FoF1-ATP Synthase of Streptomyces fradiae ATCC 19609: Structural, Biochemical, and Functional Characterization
M. G. Alekseeva1, T. A. Mironcheva1, D. A. Mavletova1, S. M. Elizarov2, N. V. Zakharevich1, and V. N. Danilenko1*
1Vavilov Institute of General Genetics, Russian Academy of Sciences, ul. Gubkina 3, 119991 Moscow, Russia; fax: +7 (499) 132-8962; E-mail: valerid@rutenia.ru2Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33/2, 119071 Moscow, Russia; fax: +7 (495) 954-2732
* To whom correspondence should be addressed.
Received July 22, 2014; Revision received October 27, 2014
The patterns of protein phosphorylation in inverted membrane vesicles from the strain Streptomyces fradiae ATCC 19609 were investigated to elucidate the mechanisms of regulation of bacterial membrane bound FoF1-ATP synthase. We found for the first time by two-dimensional gel electrophoresis and mass spectrometry that the β- and b-subunits of the FoF1-ATP synthase complex undergo phosphorylation; 20 proteins with known functions were identified. All eight subunits of FoF1-ATP synthase, i.e. α, β, γ, δ, ε, a, b, and c, were cloned into Escherichia coli and expressed as recombinant proteins. Using a crude preparation of serine/threonine protein kinases, we demonstrated the phosphorylation of recombinant γ-, β-, α- and ε-subunits. The β-subunit was phosphorylated both as a recombinant protein and in vesicles. Differential phosphorylation of membrane-bound and recombinant proteins can be attributed to different pools of protein kinases in each preparation; in addition, certain steps of FoF1-ATP synthase assembly and function might be accompanied by individual phosphorylation patterns. The structure of the operon containing all subunits and regulatory protein I was identified. The phylogenetic similarity of FoF1-ATP synthase from Streptomyces fradiae ATCC 19609 with the respective proteins in saprophytic and pathogenic (including Mycobacterium tuberculosis) bacteria was investigated. Thus, bacterial serine/threonine protein kinases are important for the regulation of FoF1-ATP synthase. From the practical standpoint, our results provide a basis for designing targeted antibacterial drugs.
KEY WORDS: FoF1-ATP synthase, Ser/Thr protein kinase, inverted membrane vesicles, StreptomycetesDOI: 10.1134/S0006297915030050
Abbreviations: a.a., amino acid residue; Bis-I, bis-indolyl-maleimide I; bp, base pairs; DTT, dithiothreitol; KN-62, inhibitor of Ca2+-dependent protein kinases; PMSF, phenylmethylsulfonyl fluoride; STPK, serine/threonine protein kinases; TCA, trichloroacetic acid.
The search of novel drugs for various human diseases is a vital problem
of modern biology and medicine. Many studies aimed at creating
innovative drugs are based on the data on molecular targets of the new
generation of drugs. One of the currently popular biotargets is
FoF1-ATP synthase, a universal molecular
machinery that performs ATP synthesis (and, under certain conditions,
hydrolysis) in eukaryotic mitochondria, plant chloroplasts, and
bacteria. FoF1-ATP synthase is a multisubunit
enzyme consisting of the soluble F1-part catalyzing ATP
synthesis from ADP and inorganic phosphate due to rotational movement
of the central core of the molecule and the associated
Fo-part submerged in the membrane of the molecule [1, 2]. The F1-part of
bacterial FoF1-ATP synthase comprises five
subunits: α, β, γ, δ, and ε. The
Fo-part contains three subunits: a, b, and c. This complex
is present in eukaryotic mitochondria and bacterial cell wall [3-5].
FoF1-ATP synthase is involved in maintaining pH level in cells, as well as in the regulation of endocytosis, proliferation, and apoptosis [6, 7]. Drugs developed in recent years can influence the function of FoF1-ATP synthase of different organisms, in first place human beings and pathogenic bacteria [8, 9]. The activity of FoF1-ATP synthase can be inhibited not only via the interaction with its own subunits, but also with coupled biotargets in subbacterial inverted membrane vesicles. A novel preparation targeted at the FoF1-ATP synthase of Mycobacterium tuberculosis was developed: the compound bedaquiline, which inhibits mycobacterial FoF1-ATP synthase and demonstrates high and selective activity in vitro and in vivo [10, 11]. The difference of actinobacterial FoF1-ATP synthases from the enzymes of other bacteria, with all their great structural similarity, is the dependence of their activity on Ca2+ [12, 13] and sensitivity to oligomycin A [14-17]. The main biotarget is the c-subunit of FoF1-ATP synthase and its homolog in eukaryotic cells [18]. Antibiotics of the oligomycin family and other human and bacterial FoF1-ATP synthase inhibitors are considered as potential drugs [19]; however, their advancement as pharmaceuticals is limited by their high toxicity. Russian scientists have obtained for the first time more than 20 semisynthetic oligomycin A derivatives with lower toxicity [20, 21].
The literature gives some indications of phosphorylation of the δ- and c-subunits in eukaryotic mitochondria [22, 23], the β-subunit in the mitochondria of human skeletal muscle [24] and chloroplasts [25], and the β-subunit in M. tuberculosis [26]. Phosphorylation of the components of the complex supposedly influences mitochondrial membrane permeability (subunit c) and the structure of the FoF1-ATP synthase complex (subunit β) and perhaps PDGF-dependent (subunit δ). Previously, we established the phenomenon of enhanced resistance of bacterial cells and FoF1-ATP synthase as a component of membrane vesicles to oligomycin A because of appearance of an oligomycin A-insensitive form of the FoF1-complex in cells and assessed the sensitivity of FoF1-ATP synthases from three streptomycete strains in vivo and in vitro to inhibitors (oligomycin A, etc.). We showed that the fraction of membrane vesicles of the actinobacterium Streptomyces fradiae ATCC 19609 contains at least two active serine/threonine protein kinases (STPK) capable of membrane protein phosphorylation, and their activity is regulated by Ca2+ [17]. The FoF1-ATP synthase operon was constructed and the subunits of proteins encoded by this operon were compared phylogenetically with analogous proteins from different groups of saprophytic and pathogenic bacteria.
The main objective of this work was to study the phosphorylation of proteins of the FoF1-ATP synthase complex from S. fradiae ATCC 19609 using two approaches: (1) as a component of membrane vesicles including membrane-bound STPK and (2) via the phosphorylation of recombinant proteins of all eight subunits obtained in Escherichia coli using the total preparation of STPK from S. fradiae ATCC 19609.
MATERIALS AND METHODS
Bacterial strains, vectors, media, and cultivation conditions. The strains used in the work were S. fradiae ATCC 19609 (the genome was sequenced at the Laboratory of Genetics of Microorganisms, Vavilov Institute of General Genetics, Russian Academy of Sciences; the genome sequence is deposited in GenBank with access number JNAD00000000) and E. coli DH5α (F–, Φ80 ΔlacZΔM15 Δ(lacZYA-argF) U169) from Promega (USA) [27] and BL21(DE3) (F– dcm ompT hsdS(rB–mB–) gal λ (DE3)), and the pET32a plasmid from Novagen (USA) [28]. The strain S. fradiae ATCC 19609 was grown in a liquid YEME medium with 25% sucrose (w/v) [29] at 28°C for 24 h (late logarithmic growth); mycelium was harvested by centrifugation at 3000g for 30 min. Escherichia coli cells were grown in Luria medium (L-broth). Solid media contained 2% agar (w/v) [30]. Ampicillin (100 µg/ml) was added for selective growth of plasmid-carrying cells.
DNA manipulations. The total DNA of the strain S. fradiae ATCC 19609 was isolated by the method described by Kieser et al. [29]. Plasmid DNA isolation, preparation of a competent E. coli culture, transformation, and analysis of recombinant plasmids were carried out by the standard methods [30]. PCR with the total DNA of the strain S. fradiae ATCC 19609 was performed with a PCK-100 kit (Dialat Ltd., Russia) using PTC-0150 (MJ Research, USA). Ten oligonucleotides constructed for amplification of the fragments of DNA from all subunits using NCBI/Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) were homologous to the flanking regions of the a-, c-, b-, δ-, α-, γ-, β-, and ε-subunits of FoF1-ATP synthases present in the NCBI database (http://www.ncbi.nlm.nih.gov/) from the strains of S. coelicolor, S. lividans, and S. avermitilis. The oligonucleotides for cloning purposes were constructed based on sequencing the amplified subunits (Table 1). The amplified subunits were cloned into pET32a expression vector at the sites of restriction endonucleases EcoRI and HindIII. The resulting recombinant clones were screened by PCR using S·Tag and T7term standard primers.
Table 1. Oligonucleotides used in the
work
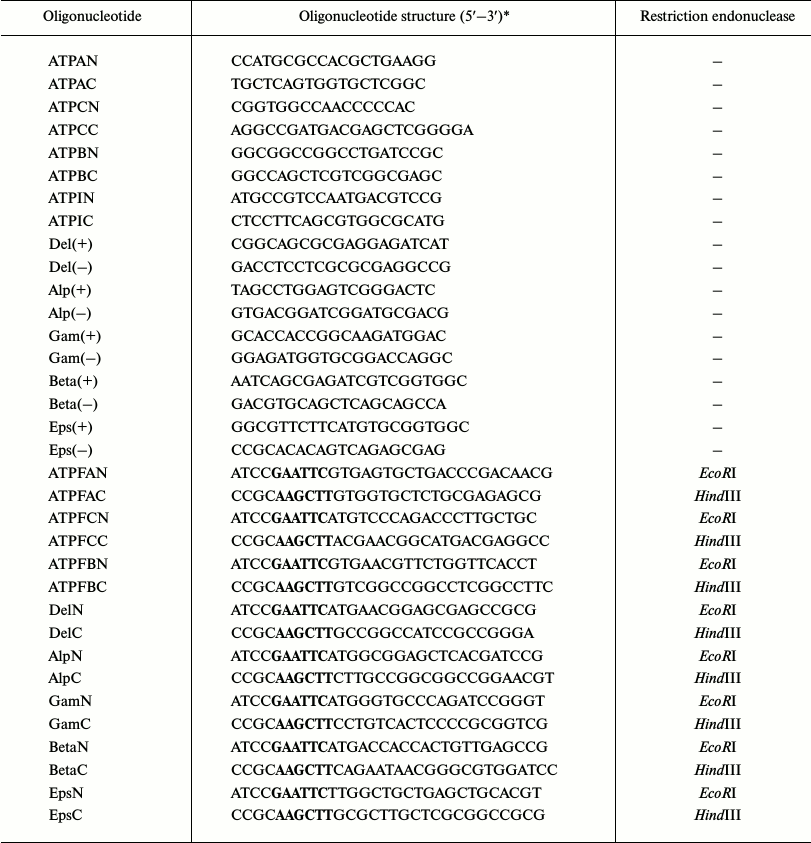
*Recognition sites for EcoRI (GAATTC) and
HindIII (AAGCTT) restriction endonucleases are in bold.
Study of FoF1-ATP synthase gene expression in E. coli. The genes of subunits of FoF1-ATP synthase from the strain S. fradiae ATCC 19609 were expressed in E. coli BL21(DE3) cells. For the study of protein expression and for biomass production, E. coli cells carrying the constructed plasmids were grown in liquid medium (L-broth) with ampicillin on a shaker at 34°C until the optical density of 0.6 (~2 h); then expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cultivation was carried out at 28°C for 4 h; the biomass was harvested and suspended in buffer of the following composition: 62.5 mM Tris-HCl (pH 6.8), 5% glycerol (w/v), 2% mercaptoethanol (w/v), 0.1% SDS, bromophenol blue. Then the cells were destroyed by boiling for 10 min. The soluble protein fraction was analyzed by SDS-PAGE. The soluble protein fraction of the strain E. coli BL21(DE3) carrying the pET32a plasmid without an insert was analyzed as a control. Recombinant proteins were isolated from extracts under native conditions on His-binding Ni-NTA columns (Qiagen, Germany) according to the QIAexpress protocol.
Membrane vesicle preparations for proteomic analysis. The mycelium of S. fradiae ATCC 19609 was washed three times in 0.1 M Tris-HCl buffer (pH 7.5). The cells were suspended in 10 volumes of buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10% glycerol (w/v), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM dithiothreitol (DTT), and protease inhibitor cocktail (Promega, USA), with the addition of lysozyme to final concentration of 1 mg/ml. The resulting suspension was incubated at 37°C for 30 min under continuous stirring. Protoplasts were broken by sonication in a Vibra Cell™ Ultrasonic Processor (Sonics, USA) at 20 kHz three times by 30 s, with a 15-s interval between the treatments at 4°C. Cell debris was precipitated by centrifugation for 20 min at 4°C and 10,000g. Vesicles were isolated as follows: supernatant proteins were centrifuged for 10 h at 4°C and 100,000g, suspended in the same buffer (without lysozyme), and centrifuged for 16 h at 4°C and 150,000g. The precipitates were dissolved in six volumes of the buffer (without lysozyme). Protein concentration was assayed with a Qubit 2.0 fluorimeter (Invitrogen, USA). Membrane vesicles were frozen in liquid nitrogen and stored at –70°C.
Proteins of membrane vesicles were phosphorylated for 10 min at 25°C in buffer containing 25 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 1 mM EDTA, and 0.1 mM PMSF. The reaction was started by adding ATP to final concentration of 100 µM, containing 10-20 µCi [γ-32P]ATP (5000 Ci/mmol; 186 PBq/mol) (Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences), in the presence of 100 µg of vesicular protein. Proteins were precipitated with five volumes of cold acetone and kept for 2 h at –20°C; the precipitates were harvested by centrifugation for 20 min at 4°C and 20,000g. Slightly dried precipitates were dissolved in buffer containing 8.5 M urea, 2% Triton X-100 (w/v), 2.2% ampholytes (w/v), and 5% β-mercaptoethanol (w/v); insoluble aggregates were removed by centrifugation, and the supernatant was used for further separation by two-dimensional electrophoresis.
Two-dimensional gel electrophoresis (SDS-PAGE) was carried out according to the method of O’Farrell [31] with minor modifications. The samples for mass spectrometry were prepared by the method recommended by Promega (USA) (In Gel Digest Protocol); mass spectra were obtained by the method recommended by Bruker Daltonics (USA) with a Bruker Daltonics analytic mass spectrometer (UltrafleXtreme MALDI TOF/TOF Ms) (Bruker Daltonics GmbH, Germany) at the Department of Proteomic Research, Orekhovich Research Institute of Biomedical Chemistry, Russian Academy of Medical Sciences. Proteins were identified using Mascot software (www.matrixscience.com). The search was carried out in the NCBI database.
Membrane vesicles for phosphorylation of FoF1-ATP synthase subunits. The strain S. fradiae ATCC 19609 was grown in basic liquid nutrient medium prepared as described previously [32, 33] to the late exponential growth phase; the mycelium was harvested by centrifugation and suspended in buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10% glycerol (w/v), 1 mM PMSF, 0.5 mM DTT, 1 mg/ml lysozyme, and 1 µg/ml DNase; protoplasts were obtained by incubation at 37°C for 30 min. The protoplasts were broken by sonication in a domestic ultrasonic disintegrator UZDN-1, U-42 at frequency of 30 kHz for 3 min; cell debris was removed by 20-min centrifugation at 4°C and 10,000g. Membrane vesicles were precipitated by 120-min centrifugation at 4°C and 180,000g, washed with the above buffer, and stored in liquid nitrogen.
Phosphorylation of FoF1-ATP synthase subunits. The subunits were phosphorylated for 10 min at 28°C in a buffer containing 50 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mM PMSF, 0.01% Tween-20, 10% glycerol (w/v) (pH 7.8), and 0.5 mM [γ-32P]ATP (5000 pulses/min per pmol; Fosfor, Russia), in the presence of 100 µg/ml protein.
Total STPK preparation from S. fradiae ATCC 19609 was isolated from bacterial extracts by chromatography on Cibacron Sepharose [34].
ATPase activity in the membrane vesicles of Streptomyces fradiae 19609 was analyzed by the release of 32Pi after its separation from [γ-32P]ATP bound with Norit A activated carbon as described previously [17]. In all cases, the average values of three independent measurements with root mean square deviations are presented.
Bioinformatic methods of analysis. The nucleotide and amino acid sequences of the subunits of bacterial FoF1-ATP synthases were searched in the nucleotide and amino acid databases available at the NCBI website (http://www.ncbi.nlm.nih.gov/). The sequences were analyzed with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The subunits were aligned using ClustalW [34] and TCOFFEE (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) [35]. The alignment was visualized with GeneDoc (http://www.nrbsc.org/gfx/genedoc/) [36]. The percentage of identity of amino acid sequences was calculated using Blastp software [37]. Phylogenetic trees were constructed using MEGA v. 5.1 [38] with the neighbor-joining algorithm [39]. The reliability of topology of the phylogenetic tree was estimated by bootstrap analysis (1000 replicas) [40]. Potential phosphorylation sites were mapped with NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) and NetPhosK (http://www.cbs.dtu.dk/services/NetPhosK/).
RESULTS
Analysis of ATPase activity of S. fradiae ATCC 19609 in membrane vesicles. Vesicles containing FoF1-ATP synthase were isolated from S. fradiae ATCC 19609; ATPase activity was determined by the cleavage of the γ-phosphoryl residue from [γ-32P]ATP in the presence and absence of STPK inhibitors. The vesicles were preincubated with ATP and with the ATPase and STPK inhibitors, and their ATPase activity was determined in the presence or absence of oligomycin A. The activity of FoF1-ATP synthase was shown to be 157 ± 19 U/mg protein in the presence of 10 mM CaCl2 and 104 ± 7 U/mg protein of membrane vesicles in the presence of 10 mM MgCl2 (1 activity unit is equal to 1 nmol of phosphate released during 1 min in the presence of 1 mg of protein under standard conditions). The differences in the efficiency of divalent cations may be accounted for by the differences in the atomic structure of their complex with the nucleotide substrate, influencing conformational changes in the catalytic center of the enzyme; earlier, they were found in ATPases from various Streptomyces species [12, 13]. The effects of increasing concentrations of the selective F-type ATPase inhibitor, oligomycin A, on ATPase activity were analyzed; it was shown that the IC50 values of oligomycin A for Ca2+- and Mg2+-dependent ATPase activity in membrane vesicle preparations were 165 ± 23 and 110 ± 11 nM, respectively. Separate experiments showed that dicyclohexylcarbodiimide (the inhibitor affecting the Fo-part at a concentration of 0.05 mM) reduces ATPase activity of the studied strain to 7-10% of the initial level. Then the vesicles of S. fradiae were preincubated with STPK inhibitors, and their ATPase activity was determined in the presence and absence of oligomycin A. The results presented in Table 2 show that the Ca2+-dependent protease inhibitor KN-62 (5 µM) and the ATP-competitive kinase inhibitor bis-indolyl-maleimide I (Bis I) (1 µM) reduce the activity by 25 and 23%, respectively. At the same time, the determined ATPase activity can be almost completely ascribed to FoF1-ATP synthase, because its specific inhibitors dicyclohexylcarbodiimide and oligomycin A suppress the ATPase activity of membrane vesicles by 83 and 56%, respectively. KN-62 and Bis I enhance the sensitivity of FoF1-ATP synthase to oligomycin A by 4-6% when added separately and nearly twofold when added together (Table 2). These data suggest that endogenous membrane-bound STPKs are involved in FoF1-ATP synthase activation and protection from antibiotic and confirm the preliminary data of the same analysis of the phosphorylation and potential regulation of activity of FoF1-ATP synthase in S. fradiae vesicles that we published previously [17].
Table 2. Effects of oligomycin A and STPK
inhibitors on the activity of ATPase from S. fradiae 19609
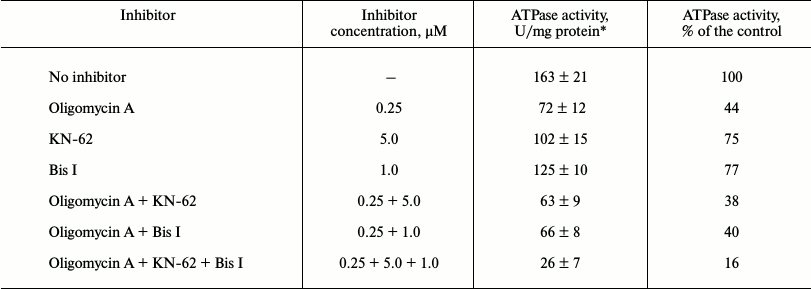
*Results of three independent measurements ± S. D. are presented.
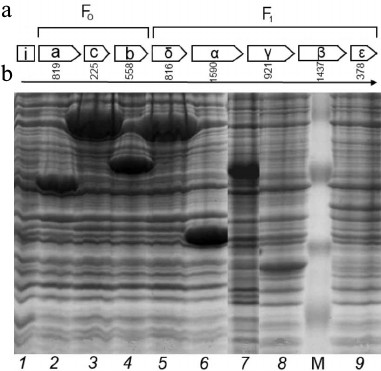
Recombinant proteins of subunits of FoF1-ATP synthase from S. fradiae ATCC 19609. The genes of FoF1-ATP synthase subunits were isolated from genomic DNA by PCR. The following oligonucleotides were used for DNA amplification: ATPAN and ATPAC for the a-subunit, ATPCN and ATPCC for the c-subunit, ATPBN and ATPBC for the b-subunit, Del(+) and Del(–) for the δ-subunit, Alp(+) and Alp(–) for the α-subunit, Gam(+) and Gam(–) for the γ-subunit, Beta(+) and Beta(–) for the β-subunit, and Eps(+) and Eps(–) for the ε-subunit (Table 1). The start codons of translation and the termination codons were determined by sequencing all of the amplified DNA fragments. The oligonucleotides synthesized for cloning all subunits were homologous to the N- and C-terminal regions of the subunit genes, respectively, and contained the EcoRI and HindIII restriction sites (Table 1). The amplified DNA fragments were sequenced and cloned into pET32a expression vector, the linker containing His-Tag and S-tag for protein isolation and purification and the Trx·Tag sequence (109 bp) in the N-terminal region. IPTG induction for 1, 2, and 4 h was used to study protein expression. The maximal level of gene expression for all subunits was achieved with 4-h induction. The cloning of δ-, α-, γ-, β-, ε-, a-, c-, and b-subunits in E. coli cells carrying the plasmids pET32aAtpH, pET32aAtpA, pET32aAtpG, pET32aAtpD, pET32aAtpC, pET32aAtpB, pET32aAtpE, and pET32aAtpF revealed the presence of additional protein fractions with respective molecular masses of 51, 80, 56, 75, 36, 53, 30, and 43 kDa (Fig. 1). These values correspond to the calculated molecular masses of proteins of the respective subunits plus the protein molecular mass of the whole linker of the plasmid pET32a containing thioredoxin. The recombinant proteins of all subunits were purified on His-bindings Ni-NTA columns and produced in preparative amounts for further study of their phosphorylation.
Fig. 1. a) Structure of the operon of FoF1-ATP synthase from the strain S. fradiae 19609: I-protein, Fo-part (subunits a, c, and b), and F1-part (subunits δ, α, γ, β, ε). b) Electrophoregram of the soluble protein fraction of the strain E. coli BL21(DE3). Escherichia coli clones containing plasmids: 1) pET32a (control); 2) pET32aAtpH (subunit δ); 3) pET32aAtpA (subunit α); 4) pET32aAtpG (subunit γ); 5) pET32aAtpD (subunit β); 6) pET32aAtpC (subunit ε); 7) pET32aAtpB (subunit a); 8) pET32aAtpE (subunit c); 9) pET32aAtpF (subunit b); M) protein marker SM0441 (Fermentas, Lithuania).
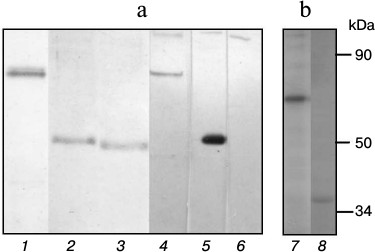
Phosphorylation of recombinant subunits of FoF1-ATP synthase from S. fradiae ATCC 19609 by total STPK preparation. The phosphorylation of recombinant subunits of FoF1-ATP synthase was analyzed in the presence of total protein kinase preparation from S. fradiae 19609 obtained by affinity chromatography on Cibacron Sepharose. To avoid misrepresentation of results because of endogenous phosphorylation in the preparation of kinases, the latter were preincubated with unlabeled ATP immediately before being added to the working mixture for analysis. Then ATP was removed by gel filtration. The analytic mixture included the labeled ATP and FoF1-ATP synthase subunits isolated on His Bind resin. The inclusion of radioactive phosphate in the subunits was detected by autoradiography. Figure 2 shows that labeled phosphate is included in the proteins with molecular weights of 75 and 56 kDa, which are thioredoxin-fused β- and γ-subunits of FoF1-ATP synthase, respectively. Specific labeling of γ-subunit is higher by approximately an order of magnitude compared to that of β-subunit. There is a minor inclusion of the label in proteins with molecular weights of 80 and 36 kDa (α- and ε-subunits). The inclusion of labeled phosphate in other subunits of the Fo- and F1-parts of FoF1-ATP synthase (a, b, c, and δ) was not observed. The absence of inclusion indicates that it is just FoF1-ATP synthase subunits, but not thioredoxin, that are modified in the recombinant hybrid proteins fused with thioredoxin of α-, β-, γ-, and ε-subunits. This is the first evidence of the fact that α-, β-, γ-, and ε-subunits of the actinomycete complex can be phosphorylated by endogenous STPKs.
Fig. 2. a) Electrophoregram of the gels after electrophoresis of recombinant subunits α (1), γ (2), δ (3). b) Autoradiograph of phosphorylated subunits α (4), γ (5), δ (6), ε (7), and β (8) of the FoF1-ATP synthase from S. fradiae ATCC 19609.
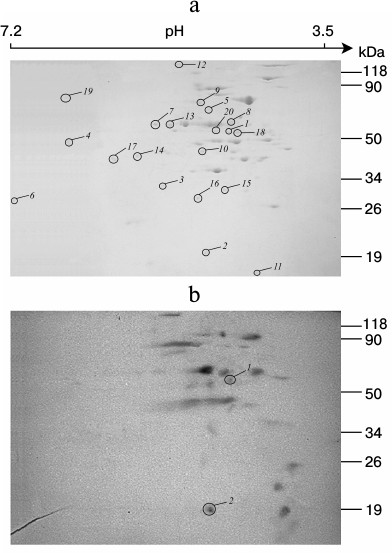
Identification of phosphorylated proteins in the membrane vesicle fraction of S. fradiae ATCC 19609. The vesicles isolated from the culture of S. fradiae ATCC 19609 were incubated in the presence of [γ-32P]ATP. Phosphorylated proteins separated by 2D electrophoresis are shown in Fig. 3. Mass spectrometry revealed two proteins identical to the β-subunit of the F1-part and the b-subunit of the Fo-part of FoF1-ATP synthase. The other 18 identified proteins can be divided into seven functional groups (Table 3). The functional group of “energy conversion and fatty acid metabolism”, in addition to β- and b-subunits, contains proteins probably associated with the function of FoF1-ATP synthase. The absence of γ-subunit in the preparations might be accounted for by its selective loss when preparing for electrophoresis.
Fig. 3. 2D SDS-PAGE of proteins of membrane vesicles from S. fradiae 19609. a) Electrophoregram of Coomassie G250 stained gel: 1-20 are the proteins identified by mass spectrometry and presented in Table 3. b) Autoradiogram. Mass-spectrometric identification of the marked polypeptides: 1) F1 part of FoF1-ATP synthase, subunit β; 2) Fo part of FoF1-ATP synthase, subunit b.
Table 3. Major proteins with established
functions identified in the membrane vesicle fraction of
Streptomyces fradiae 19609 presented in Fig. 3
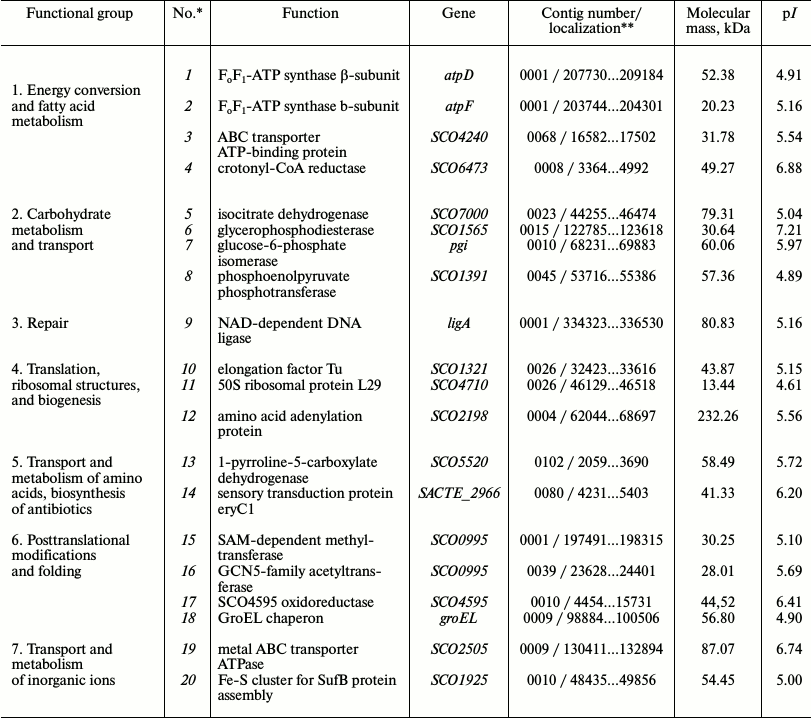
*Protein numbers indicated in Fig. 3a.
**Localization of nucleotide sequences in the NCBI database
for S. fradiae 19609.
Determination of operon structure of FoF1-ATP synthase from strain S. fradiae ATCC 19609. The sequencing of amplified DNA fragments of all subunits showed that the operon of FoF1-ATP synthase from strain S. fradiae 19609 has a standard structure: subunits a, c, and b (the Fo-part) and subunits δ, α, γ, β, and ε (the F1-part) (Table 4). Comparative analysis showed that the nucleotide sequences of all subunits of FoF1-ATP synthase from S. fradiae ATCC 19609 have high homology with the genes of the respective subunits from strains of S. coelicolor, S. lividans, and S. avermitilis. The nucleotide sequences of the genes of all subunits of FoF1-ATP synthase from S. fradiae ATCC 19609 are registered in GenBank (KC169996, KC169997, KC169998, KC169999, KC170000, KC170001, KC170002, and KC170003). The operon of FoF1-ATP synthase from the strain S. fradiae ATCC1 9609 is shown in Fig. 1a. The whole genome of S. fradiae 19609 was sequenced at the Laboratory of Genetics of Microorganisms (Vavilov Institute of General Genetics, Russian Academy of Sciences), and the amino acid sequences were deposited in GenBank (JNAD00000000). The FoF1-ATP synthase genes were annotated; the nucleotide sequences of all subunits coincided with those sequenced previously.
Table 4. Operon structure of
FoF1-ATP synthase from S. fradiae ATCC
19609
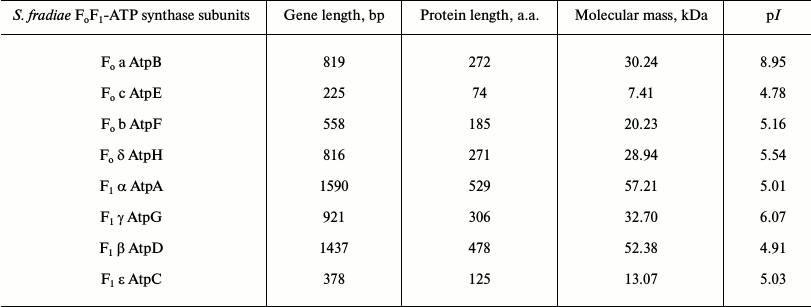
Comparative analysis of subunits of FoF1-ATP synthases from S. fradiae 19609, S. lividans TK24, and S. avermitilis MA-4680. The genetically best-studied strains S. lividans TK24 and S. avermitilis MA-4680 were chosen for this experiment. The alignment of amino acid sequences of eight subunits of FoF1-ATP synthase from strain S. fradiae 19609 with subunits from strain S. lividans TK24 (oligomycin A sensitive) and strain S. avermitilis MA-4680 producing oligomycin A (oligomycin A resistant) showed that subunits a, c, b, α, and β from strain 19609 have high similarity to the analogous subunits from strain TK24, while subunits δ and γ from strain 19609 have high similarity to subunits from strain MA-4680. The sequence of subunit ε is similar in all three strains. The results of comparative analysis of the operon of FoF1-ATP synthase from S. fradiae 19609 with operons of FoF1-ATP synthases from S. lividans TK24 and S. avermitilis MA-4680 are given in Table 5.
Table 5. Comparative analysis of the operon
of FoF1-ATP synthase from S. fradiae ATCC
19609 with operons of FoF1-ATP synthases from
S. lividans TK24 and S. avermitilis MA-4680
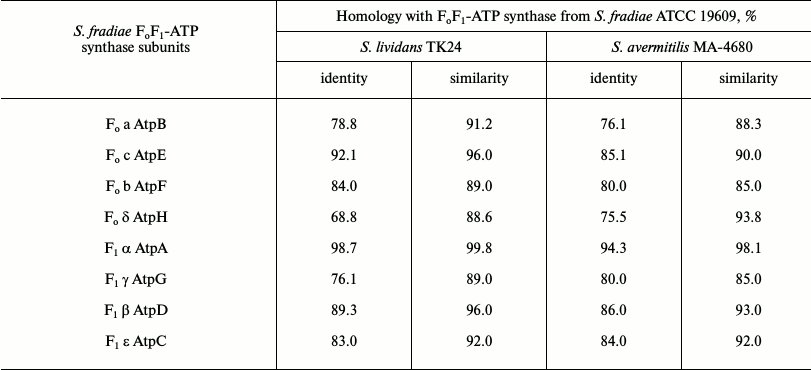
Comparative analysis of nucleotide sequences located upstream and downstream of the FoF1-ATP synthase genes on the chromosome showed the presence of I-protein; the identity (similarity) with the respective amino acid sequences of strains S. lividans TK24 and S. avermitilis MA-4680 was 64 (77) and 64 (80%), respectively; the nucleotide sequence homology was no more than 35-38%. The neighborhoods of the genes of FoF1-ATP synthase from S. fradiae 19609 completely coincide with the neighborhoods typical of most strains of the genus Streptomyces (with the exception of S. avermitilis strains): at the 5′-end of the chromosome, immediately before the operon, there are genes of tyrosine phosphatase, serine hydroxymethyl transferase, and transferase; after the operon, there are genes of secreted protein and chitinase C. In S. avermitilis strains, the genes for the synthesis of oligomycin A (FoF1-ATP synthase inhibitor) are located before the operon.
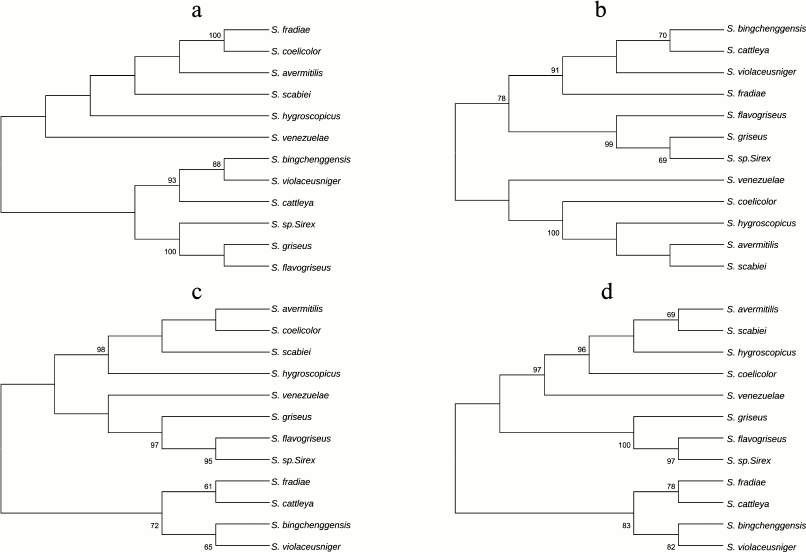
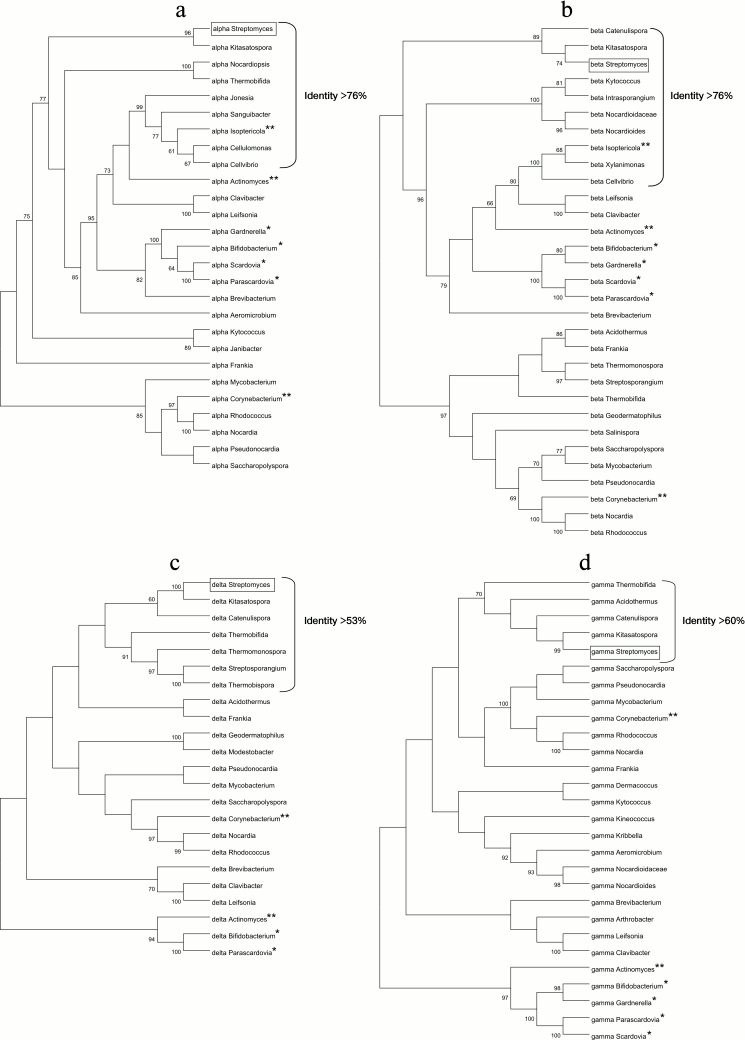
Phylogenetic analysis of subunits of the FoF1-ATP synthase operon from strains of genus Streptomyces. Phylogenetic relationships between the F1-part subunits of the FoF1-ATP synthase operon were established by aligning the amino acid sequences of α-, β-, δ-, and γ-subunits for different representatives of the genus Streptomyces and by assessing sequence identity. Four phylogenetic trees were constructed for Streptomyces bacteria based on the alignment of amino acid sequences of four subunits (α, β, δ, and γ) of the F1-part of the FoF1-ATP synthase operon (Fig. 4).
Fig. 4. Phylogenetic trees of representatives of the Streptomyces genus based on comparison of four subunits of the F1-part of the FoF1-ATP synthase operon: a) α-subunit; b) β-subunit; c) δ-subunit; d) γ-subunit. The trees were constructed using the MEGA 5 software package by alignment of amino acid sequences using the neighbor-joining (NJ) algorithm and the p-distance model. The numerals show the stability of branches calculated for bootstrap NJ analysis (1000 replicas). The branches reproduced in less than 60% bootstrap replicas are marked.
As Fig. 4 shows, the closest neighbors of S. fradiae on three trees constructed for subunits β, δ, and γ are S. violaceusniger, S. bingchenggensis, and S. cattleya, in contrast to the tree constructed for subunit α. On the α-subunit tree, the closest neighbors of S. fradiae are S. coelicolor, S. avermitilis, and S. scabiei. This suggests that subunits of the F1-part of the FoF1-ATP synthase operon evolved separately.
Comparative analysis of subunits of the FoF1-ATP synthase operon from strains of genus Streptomyces. The alignment of amino acid sequences showed a high degree of conservatism for subunits α and β. The identity of these subunits was 88-99%. Subunits δ and γ were less conservative; their identity was 75-92%. Conservative regions in these subunits cover almost the entire amino acid sequence (Figs. S1-S4 in the Supplement to this paper on the website of the journal (http://protein.bio.msu.ru/biokhimiya), in black). However, the sequence of the γ-subunit (Fig. S4 in the Supplement) of the completely sequenced genome of S. bingchenggensis is shorter by 56 a.a. than its homologs from other streptomycete species. It is unknown why this subunit has lost some the amino acids in its N-terminal part. Inserts of different length were found in the γ-subunit of plants and the bacterium M. tuberculosis. These variations are considered as adaptations of the subunit to the function of ATP synthesis in different organisms [9].
Comparative analysis of subunits of the FoF1-ATP synthase operon in actinobacteria. The analytical methods of bioinformatics were used to compare the eight subunits of FoF1-ATP synthases from different actinobacteria. The results of this analysis showed similarity of operon structure for FoF1-ATP synthases from all actinobacteria. The alignment of amino acid sequences of subunits α, β, δ, and γ from different species of the type Actinobacteria (Figs. S5-S8 of the Supplement) showed that subunits α and β are conservative nearly along the entire length (>70% identity). The most variable regions are observed in subunits α and β only in the C-terminal part of the sequence; in subunit β, the first 40 a.a. are variable too.
Subunits δ and γ are less conservative (identity of subunits γ and δ is within the range of 78-33 and 71-30%, respectively). Subunit γ has a region of elongated deletions approximately in the middle of the amino acid sequence. In subunit δ, the region of deletions is located in the N-terminal part of the sequence, while identical regions are represented by single conservative amino acid residues located mainly in the center and at the end of the sequence.
It is interesting to note that the amino acid sequences of a-, β-, and γ-subunits from the strains of probiotic anaerobic actinobacteria of the genus bifidobacterium contain additional amino acid regions, so-called inversions: 222-226 a.a. in subunit a; 53-56 and 119-121 a.a in subunit β; and 205-211 a.a in subunit γ. Investigation of the significance of these differences for the function of FoF1-ATP synthase in human microbiota under anaerobic conditions is a subject for further research.
Phylogenetic analysis of FoF1-ATP synthases from actinobacteria. Four phylogenetic trees were constructed based on alignment of the amino acid sequences of the four subunits α, β, δ, and γ of the F1-part of the FoF1-ATP synthase operon from different species of actinobacteria (Fig. 5). All the trees reflect evolutionary divergence of the species. The identity percentage of each subunit is indicated for the bacterial genera closest to the genus Streptomyces (Fig. 5). The findings show that subunits α and β have been most conservative during evolution (as demonstrated above), because the identity percentage of these subunits is higher compared to subunits δ and γ.
Fig. 5. Phylogenetic trees based on the comparison of amino acid sequences of four subunits of the F1-part of the FoF1-ATP synthase operon of actinobacteria: a) α-subunit; b) β-subunit; c) δ-subunit; d) γ-subunit.
Comparative analysis of subunits of FoF1-ATP synthase complex from S. fradiae ATCC 19609 and ATPases from pathogenic microorganisms. The strain S. fradiae 19609 is considered as a convenient test system for studying inhibitors of bacterial FoF1-ATP synthase, including oligomycin derivatives [17, 20, 21]. Therefore, it was interesting to examine the similarity between the proteins of this operon and the respective proteins of some pathogenic bacteria. We aligned the amino acid sequences of eight subunits of FoF1-ATP synthases from S. fradiae with the subunits of FoF1-ATPases from M. tuberculosis (type (division) Actinobacteria) and C. difficile (type (division) Firmicutes). These alignments showed that: (1) subunits α and β remained rather conservative and are aligned along the entire length at 52-70% identity; (2) subunits δ and γ are less conservative, have low homology (25-48%), and are aligned not by the entire length; (3) in subunit ε from S. fradiae (the full length of subunit ε is 121 a.a. on average), only a 46-a.a. region was aligned with the analogous region from M. tuberculosis and C. difficile, with alignment identity of 43 and 35%, respectively; and (4) subunits a, b, and c from S. fradiae either are very poorly aligned with the homologs from M. tuberculosis and C. difficile (~26% identity) and not along the entire length, or there are no similar regions at all; e.g. all other conditions being equal, Blastp did not align whatsoever the amino acid sequence of subunit b from S. fradiae with the amino acid sequence of the same subunit from C. difficile.
Oligomycin A is known to bind only to certain amino acids in the c-subunit. Table 6 shows the comparison of oligomycin-binding domains of the c-subunit of FoF1-ATP synthase in oligomycin sensitive and resistant organisms, demonstrating that S. fradiae ATCC19609 has the highest similarity with Homo sapiens sapiens by these amino acids.
Table 6. Comparison of oligomycin-binding
domains of the FoF1-ATP synthase c-subunit in
oligomycin sensitive and resistant organisms (the table is taken from
Ko et al. [22], with modifications)
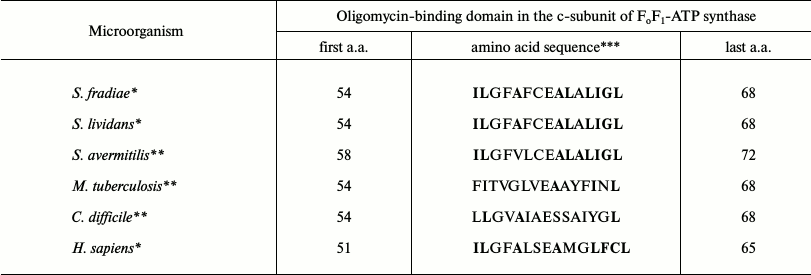
*Oligomycin A sensitive organisms.
**Oligomycin A resistant organisms.
***Amino acids involved in oligomycin A binding are in bold.
DISCUSSION
We show for the first time that the recombinant proteins of the γ-, β-, α-, and ε-subunits of the F1-part of the ATPase complex from the bacterium S. fradiae ATCC 19609 are phosphorylated by the STPK complex as a component of the cell extract. Two-dimensional gel electrophoresis and mass spectrometry (MALDI-TOF) shows for the first time the phosphorylation of the β- and b-subunits of the FoF1-ATP synthase complex in the membrane vesicle fraction. Phosphorylation of the β-subunit is observed in both types of experiments. The phosphorylation of the β-subunit was found in eukaryotic mitochondria [22-24] and plant chloroplasts [25], as well as in M. tuberculosis [26]. The ability to phosphorylate γ- and α-subunits in one type and b-subunit in the other type of experiments might be indicative of the importance of phosphorylation of γ- and α-subunits for the assembly of FoF1-ATP synthase complex and for its functioning at the stage of ATP synthesis or hydrolysis. Differences in phosphorylation conditions between different experiments in vitro and in vivo might also be significant.
Ser-Thr protein kinases are present and have been characterized in many bacteria; they have diverse functions including viability and virulence in pathogenic bacteria [41, 42]. The sequenced genome of S. fradiae 19609 (JNAD00000000) was shown to contain 30 STPKs; some are membrane-bound and can participate in the phosphorylation of FoF1-ATP synthase complex and associated proteins involved in cell energy supply and regulation of the function of this complex. In recent years, the FoF1-ATP synthase complex and the coupled proteins from bacteria and human mitochondria have become objects of keener interest as biotargets for drug production [9, 43, 44], as well as key objects of research, since the normal function of this complex is of practical importance for human health and longevity [45-47]. The universal nature of FoF1-ATP synthase in all living organisms, including bacteria and human beings, and its role as a biological nanomotor, require detailed studies of the function of this complex in all organisms. More profound understanding of the mechanisms of bacterial FoF1-ATP synthase functioning, the role of STPKs in this process, the differences of its structure and work from the human FoF1-ATP synthase can contribute to the development of novel drugs. Previously we showed that strain S. fradiae 19609 is supersensitive to oligomycin A and its semisynthetic derivatives, as well as more sensitive to a number of compounds of other chemical classes [17, 20]. It is important to note the fact that the sensitivity of S. fradiae 19609 and S. lividans TK24 to oligomycin A increases during inhibition of STPKs [17, 48]. This opens new possibilities for the development of anti-actinobacterial drugs with synergistic effects based on FoF1-ATP synthase inhibitors and STPKs. The FoF1-ATP synthase from S. fradiae 19609, due to similarity of its c-subunit (proton transport system) with the c-subunit of human FoF1-ATP synthase, as well as the high sensitivity of this strain to oligomycin A and its derivatives, can be a convenient model for studying human FoF1-ATP synthase.
These findings provide for scientific substantiation and expansion of prospects for creating a highly sensitive test system based on strain S. fradiae 19609 for selection of inhibitors of human and bacterial ATP synthases using approaches that we have developed [49, 50].
This work was supported by the Federal Targeted Program for Research and Development in Priority Areas of Development of the Russian Scientific and Technological Complex for 2007-2013 (State contract No. 02.512.12.2056 of May 20, 2009).
REFERENCES
1.Capaldi, R. A., and Aggeler, R. (2002) Mechanism of
the F(1)F(0)-type ATP synthase, a biological rotary motor, Trends
Biochem. Sci., 27, 154-160.
2.Senior, A. E. (2007) ATP synthase: motoring to the
finish line, Cell, 130, 220-201.
3.Watanabe, R., and Noji, H. (2013) Chemomechanical
coupling mechanism of F(1)-ATPase: catalysis and torque generation,
FEBS Lett., 587, 1030-1035.
4.Okuno, D., Iino, R., and Noji, H. (2011) Rotation
and structure of FoF1-ATP synthase, J.
Biochem., 149, 655-664.
5.Walker, J. E. (2013) The ATP synthase: the
understood, the uncertain and the unknown, Biochem. Soc. Trans.,
41, 1-16.
6.Chi, S. L., and Pizzo, S. V. (2006) Cell surface
F1Fo ATP synthase: a new paradigm, Ann.
Med., 38, 429-438.
7.Skulachev, V. P. (1999) Bacterial energetics at
high pH: what happens to the H+ cycle when the extracellular
H+ concentration decreases, Novartis Found. Symp.,
221, 200-213.
8.Ahmad, Z., Okafor, F., Azim, S., and Laughlin, T.
F. (2013) ATP synthase: a molecular therapeutic drug target for
antimicrobial and antitumor peptides, Curr. Med. Chem.,
20, 1956-1973.
9.Lu, P., Lill, H., and Bald, D. (2014) ATP synthase
in mycobacteria: special features and implications for a function as
drug target, Biochim. Biophys. Acta, 1837, 1208-1218.
10.Gras, J. (2013) Bedaquiline for the treatment of
pulmonary, multidrug-resistant tuberculosis in adults, Drugs Today
(Barcelona), 49, 353-361.
11.Biukovic, G., Basak, S., Manimekalai, M. S.,
Rishikesan, S., Roessle, M., Dick, T., Rao, S. P., Hunke, C., and
Gruber, G. (2013) Variations of subunit {varepsilon} of the
Mycobacterium tuberculosis F1Fo ATP
synthase and a novel model for mechanism of action of the tuberculosis
drug TMC207, Antimicrob. Agents Chemother., 57,
168-176.
12.Hensel, M., Deckers-Hebestreit, G., and
Altendorf, K. (1991) Purification and characterization of the
F1 portion of the ATP synthase (F1Fo)
of Streptomyces lividans, Eur. J. Biochem., 202,
1313-1319.
13.Hensel, M., Ahmus, H., and Deckers-Hebestreit, G.
(1991) The ATP synthase of Streptomyces lividans:
characterization and purification of the F1F0
complex, Biochim. Biophys. Acta, 1274, 101-108.
14.Pagliarani, A., Nesci, S., and Ventrella, V.
(2013) Modifiers of the oligomycin sensitivity of the mitochondrial
F1F0-ATPase, Mitochondrion, 13,
312-319.
15.Shchepina, L. A., Pletjushkina, O. Y., Avetisyan,
A. V., Bakeeva, L. E., Fetisova, E. K., Izyumov, D. S., Saprunova, V.
B., Vyssokikh, M. Y., Chernyak, B. V., and Skulachev, V. P. (2002)
Oligomycin, inhibitor of the F0 part of
H+-ATP-synthase, suppresses the TNF-induced apoptosis,
Oncogene, 21, 8149-8157.
16.Symersky, J., Osowski, D., Walters, D. E., and
Mueller, D. M. (2012) Oligomycin frames a common drug-binding site in
the ATP synthase, Proc. Natl. Acad. Sci. USA, 109,
13961-13965.
17.Alekseeva, M. G., Elizarov, S. M., Bekker, O. B.,
Lyubimova, I. K., and Danilenko, V. N. (2009)
F0F1-ATP synthase of streptomycetes: modulation
of activity and sensitivity to oligomycin by serine/threonine protein
kinases, Biol. Membr. (Moscow), 26, 41-49.
18.Symersky, J., Pagadala, V., Osowski, D., Krah,
A., Meier, T., Faraldo-Gomez, J. D., and Mueller, D. M. (2012)
Structure of the c(10) ring of the yeast mitochondrial ATP synthase in
the open conformation, Nature Struct. Mol. Biol., 19,
485-491, S1.
19.Hong, S., and Pedersen, P. L. (2008) ATP synthase
and the actions of inhibitors utilized to study its roles in human
health, disease, and other scientific areas, Microbiol. Mol. Biol.
Rev., 72, 590-641.
20.Lysenkova, L. N., Turchin, K. F., Korolev, A. M.,
Dezhenkova, L. G., Bekker, O. B., Shtil, A. A., Danilenko, V. N., and
Preobrazhenskaya, M. N. (2013) Synthesis and cytotoxicity of oligomycin
A derivatives modified in the side chain, Bioorg. Med. Chem.,
21, 2918-2924.
21.Lysenkova, L. N., Turchin, K. F., Korolev, A. M.,
Danilenko, V. N., Bekker, O. B., Dezhenkova, L. G., Shtil, A. A., and
Preobrazhenskaya, M. N. (2014) Study on retroaldol degradation products
of antibiotic oligomycin A, J. Antibiot. (Tokyo),
67, 153-158.
22.Ko, Y. H., Pan, W., Inoue, C., and Pedersen, P.
L. (2002) Signal transduction to mitochondrial ATP synthase: evidence
that PDGF-dependent phosphorylation of the delta-subunit occurs in
several cell lines, involves tyrosine, and is modulated by
lysophosphatidic acid, Mitochondrion, 1, 33-48.
23.Azarashvily, T. S., Tyynela, J., Baumann, M.,
Evtodienko, Y. V., and Saris, N. E. (2000) Ca2+-modulated
phosphorylation of a low-molecular-mass polypeptide in rat liver
mitochondria: evidence that it is identical with subunit c of
F0F1-ATPase, Biochem. Biophys. Res.
Commun., 270, 741-744.
24.Hojlund, K., Wrzesinsk, K., Larsen, P. M., Fey,
S. J., Roepstorff, P., Handberg, A., Dela, F., Vinten, J., McCormack,
J. G., Reynet, C., and Beck-Nielsen, H. (2003) Proteome analysis
reveals phosphorylation of ATP synthase beta-subunit in human skeletal
muscle and proteins with potential roles in type 2 diabetes, J.
Biol. Chem., 278, 10436-10442.
25.Kanekatsu, M., Saito, H., Motohashi, K., and
Hisabori, T. (1998) The beta subunit of chloroplast ATP synthase
(CF0CF1-ATPase) is phosphorylated by casein kinase II, Biochem. Mol.
Biol. Int., 46, 99-105.
26.Prisic, S., Dankwa, S., Schwartz, D., Chou, M.
F., Locasale, J. W., Kang, C. M., Bemis, G., Church, G. M., Steen, H.,
and Husson, R. N. (2010) Extensive phosphorylation with overlapping
specificity by Mycobacterium tuberculosis serine/threonine
protein kinases, Proc. Natl. Acad. Sci. USA, 107,
7521-7526.
27.Inoue, H., Nojima, H., and Okayama, H. (1990)
High efficiency transformation of Escherichia coli with
plasmids, Gene, 96, 23-28.
28.Mierendorf, R., Yeager, K., and Novy, R. (1994)
Innovations, Newsletter Novagen, 1, 1-3.
29.Kieser, T., Bibb, M. J., Buttner, M. J., Chater,
K. F., and Hopwood, D. A. (2000) Practical Streptomyces
Genetics, The John Innes Foundation, Norwich, United Kingdom.
30.Sambrook, J., Fritsch, E. E., and Maniatis, T.
(1989) Molecular Cloning: a Laboratory Manual, Cold Spring
Harbor Laboratory Press.
31.O’Farrell, P. H. (1975) High resolution
two-dimensional electrophoresis of proteins, J. Biol. Chem.,
250, 4007-4021.
32.Elizarov, S. M., and Danilenko, V. N. (2001)
Multiple phosphorylation of membrane-associated calcium-dependent
protein serine/threonine kinase in Streptomyces fradiae, FEMS
Microbiol Lett., 202, 135-138.
33.Elizarov, S. M., Mironov, V. A., and Danilenko,
V. N. (2000) Calcium-induced alterations in the functioning of protein
serine/threonine and tyrosine kinases in Streptomyces fradiae
cells, IUBMB Life, 50, 139-143.
34.Thompson, J. D., Higgins, D. G., and Gibson, T.
J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific gap
penalties and weight matrix choice, Nucleic Acids Res.,
22, 4673-4680.
35.Notredame, C., Higgins, D. G., and Heringa, J.
(2000) T-Coffee: a novel method for fast and accurate multiple sequence
alignment, J. Mol. Biol., 302, 205-217.
36.Nicholas, K. B., Nicholas, H. B., Jr., and
Deerfield, D. W. (1997) II. GeneDoc: analysis and visualization of
genetic variation, EMB News, 4, 14.
37.Altschul, S. F., Gish, W., Miller, W., Myers, E.
W., and Lipman, D. J. (1990) Basic local alignment search tool, J.
Mol. Biol., 215, 403-410.
38.Tamura, K., Peterson, D., Peterson, N., Stecher,
G., Nei, M., and Kumar, S. (2011) MEGA5: molecular evolutionary
genetics analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods, Mol. Biol. Evol., 28,
2731-2739.
39.Saitou, N., and Nei, M. (1987) The
neighbor-joining method: a new method for reconstructing phylogenetic
trees, Mol. Biol. Evol., 4, 406-425.
40.Felsenstein, J. (1985) Confidence limits on
phylogenies: an approach using the bootstrap, Evolution,
39, 783-791.
41.Cousin, C., Derouiche, A., Shi, L., Pagot, Y.,
Poncet, S., and Mijakovic, I. (2013) Protein-serine/threonine/tyrosine
kinases in bacterial signaling and regulation, FEMS Microbiol.
Lett., 346, 11-19.
42.Pereira, S. F., Goss, L., and Dworkin, J. (2011)
Eukaryote-like serine/threonine kinases and phosphatases in bacteria,
Microbiol. Mol. Biol. Rev., 75, 192-212.
43.Lu, P., Villellas, C., Koul, A., Andries, K.,
Lill, H., and Bald, D. (2014) The ATP synthase inhibitor bedaquiline
interferes with small-molecule efflux in Mycobacterium
smegmatis, J. Antibiot. (Tokyo), DOI: 10.1038/ja.
2014.74.
44.Bald, D., and Koul, A. (2013) Advances and
strategies in discovery of new antibacterials for combating
metabolically resting bacteria, Drug Discov. Today, 18,
250-255.
45.Plotnikov, E. Y., Morosanova, M. A., Pevzner, I.
B., Zorova, L. D., Manskikh, V. N., Pulkova, N. V., Galkina, S. I.,
Skulachev, V. P., and Zorov, D. B. (2013) Protective effect of
mitochondria-targeted antioxidants in an acute bacterial infection,
Proc. Natl. Acad. Sci. USA, 110, 3100-3108.
46.Skulachev, V. P., Anisimov, V. N., Antonenko, Y.
N., Bakeeva, L. E., Chernyak, B. V., Erichev, V. P., Filenko, O. F.,
Kalinina, N. I., Kapelko, V. I., Kolosova, N. G., Kopnin, B. P.,
Korshunova, G. A., Lichinitser, M. R., Obukhova, L. A., Pasyukova, E.
G., Pisarenko, O. I., Roginsky, V. A., Ruuge, E. K., Senin, I. I.,
Severina, I. I., Skulachev, M. V., Spivak, I. M., Tashlitsky, V. N.,
Tkachuk, V. A., Vyssokikh, M. Y., Yaguzhinsky, L. S., and Zorov, D. B.
(2009) An attempt to prevent senescence: a mitochondrial approach,
Biochim. Biophys. Acta, 1787, 437-461.
47.Skulachev, V. P. (2002) Programmed death
phenomena: from organelle to organism, Ann. NY Acad.
Sci., 959, 214-237.
48.Bekker, O. B., Mavletova, D. A., Lyubimova, I.
K., Mironcheva, T. A., Shtil, A. A., and Danilenko, V. N. (2012)
Induction of programmed lysis of the Streptomyces lividans
culture by the inhibitors of eukaryotic-type serine/threonine protein
kinases, Mikrobiologiya, 81, 177-184.
49.Bekker, O. B., Alekseeva, M. G., Osolodkin, D.
I., Palyulin, V. A., Elizarov, S. M., Zefirov, N. S., and Danilenko, V.
N. (2010) Novel test system for the screening of serine-threonine
protein kinase inhibitors: E. coli APHVIII/Pk25 construct,
Acta Naturae, 2, 126-139.
50.Elizarov, S. M., Alekseeva, M. G., Novikov, F.
N., Chilov, G. G., Maslov, D. A., Shtil, A. A., and Danilenko, V. N.
(2012) Identification of the sites of phosphorylation of aminoglycoside
phosphotransferase VIII from Streptomyces rimosus,
Biochemistry (Moscow), 77, 1258-1265.
Supplementary FIGS S1-S4 (PDF)
Supplementary FIGS S5-S8 (PDF)