Nonhydrolyzable ATP Analog 5′-Adenylyl-imidodiphosphate (AMP-PNP) Does Not Inhibit ATP-Dependent Scanning of Leader Sequence of mRNA
P. A. Sakharov, A. S. Sokolov, and S. Ch. Agalarov*
Institute of Protein Research, Russian Academy of Sciences, ul. Institutskaya 4, 142290 Pushchino, Moscow Region, Russia; fax: +7 (495) 514-0218; E-mail: sultan@vega.protres.ru* To whom correspondence should be addressed.
Received June 14, 2014; Revision received July 22, 2014
The objective of the present work was to determine whether it is possible to use a nonhydrolyzable analog of ATP (AMP-PNP) as an inhibitor of ATP-dependent scanning of the leader sequence of eukaryotic mRNA in translation initiation. The formation of ribosomal 48S initiation complexes at the start codon of the capped mRNA leader sequence of rabbit β-globin mRNA was studied. The study was carried out in a system composed of individual components of translation initiation. The dependences of the efficiency of formation of 48S initiation complexes on ATP concentration and incubation time were obtained in the absence and presence of AMP-PNP. It was found that AMP-PNP did not affect the efficiency of formation of 48S initiation complexes in all cases under study. We conclude that the uncleavable analog of ATP, AMP-PNP, is not an inhibitor of translation initiation in eukaryotes.
KEY WORDS: translation initiation, 48S ribosomal initiation complex, AMP-PNP, toeprintingDOI: 10.1134/S0006297915010058
Abbreviations: AMP-PNP, 5′-adenylyl-imidodiphosphate; CXR, carboxy-X-rhodamine.
Translation initiation is the most important stage of protein
biosynthesis in eukaryotes: the basic mechanisms of regulation of
protein synthesis in cells work at this very stage. According to the
classical mechanism of initiation (the scanning model of Kozak [1]), the 40S ribosomal subunit in complex with protein
initiation factors binds to the 5′-end cap structure of the mRNA
leader sequence and begins ATP-dependent movement (scanning) from the
5′-end to the 3′-end along the mRNA chain until reaching
the initiation codon. The unidirectionality of this movement has
recently been proved experimentally [2]. According
to the diffusion–ratchet model proposed earlier, the energy of
ATP hydrolysis provides unidirectional movement via cyclic variations
in the affinity of translation initiation factor eIF4A bound to the 40S
ribosomal particle towards the RNA-binding protein eIF4B and towards
mRNA; as a result, the backward movement of the 43S initiation complex
is limited and the forward stochastic (diffusion) movement along the
mRNA chain is allowed [3]. The initiation factor
eIF4A is an RNA-dependent ATPase catalyzing ATP hydrolysis with the
formation of ADP and orthophosphate [4].
Consequently, if the model is correct, an ATP analog such as
5′-adenylyl-imidodiphosphate (AMP-PNP), where the pyrophosphate
bond between β- and γ-phosphates hydrolyzed by ATPase is
replaced by a nonhydrolyzable bond, might seem to inhibit the
energy-dependent unidirectional movement of the 43S initiation complex
along the mRNA chain and, accordingly, the formation of the 48S
initiation complex at the start codon. However, according to some
literature data, ATP hydrolysis catalyzed by the eIF4A factor is not
inhibited in the presence of AMP-PNP [5].
The possibility of inhibition of unidirectional scanning of the eukaryotic mRNA leader sequence by the uncleavable analog of ATP (AMP-PNP) and further use of this reagent in translation initiation studies was verified by the method of inhibition of primer extension (so-called “toeprinting”) [6] modified in our laboratory [7]. The well-studied leader sequence of globin mRNA was used as a scanned mRNA leader sequence. The efficiency of formation of the 48S initiation complex depending on ATP concentration and incubation time was assessed in the absence and presence of AMP-PNP. Surprisingly, the nonhydrolyzable analog of ATP (AMP-PNP) did not inhibit the formation of the 48S initiation complex at the start codon in all studied cases (under optimal and suboptimal conditions).
MATERIALS AND METHODS
Components of system for 48S initiation complex assembly. Native and recombinant translation initiation factors, ribosomal subunits, and native mRNA of rabbit β-globin were obtained as described previously [7].
AMP-PNP quality control. AMP-PNP preparation (Sigma, USA) was used in this work. Since it was the key reagent of the present study, it was subjected to quality testing. The AMP-PNP mass spectrum showed complete correspondence between the measured molecular mass and chemical composition of the preparation. In addition, the luciferin–luciferase reaction was performed in the presence of AMP-PNP (this reaction proceeds in the presence of ATP with the hydrolysis of its α–β bond). AMP-PNP was also be reactive in this test; hence, it has been concluded that the nonhydrolyzable bond in this preparation is in the β–γ position, i.e. as should be the case.
Formation of ribosomal initiation complexes. The 48S initiation complexes were assembled as follows: 0.5 pmol of mRNA was added to a mixture cooled on ice (40S ribosomal subunits, 4 pmol; eIF2, 5 pmol; eIF3, 3 pmol; eIF4F, 3 pmol; eIF1, 15 pmol; eIF1A, 15 pmol; eIF4A, 3 pmol; eIF4B, 10 pmol; and Met-tRNAi, 1 pmol) in buffer containing 40 mM Tris-OAc (pH 7.5), 3.7 mM Mg(OAc)2, 2 mM DTT, 0.25 mM spermidine, 0.2 mM GMP-PNP, 0.1 mM EDTA, 120 mM KCl, and 0.3 U/µl RiboLock RNase inhibitor (Fermentas, Lithuania). The mixture also contained ATP and, where necessary, AMP-PNP (concentrations are indicated in the text and figure captions). The reaction mixture (20 µl) was incubated at 37°C for 1, 2, 5, or 15 min.
Inhibition of primer extension (toeprinting). The Mg2+ concentration in the mixture with preformed initiation complexes was brought to 7 mM. As a result, the reaction was stopped, because such concentration of magnesium ions prevents complex formation. dATP, dCTP, dGTP, and dTTP (0.5 mM each), DNA primer with fluorescent label (2 pmol/µl), and AMV reverse transcriptase (0.3 U/µl) (Promega, USA) were added to the mixture to carry out the reaction of primer elongation. The reaction mixture was incubated at 37°C for 30 min. The products of reverse transcription were purified by phenol extraction, precipitated with 70% ethanol and 0.7 M NH4OAc, and dissolved in 20 µl of 90% formamide in TBE buffer. The aliquots (0.5 µl) of fluorescently labeled DNA size standards (CXR, carboxy-X-rhodamine; 60-400 nucleotides) (Promega) were added to each sample for capillary electrophoresis. The nucleotide sequence of DNA primer for the annealing to mRNA was 5′-[6-carboxyfluorescein]-GGACTCGAAGAACCTCTG-3′.
Capillary electrophoresis of reaction products of primer extension. The cDNA fragments formed in the reaction of primer extension were analyzed by capillary gel electrophoresis in an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, USA) according to the manufacturer’s instructions. The data were processed with GeneMarker 1.5 (SoftGenetics, USA). The distribution profiles of cDNA fluorescence were correlated with the mRNA sequence using fluorescently labeled DNA size standards (CXR) of 60-400 nucleotides.
RESULTS AND DISCUSSION
As mentioned above, according to the classical model, the 40S ribosomal subunit in complex with protein initiation factors (the 43S initiation complex) scans the mRNA chain in the 5′-3′ direction until reaching the initiation codon. The ribosomal complex is fixed at the AUG start codon, forming the so-called 48S initiation complex. Thus, the efficiency of scanning can be judged by the efficiency of formation of this 48S complex at the initiation codon. The efficiency of formation of the 48S ribosomal initiation complex is usually assessed by the inhibition of primer extension, or toeprinting [6]. Here, we have used a modification of this method with the fluorescently labeled 5′-end of the primer; fluorescent products were analyzed by capillary electrophoresis with optical recording of the samples [7].
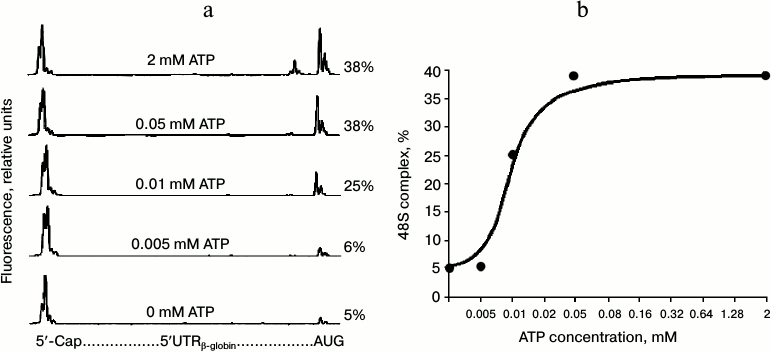
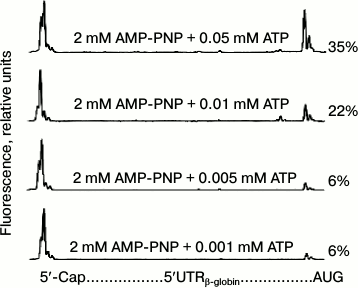
Since the dependence of the yield of 48S initiation complexes on ATP concentration had not been estimated previously, we first carried out the appropriate experiments (the results are shown in Fig. 1). The electrophoregrams in Fig. 1a demonstrate the formation of 48S initiation complexes on the mRNA over a wide range of ATP concentrations. The characteristic “trident” on the right side of the electrophoregrams indicates the position of the 48S ribosomal complex at the AUG codon, while the intensity of trident peaks represents the number of ribosomal initiation complexes that have reached the start codon due to scanning. Figure 1b demonstrates the dependence of the yield of 48S initiation complexes on ATP concentration: the level of half-saturation with the substrate is reached at a concentration of approximately 0.01 mM, and the maximum is reached at the ATP concentration of 0.05 mM. We checked the effect of AMP-PNP on the formation of 48S initiation complexes in the 0-0.05 mM range of ATP concentrations, i.e. in the most sensitive points, using the dependence from Fig. 1b. Figure 2 presents the electrophoregrams showing the yield of 48S initiation complexes in the presence of 2 mM AMP-PNP at different ATP concentrations. One can see that in spite of the predominance of AMP-PNP compared to ATP, the efficiency of formation of 48S initiation complexes does not vary and remains at the same level as in the absence of AMP-PNP (cf. Figs. 1a and 2). Consequently, AMP-PNP had no effect on the formation of 48S initiation complexes under the above conditions.
Fig. 1. Formation of 48S ribosomal initiation complexes at the initiation codon of native capped β-globin mRNA at different ATP concentrations. a) Electrophoregram representing the formation of 48S complexes at the AUG initiation codon. Relative fluorescence of cDNA products produced in the reaction of reverse transcription is correlated to the globin leader sequence on the graph. Total fluorescence of the major peaks on the left represents the amount of full-sized product when mRNA is read up to the 5′-end without interruption. Total fluorescence of the peaks (the trident) on the right corresponds to the product of reverse transcription stopped by the 48S ribosomal complex formed at the AUG initiation codon. Numerals in the figure indicate the percentage yield of initiation complexes calculated by fluorescence intensity of the peaks corresponding to the 48S complex, which is related to total fluorescence of all cDNA products formed at mRNA. b) Dependence of yield of 48S complex on ATP concentration according to the results of Fig. 1a.
Fig. 2. Formation of 48S ribosomal initiation complexes in the presence of 2 mM AMP-PNP and at different ATP concentrations (details in the legend to Fig. 1a).
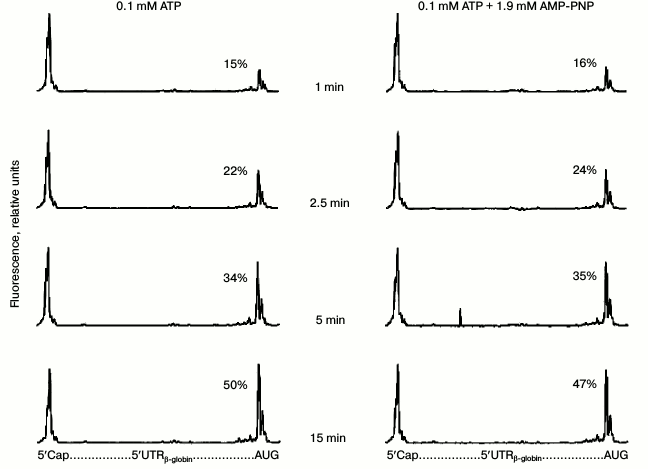
However, AMP-PNP could probably somehow slow the formation of 48S complexes. In other words, the chosen incubation time (15 min) is rather long, while the potential effect of AMP-PNP may be manifested at much shorter reaction times. There are no data in the literature that would demonstrate the time dependence of 48S ribosomal complex formation. We carried out the appropriate experiments (Fig. 3). Although a well-detected quantity of 48S initiation complexes appeared as early as after the first minute of incubation, the yield of these complexes was substantially dependent on time but completely independent of AMP-PNP predominance in the reaction mixture.
Fig. 3. Time dependence of formation of 48S ribosomal initiation complexes in the presence of 0.1 mM ATP (left panel) and 0.1 mM ATP + 1.9 mM AMP-PNP (right panel). Other details are in legend to Fig. 1a.
All our data convincingly show that AMP-PNP is not an inhibitor of ATP-dependent scanning of the mRNA leader sequence. It is known that the binding of the initiation factor eIF4A to mRNA is ATP-dependent [8]. In a review [4], with reference to the same work [8], it is mentioned that eIF4A does not bind to mRNA if ATP is replaced by its nonhydrolyzable analog AMP-PNP. Unfortunately, we failed to find any experimental evidence of this in the cited work [8]. However, this statement is in agreement with another observation, according to which the ATP hydrolysis catalyzed by the eIF4A factor is not inhibited in the presence of AMP-PNP [5]. It has been shown in a quite recently published work that AMP-PNP does not promote the conversion of eIF4A into active conformation: apparently, it simply does not bind to this factor [9]. It should be noted that our work, where AMP-PNP has been used not at the level of partial reactions but in the complete system of assembly of initiation complexes, rigorously demonstrates the absence of its influence on initiation altogether. Consequently, the uncleavable analog of ATP, AMP-PNP, cannot be used as an inhibitor in experimental studies of translation initiation in eukaryotes.
Finally, it should be added that our data cast doubt on the conclusion made in work [10] about ATP-independent translation reinitiation on polyribosomes. The authors of that work used AMP-PNP to prove their point of view, considering this reagent to be an inhibitor of ATP-dependent RNA helicase eIF4A, while it is now clear that it is not one. In light of the new data, the problem of the mechanism of translation reinitiation on polyribosomes is still open and needs further experiments.
The authors are grateful to A. S. Spirin for his support and constant interest in the work, his critical comments, and useful advice; to K. S. Vasilenko, V. A. Kolb, and E. A. Sogorin for discussion of the results; and to A. K. Surin for mass-spectrometry experiments.
This work was supported by the Russian Foundation for Basic Research (projects No. 12-04-01179-a and No. 13-04-40213-H) and by a grant of the Program “Molecular and Cell Biology” of the Presidium of the Russian Academy of Sciences.
REFERENCES
1.Kozak, M. (1978) How do eucaryotic ribosomes select
initiation regions in messenger RNA? Cell, 15,
1109-1123.
2.Vassilenko, K. S., Alekhina, O. M., Dmitriev, S.
E., Shatsky, I. N., and Spirin, A. S. (2011) Unidirectional constant
rate motion of the ribosomal scanning particle during eukaryotic
translation initiation, Nucleic Acids Res., 39,
5555-5567.
3.Spirin, A. S. (2009) How does a scanning ribosomal
particle move along the 5′-untranslated region of eukaryotic
mRNA? Brownian Ratchet model, Biochemistry, 48,
10688-10692.
4.Rogers, G. W., Jr., Komar, A. A., and Merrick, W.
C. (2002) eIF4A: the godfather of the DEAD box helicases, Nucleic
Acid Res. Mol. Biol., 72, 307-331.
5.Bi, X., Ren, J., and Goss, D. J. (2000) Wheat germ
translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase
activity by increasing the ATP binding affinity of eIF4A,
Biochemistry, 39, 5758-5765.
6.Hartz, D., McPheeters, D. S., Traut, R., and Gold,
L. (1988) Extension inhibition analysis of translation initiation
complexes, Methods Enzymol., 164, 419-425.
7.Shirokikh, N. E., and Spirin, A. S. (2008) Poly(A)
leader of eukaryotic mRNA bypasses the dependence of translation on
initiation factors, Proc. Natl. Acad. Sci. USA, 2105,
10738-10743.
8.Abramson, R. D., Dever, T. E., Lawson, T. G., Ray,
B. K., Thach, R. E., and Merrick, W. C. (1987) The ATP-dependent
interaction of eukaryotic initiation factors with mRNA, J. Biol.
Chem., 262, 3826-3832.
9.Harms, U., Andreou, Z. A., Gubaev, A., and
Klostermeier, D. (2014) eIF4B, eIF4G and RNA regulate eIF4A activity in
translation initiation by modulating the eIF4A conformational cycle,
Nucleic Acids Res., 42, 7911-7922.
10.Kopeina, G. S., Afonina, Z. A., Gromova, K. V.,
Shirokov, V. A., Vasiliev, V. D., and Spirin, A. S. (2008) Step-wise
formation of eukaryotic double-row polyribosomes and circular
translation of polysomal mRNA, Nucleic Acids Res., 36,
2476-2478.