of EC and GC nuclei in pea leaf epidermis in a broad range of its concentrations and under different conditions of treatment (Fig. 5b). SkQ1 prevented the LPS-induced destruction of EC nuclei (Fig. 5c).
Fig. 5. Effects of LPS from E. coli (a) and PG from B. subtilis (b) on GC and EC nuclei; effect of SkQ1 on LPS-induced destruction of EC nuclei (c) in pea leaf epidermis. a, b) Epidermal peels with the added LPS or PG were incubated for 22 h in the dark without mixing (a, b) or with mixing (b) on a magnetic stirrer 10 times for 2 min during 1 h for each experimental variant. c) Epidermal peels with the added SkQ1 were incubated for 2 h and then, after the addition of 0.2 mg/ml LPS, incubated for 22 h in the dark without mixing.
The interactions between the plants and the viral elicitor were simulated as follows: the tobacco (N. tabacum) leaves carrying the N-gene of TMV resistance were transformed by agrobacteria. Agrobacterium tumefaciens contained the pLH plasmid with the p50 gene encoding the helicase domain of TMV replicase. The transformation of tobacco cells by A. tumefaciens with the p50 gene resulted in the conditions for recognition of the viral protein p50 by the N-gene product of the plant and for HR development. Agrobacterium tumefaciens with the pLH plasmid but without the p50 gene was used as a control.
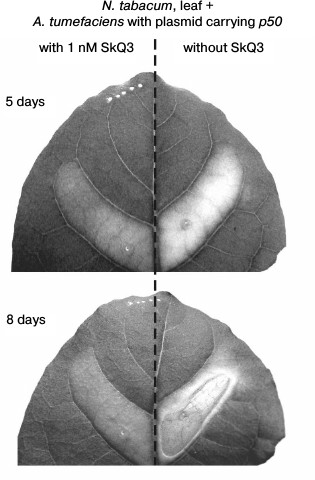
The cell suspension of A. tumefaciens with the pLH plasmid carrying the p50 gene injected into a tobacco leaf caused, after 5 days, the yellowing of the treated section of a leaf due to chlorophyll degradation – the common symptom of leaf aging that precedes its death [37, 38]. After 8 days, the section of the leaf treated with A. tumefaciens cells died off – it thinned and was dehydrated, turning into a fragile light-brown film (Fig. 6).
Fig. 6. Effect of SkQ3 on N. tabacum leaf tissue die-off caused by the treatment with the suspension of A. tumefaciens cells with the pLH plasmid carrying the gene of the helicase domain of TMV replicase (p50). Time of incubation with A. tumefaciens is indicated in the figure. Preincubation with 1 nM SkQ3 (before introduction of A. tumefaciens) lasted 2 h. The suspension of A. tumefaciens cells (optical density 0.5 at 600 nm) was injected into leaf through the lower epidermis with a syringe.
In this experiment, the mitochondria-targeted quinone was SkQ3 containing methylated plastoquinone, in contrast to the ordinary plastoquinone in SkQ1. The concentrations of SkQ3, when it protects plant cells from cyanide- or chitosan-induced death, are lower than the concentrations of other tested mitochondria-targeted quinones [36]. Pretreatment of a section of a tobacco leaf with SkQ3 at a concentration of 1 nM for 2 h before injection of the suspension of A. tumefaciens cells with the pLH plasmid carrying the p50 gene inhibited its yellowing and prevented its die-off (Fig. 6).
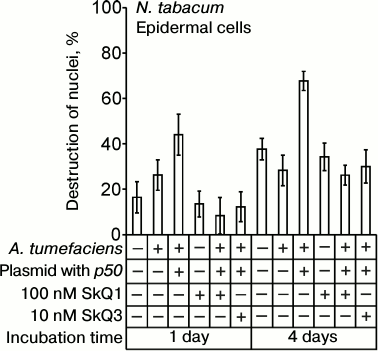
The p50 protein induced the destruction of EC nuclei in epidermal peels from N. tabacum leaves with the N-gene on application of the suspension of A. tumefaciens cells carrying the p50 gene (Fig. 7). In the control experiments, epidermal peels were treated with A. tumefaciens cells containing the pLH plasmid without the p50 gene. The difference between the control and the variant with p50 is statistically significant. It is greater when the incubation time is longer. GC nuclei were not destroyed in epidermis isolated from the leaves after the treatment with A. tumefaciens cells. SkQ1 at a concentration of 100 nM per se had no effect on cell nuclei but prevented the destruction of tobacco EC nuclei induced by A. tumefaciens with the p50 gene (Fig. 7). SkQ3 prevented the destruction of EC nuclei at a concentration of 10 nM.
Fig. 7. Effect of SkQ1 on destruction of EC nuclei in the epidermis of N. tabacum leaves caused by treatment of epidermis with the suspension of A. tumefaciens cells with the pLH plasmid carrying the gene of the helicase domain of TMV replicase (p50). Epidermal peels were put into the transformation buffer, 100 nM SkQ1 or 10 nM SkQ3 was added, and the suspension of A. tumefaciens cells in the same buffer was introduced (optical density 2 at 600 nm), followed by incubation in the dark. Incubation time is indicated on the diagram. Cells of A. tumefaciens contained the pLH plasmid with or without the p50 gene. In the control, epidermal peels were put into the buffer without A. tumefaciens.
DISCUSSION
The effects of mitochondria-targeted quinones on cyanide- or chitosan-induced destruction of pea GC and EC nuclei have been studied previously [36]. These quinones at nanomolar concentrations prevented the destruction of cell nuclei in the dark but not in the light. Their effects were eliminated by carbonyl cyanide m-chlorophenylhydrazone, an uncoupler of oxidative phosphorylation. Mitochondria-targeted quinones accumulated in the mitochondria of pea cells and prevented ROS production. The quinones reduced menadione-induced ROS generation but had no effect on H2O2-dependent ROS generation. Mitochondria-targeted quinones are antioxidants that selectively accumulate in mitochondria [36]. Predictably, SkQ1 prevented chitosan-induced destruction of nuclei in tobacco EC (Fig. 1).
The protein kinase cascade in plant cells is activated under the influence of factors of different nature, including abiotic factors such as high salinity, drought, cold, mechanic damage of plant tissues, or biotic factors (phytohormones, pathogen elicitors) [39, 40]. The best-studied of pathogen elicitors that can induce the activation of MAP kinase cascade are the protein of the bacteria flagellum (flagellin), bacterial elongation factor EF-Tu, chitin oligosaccharides, and the helicase domain of TMV replicase [41, 42].
The relationship between the MAP kinase cascade and ROS generation in HR is still unclear. ROS (H2O2, ozone) activated the MAP kinase cascade in plant cells. Diphenyleneiodonium, the inhibitor of plasma membrane NADPH oxidase, suppressed ROS production induced by the fungal elicitor but did not prevent the activation of MAP kinases. The activation and inactivation of MAP kinases stimulated and suppressed ROS generation, respectively. Thus, HR with the involvement of plasma membrane NADPH oxidase and the MAP kinase cascade in plants after the reception of pathogen elicitor proceed independently of each other but may be cross-regulated [39, 40, 42]. SkQ1-dependent suppression of ROS production in plant mitochondria apparently results in the inactivation of chitosan-induced protein kinase cascade (Fig. 2). The activation of protein kinases is probably initiated by the ROS formed as a result of the action of chitosan on plant cells (Fig. 3). Chitosan-mediated stimulation of ROS production is in agreement with the data that chitosan initiated H2O2 production in rice cell suspension culture [43].
The results demonstrate the similar effects of chitosan and LPS on plant cells as PCD inducers. The identical effects of LPS and chitin oligosaccharides on plant cells were shown previously [44]. Both chitosan- and LPS-dependent cell death was suppressed by SkQ1 ([36], Figs. 1 and 5c).
PG did not demonstrate the properties of a cell death inducer in our experiments (Fig. 5b), probably due to the lesser availability of PG for recognition by cell receptors because of its insolubility in water. PG can show antiapoptotic properties with animal cells: PG stimulated the production of the inhibitor of apoptosis cIAP-2 in keratinocytes and inhibited their death induced by the tumor necrosis factor TNFα [45]. PG recognition seems to involve the same system (with the receptor-like kinase CERK1), which recognizes also chitin oligosaccharides [46].
The absence of destruction of pea EC nuclei after the treatment with LPS at a high concentration (1-2 mg/ml) may be due to switching the mode of cell death from programmed to non-programmed, i.e. necrosis, when the permeability barrier of the plasma membrane is disrupted and cell nuclei remain intact. Such disruption of the plasma membrane permeability barrier in pea GC was induced by chitosan at a concentration above 0.1 mg/ml or H2O2 at a concentration above 0.3 mM (Fig. 4). Ca2+ (1-2 mM) and Triton X-100 had similar effects on plant cells [47].
In cultured cells of soybean (Glycine max) and haricot (Phaseolus vulgaris), chitosan at a concentration of 0.1 mg/ml and more induced disruption of the plasma membrane permeability barrier detected by the release of electrolytes (conductometrically) and proteins from the cells. Polycations (polylysine, diethylaminoethyl dextran) had the same effect on plant cells. Divalent cations, including Ca2+, suppressed polycation-induced disruption of the permeability barrier of the plasma membrane. This property of chitosan located on the outer surface of cell walls of some phytopathogenic fungi, e.g. Fusarium solani (the pathogen of pea fusarial wilt) or Puccinia striiformis (the pathogen of wheat rust), is supposed to contribute to the outflow of nutrients from plant cells into fungal cells [48]. Thus, the effect of chitosan at concentrations above 0.1 mg/ml on the state of GC plasma membrane seems to be determined not by the specific reaction of its recognition by a plant cell receptor but by its properties as a polycation. That is why SkQ1 will certainly have no effect on chitosan-induced disruption of the permeability barrier of the plasma membrane in GC, which is in agreement with our observations (Fig. 4c).
H2O2 can induce both apoptosis and necrosis of cells depending on its concentration. Human oocytes were treated for 1 h with H2O2 at different concentrations: at 50 µM, the cells died mainly through apoptosis; at 100 µM, both apoptosis and necrosis were observed; at 1 mM, mainly necrosis was observed [49]. Similar to chitosan, H2O2 per se did not stimulate the destruction of nuclei (PCD) in pea GC, but already at a concentration of 10 µM it significantly stimulated the destruction of GC nuclei induced by cyanide, the inhibitor of catalase and peroxidases [50]. It can be supposed that the photosynthesizing GC have an antioxidant protection (including catalase and peroxidases), which copes with the low concentrations of H2O2. When H2O2 concentrations exceeded the capabilities of antioxidant protection of the cells, it induced disruption of the permeability barrier of the plasma membrane, i.e. necrosis (Fig. 4d).
The N-gene imparts TMV resistance to tobacco plants. In tobacco cultivars without the N-gene, the replicating TMV induces plant diseases (a mosaic of light and dark green spots on leaves) and suppresses plant growth. In cultivars with the N-gene, TMV induces HR, which limits further spread of the virus. The helicase domain of the TMV replicase (protein p50) is sufficient to induce the N-gene-dependent HR in tobacco. The synthesis of p50 protein was achieved by transgene expression through the introduction of A. tumefaciens cells carrying the plasmid with the p50 gene into the mesophyll of N. tabacum leaves [9].
The introduction of A. tumefaciens cells with the p50 gene into a tobacco leaf initiated the development of HR manifesting itself in the wilting and dying-off section of the leaf where the A. tumefaciens cells with the p50 gene were introduced. The treatment of a section of the leaf with A. tumefaciens cells containing the plasmid pLH without the p50 gene did not result in its wilting and dying-off. SkQ1 at the concentrations of 0.1 and 1 nM prevented HR development [32]. In agreement with these data, the treatment of tobacco with the suspension of A. tumefaciens cells carrying the p50 gene induced HR, while SkQ3 at a concentration of 1 nM suppressed HR (Fig. 6). The treatment of tobacco epidermal peels with A. tumefaciens cells carrying the p50 gene induced destruction of EC nuclei, which was prevented by SkQ1 or SkQ3 (Fig. 7). In this respect, the effect of the viral protein p50 is similar to the effects of chitosan and LPS (Figs. 1 and 5c).
The data obtained show that PCD in plant EC is initiated by fungal, bacterial, and viral elicitors – chitosan, LPS, and the p50 protein of TMV. PCD occurs not only during ETI induced by the viral effector p50 but also during PTI (the resistance ensured by the recognition of PAMPs, including chitosan and LPS). In both variants, the mitochondria-targeted quinone SkQ1 had a protective effect, which is evidence of the involvement of mitochondrial ROS in PCD induced by pathogen elicitors.
The authors are grateful to the Academician V. P. Skulachev and researchers of the Institute of Mitoengineering, Lomonosov Moscow State University, for providing the mitochondria-targeted quinones. Equipment used in the work is the property of the Center for Collective Use of the Lomonosov Moscow State University.
The study was supported by the Russian Foundation for Basic Research (projects No. 12-04-31622 mol_a and No. 14-04-00507 a).
REFERENCES
1.Medzhitov, R., and Janeway, C. A. (1997) Innate
immunity: the virtues of a nonclonal system of recognition,
Cell, 91, 295-298.
2.Medzhitov, R. (2007) Recognition of microorganisms
and activation of the immune response, Nature, 449,
819-826.
3.Chisholm, S. T., Coaker, G., Day, B., and
Staskawicz, B. J. (2006) Host-microbe interactions: shaping the
evolution of the plant immune response, Cell, 124,
803-814.
4.Ding, S.-W., and Voinnet, O. (2007) Antiviral
immunity directed by small RNAs, Cell, 130, 413-426.
5.Boller, T., and Felix, G. (2009) A renaissance of
elicitors: perception of microbe-associated molecular patterns and
danger signals by pattern-recognition receptors, Annu. Rev. Plant
Biol., 60, 379-406.
6.Zvereva, A. S., and Pooggin, M. M. (2012) Silencing
and innate immunity in plant defense against viral and non-viral
pathogens, Viruses, 4, 2578-2597.
7.Muthamilarasan, M., and Prasad, M. (2013) Plant
innate immunity: an updated insight into defense mechanism, J.
Biosci., 38, 433-449.
8.Dangl, J. L., and Jones, J. D. (2001) Plant
pathogens and integrated defense responses to infection, Nature,
411, 826-833.
9.Erickson, F. L., Holzberg, S., Calderon-Urrea, A.,
Handley, V., Axtell, M., Corr, C., and Baker, B. (1999) The helicase
domain of the TMV replicase proteins induces the N-mediated defense
response in tobacco, Plant J., 18, 67-75.
10.Hofius, D., Tsitsigiannis, D. I., Jones, J. D.
G., and Mundy, J. (2007) Inducible cell death in plant immunity,
Semin. Cancer Biol., 17, 166-187.
11.Mur, L. A. J., Kenton, P., Lloyd, A. J., Ougham,
H., and Prats, E. (2008) The hypersensitive response; the centenary is
upon us but how much do we know? J. Exp. Bot., 59,
501-520.
12.Van Doorn, W. G., Beers, E. P., Dangl, J. L.,
Franklin-Tong, V. E., Gallois, P., Hara-Nishimura, I., Jones, A. M.,
Kawai-Yamada, M., Lam, E., Mundy, J., Mur, L. A. J., Petersen, M.,
Smertenko, A., Taliansky, M., Van Breusegem, F., Wolpert, T.,
Woltering, E., Zhivotovsky, B., and Bozhkov, P. V. (2011) Morphological
classification of plant cell deaths, Cell Death Differ.,
18, 1241-1246.
13.Jones, J. D. G., and Dangl, J. L. (2006) The
plant immune system, Nature, 444, 323-329.
14.Coll, N. S., Epple, P., and Dangl, J. L. (2011)
Programmed cell death in the plant immune system, Cell Death
Differ., 18, 1247-1256.
15.Lamb, C., and Dixon, R. A. (1997) The oxidative
burst in plant disease resistance, Annu. Rev. Plant Physiol. Plant
Mol. Biol., 48, 251-275.
16.Wojtaszek, P. (1997) Oxidative burst: an early
plant response to pathogen infection, Biochem. J., 322,
681-692.
17.Sagi, M., and Fluhr, R. (2006) Production of
reactive oxygen species by plant NADPH oxidases, Plant Physiol.,
141, 336-340.
18.Imlay, J. A. (2013) The molecular mechanisms and
physiological consequences of oxidative stress: lessons from a model
bacterium, Nat. Rev. Microbiol., 11, 443-454.
19.Ravagnan, L., Roumier, T., and Kroemer, G. (2002)
Mitochondria, the killer organelles and their weapons, J. Cell.
Physiol., 192, 131-137.
20.Skulachev, V. P. (2006) Bioenergetic aspects of
apoptosis, necrosis and mitoptosis, Apoptosis, 11,
473-485.
21.Orrenius, S. (2007) Reactive oxygen species in
mitochondria-mediated cell death, Drug Metab. Rev., 39,
443-455.
22.Moller, I. M. (2001) Plant mitochondria and
oxidative stress: electron transport, NADPH turnover, and metabolism of
reactive oxygen species, Annu. Rev. Plant Physiol. Plant Mol.
Biol., 52, 561-591.
23.Lam, E., Kato, N., and Lawton, M. (2001)
Programmed cell death, mitochondria and the plant hypersensitive
response, Nature, 411, 848-853.
24.Samuilov, V. D., Lagunova, E. M., Kiselevsky, D.
B., Dzyubinskaya, E. V., Makarova, Ya. V., and Gusev, M. V. (2003)
Participation of chloroplasts in plant apoptosis, Biosci. Rep.,
23, 103-117.
25.De Souza, W. R., Vessecchi, R., Dorta, D. J.,
Uyemura, S. A., Curti, C., and Vargas-Rechia, C. G. (2011)
Characterization of Rubus fruticosus mitochondria and salicylic
acid inhibition of reactive oxygen species generation at Complex III/Q
cycle: potential implications for hypersensitive response in plants,
J. Bioenerg. Biomembr., 43, 237-246.
26.Cvetkovska, M., and Vanlerberghe, G. C. (2013)
Alternative oxidase impacts the plant response to biotic stress by
influencing the mitochondrial generation of reactive oxygen species,
Plant Cell Environ., 36, 721-732.
27.Kelso, G. F., Porteous, C. M., Coulter, C. V.,
Hughes, G., Porteous, W. K., Ledgerwood, E. C., Smith, R. A. J., and
Murphy, M. P. (2001) Selective targeting of a redox-active ubiquinone
to mitochondria within cells: antioxidant and antiapoptotic properties,
J. Biol. Chem., 276, 4588-4596.
28.Skulachev, V. P. (2007) The attempt of
biochemists to attack the problem of ageing: a
“megaproject” on penetrating ions. First results and
prospects, Biochemistry (Moscow), 72, 1385-1398.
29.Antonenko, Yu. N., Avetisyan, A. V., Bakeeva, L.
E., Chernyak, B. V., Chertkov, V. A., Domnina, L. V., Ivanova, O. Yu.,
Izyumov, D. S., Khaylova, L. S., Klishin, S. S., Kurshunova, G. A.,
Lyamzayev, K. G., Muntyan, M. S., Nepryakhina, O. K., Pashkovskaya, A.
A., Pletyushkina, O. Yu., Pustovidko, A. V., Roginsky, V. A.,
Rokitskaya, T. I., Ruuge, E. K., Saprunova, V. B., Severina, I. I.,
Simonyan, R. A., Skulachev, I. V., Skulachev, M. V., Sumbatyan, N. V.,
Sviryaeva, I. V., Tashlitsky, V. N., Vasilyev, Yu. N., Vysokikh, M.
Yu., Yaguzhinsky, L. S., Zamyatnin, A. A., and Skulachev, V. P. (2008)
Plastoquinone derivative addressed to mitochondria as an agent that
interrupts the aging program. 1. Cationic plastoquinone derivatives:
synthesis and in vitro study, Biochemistry (Moscow),
73, 1273-1287.
30.LeBel, C. P., Ischiropoulos, H., and Bondy, S. C.
(1992) Evaluation of the probe 2′,7′-dichlorofluorescin as
an indicator of reactive oxygen species formation and oxidative stress,
Chem. Res. Toxicol., 5, 227-231.
31.Karlsson, M., Kurz, T., Brunk, U. T., Nilsson, S.
E., and Frennesson, C. I. (2010) What does the commonly used DCF test
for oxidative stress really show? Biochem. J., 428,
183-190.
32.Solovieva, A. D., Frolova, O. Yu., Solovyev, A.
G., Morozov, S. Yu., and Zamyatnin, A. A., Jr. (2013) The effect of
mitochondria-targeted antioxidant SkQ1 on programmed cell death induced
by viral proteins in tobacco plants, Biochemistry (Moscow),
78, 1006-1012.
33.Turk, S. C. H. J., van Lange, R. P.,
Regensburg-Tuink, T. J. G., and Hooykaas, P. J. J. (1994) Localization
of the VirA domain involved in acetosyringone-mediated vir gene
induction in Agrobacterium tumefaciens, Plant Mol. Biol.,
25, 899-907.
34.Gelvin, S. B. (2000) Agrobacterium and
plant genes involved in T-DNA transfer and integration, Annu. Rev.
Plant Physiol. Plant Mol. Biol., 51, 223-256.
35.Vasilyev, L. A., Dzyubinskaya, E. V., Zinovkin,
R. A., Kiselevsky, D. B., Lobysheva, N. V., and Samuilov, V. D. (2009)
Chitosan-induced programmed cell death in plants, Biochemistry
(Moscow), 74, 1035-1043.
36.Vasilyev, L. A., Dzyubinskaya, E. V., Kiselevsky,
D. B., Shestak, A. A., and Samuilov, V. D. (2011) Programmed cell death
in plants: protective effect of mitochondria-targeted quinones,
Biochemistry (Moscow), 76, 1120-1130.
37.Lee, I. C., Hong, S. W., Whang, S. S., Lim, P.
O., Nam, H. G., and Koo, J. C. (2011) Age-dependent action of an
ABA-inducible receptor kinase, RPK1, as a positive regulator of
senescence in Arabidopsis leaves, Plant Cell Physiol.,
52, 651-662.
38.Dzyubinskaya, E. V., Ionenko, I. F., Kiselevsky,
D. B., Samuilov, V. D., and Samuilov, F. D. (2013)
Mitochondria-targeted cations slow down the aging and death of
Arabidopsis thaliana leaves, extend the vegetation period, and
improve the yield structure of the wheat Triticum aestivum,
Biochemistry (Moscow), 78, 68-74.
39.Zhang, S., and Klessig, D. F. (2001) MAPK
cascades in plant defense signaling, Trends Plant Sci.,
6, 520-527.
40.Pitzschke, A., and Hirt, H. (2006)
Mitogen-activated protein kinases and reactive oxygen species signaling
in plants, Plant Physiol., 141, 351-356.
41.Miya, A., Albert, P., Shinya, T., Desaki, Y.,
Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and
Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for
chitin elicitor signaling in Arabidopsis, Proc. Natl. Acad.
Sci. USA, 104, 19613-19618.
42.Meng, X., and Zhang, S. (2013) MAPK cascades in
plant disease resistance signaling, Annu. Rev. Phytopathol.,
51, 245-266.
43.Lin, W., Hu, X., Zhang, W., Rogers, W. J., and
Cai, W. (2005) Hydrogen peroxide mediates defense responses induced by
chitosans of different molecular weights in rice, J. Plant
Physiol., 162, 937-944.
44.Desaki, Y., Miya, A., Venkatesh, B., Tsuyumu, S.,
Yamane, H., Kaku, H., Minami, E., and Shibuya, N. (2006) Bacterial
lipopolysaccharides induce defense responses associated with programmed
cell death in rice cells, Plant Cell Physiol., 47,
1530-1540.
45.Vazquez-Sanchez, E. A., Rodriguez-Romero, M.,
Sanchez-Torres, L. E., Rodriguez-Martinez, S., Cancino-Diaz, J. C.,
Rodriguez-Cortes, O., Garcia-Lopez, E. S., and Cancino-Diaz, M. E.
(2014) Peptidoglycan from Staphylococcus aureus has an
anti-apoptotic effect in HaCaT keratinocytes by the production of the
cellular inhibitor of apoptosis protein-2 (cIAP-2), Microbiol.
Immunol., 58, 87-95.
46.Willmann, R., Lajunen, H. M., Erbs, G., Newman,
M.-A., Kolb, D., Tsuda, K., Katagiri, F., Fliegmann, J., Bono, J.-J.,
Cullimore, J. V., Jehle, A. K., Gotz, F., Kulik, A., Molinaro, A.,
Lipka, V., Gust, A. A., and Nurnberger, T. (2011) Arabidopsis
lysine-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan
sensing and immunity to bacterial infection, Proc. Natl. Acad. Sci.
USA, 108, 19824-19829.
47.Kiselevsky, D. B., Kuznetsova, Yu. E., Vasilyev,
L. A., Lobysheva, N. V., Zinovkin, R. A., Nesov, A. V., Shestak, A. A.,
and Samuilov, V. D. (2010) The effect of Ca2+ on the
programmed cell death of guard and epidermal cells in pea leaves,
Biochemistry (Moscow), 75, 614-622.
48.Young, D. H., Kohle, H., and Kauss, H. (1982)
Effect of chitosan on membrane permeability of suspension-cultured
Glycine max and Phaseolus vulgaris cells, Plant
Physiol., 70, 1449-1454.
49.Zhang, X., Li, X. H., Ma, X., Wang, Z. H., Lu,
S., and Guo, Y. L. (2006) Redox-induced apoptosis of human oocytes in
resting follicles in vitro, J. Soc. Gynecol. Invest.,
13, 451-458.
50.Samuilov, V. D., Kiselevsky, D. B., Sinitsyn, S.
V., Shestak, A. A., Lagunova, E. M., and Nesov, A. V. (2006)
H2O2 intensifies CN–-induced
apoptosis in pea leaves, Biochemistry (Moscow), 71,
384-394.