Influence of SkQ1 on Expression of Nrf2 Transcription Factor Gene, ARE-Controlled Genes of Antioxidant Enzymes and Their Activity in Rat Blood Leukocytes
V. V. Vnukov, O. I. Gutsenko, N. P. Milutina*, A. A. Ananyan, A. O. Danilenko, S. B. Panina, and I. V. Kornienko
Southern Federal University, Academy of Biology and Biotechnology, Department of Biochemistry and Microbiology, pr. Stachki 194/1, 344090 Rostov-on-Don, Russia; E-mail: natmilut@rambler.ru* To whom correspondence should be addressed.
Received October 23, 2014; Revision received January 16, 2015
This study demonstrated that pretreatment of rats with mitochondria-targeted antioxidant SkQ1 (50 nmol/kg during 5 days) significantly increased the mRNA levels of Nrf2 transcription factor and Nrf2-induced genes encoding antioxidant enzymes SOD1, SOD2, CAT, and GPx4 in rat peripheral blood leukocytes. The increase in expression of these genes with SkQ1 addition was accompanied by increased activities of catalase, glutathione peroxidase, and glutathione-S-transferase in leukocytes. These results indicate that antioxidant properties of SkQ1 might be realized via induction of expression of the genes regulating activity of antioxidant system elements.
KEY WORDS: gene expression, antioxidant enzymes, leukocytes, mitochondria-targeted antioxidantDOI: 10.1134/S0006297915050107
Abbreviations: ARE, antioxidant responsive element; bZip, basic region and leucine zipper; CAT, catalase; Cu,Zn-SOD, Cu,Zn-superoxide dismutase; ECSOD, extracellular superoxide dismutase; GPx, glutathione peroxidase; GR, glutathione reductase; GST, glutathione-S-transferase; Keap 1, Kelch-like association protein 1; MnSOD, Mn-superoxide dismutase; NF-E2, nuclear factor erythroid 2; Nrf2, NF-E2-related factor 2.
The mechanisms of cytoprotective effects of a new class of organic
molecules have been intensively studied in the recent years –
mitochondria-targeted antioxidants composed of a penetrating cation
(Skulachev ions) with plastoquinone attached to it [1, 2]. Among these substances, SkQ
derivatives and the first synthesized substance SkQ1 have been
confirmed to be highly efficient. SkQ1
(10-(6′-plastoquinonyl)decyltriphenylphosphonium) is a conjugate
of the penetrating cation decyltriphenylphosphonium and plastoquinone,
a very effective electron carrier and an antioxidant of chloroplasts
[2]. It was demonstrated that SkQ1 specifically
accumulates in the inner mitochondrial membrane, and it was shown to be
a renewable antioxidant that can be used in the nanomolar concentration
range [2]. Antioxidant properties of SkQs are based
on the ability of this substance to prevent oxidation of mitochondrial
cardiolipin [2], to scavenge generated reactive
oxygen species (ROS) [1, 3],
and to induce mild uncoupling of oxidative phosphorylation via
protonophorous effect or intensification of fatty-acid cycling that
inhibits the formation of ROS in mitochondria [4-6].
To a great extent, complex cellular mechanisms of protection against oxidative/electrophilic stress have been established to be linked with the Keap1/Nrf2 redox sensitive signaling system that regulates the expression of more than 200 genes, including the genes for antioxidant enzymes, detoxification enzymes, etc. [7-9]. Moreover, evidences indicate that Nrf2 influences intermediary metabolism and mitochondrial functions [9]. Compounds activating this transcription pathway, Nrf2 in particular, have been designed; para- and ortho-hydroquinone are significant among them [10]. The purpose of this study was to test whether SkQ1 with plastoquinone affects expression levels of Nrf2 transcription factor and Nrf2-induced genes encoding antioxidant enzymes under physiological conditions.
MATERIALS AND METHODS
Mature male rats Rattus norvegicus weighing 180-200 g were used for this study. The experimental animals were divided into two groups (n = 10-23). The rats were pretreated with 50 nmol SkQ1 per kg body weight daily during 5 days. The calculated SkQ1 dose was diluted in 100 µl 0.2% ethanol in distilled water and administered into the cheek pouch of rats. The animals of the control group were given 100 µl 0.2% ethanol daily during 5 days. All experiments on the laboratory animals were performed in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (Strasbourg, March 18, 1986).
The mRNA expression was analyzed by reverse transcription and following polymerase chain reaction (RT-PCR) with gene specific primers. Blood (50 µl) was sampled for the analysis. Total RNA was isolated by guanidinium thiocyanate–phenol–chloroform extraction using the Ribo-zol-B kit (InterLabService, Russia). The quality of extracted RNA was assessed using electrophoresis in a 1.2% agarose gel; the quantity of RNA was assessed using absorbance at 260 nm. Reverse transcription was performed using the cDNA synthesis kit (Syntol, Russia) containing MMLV-RT (Moloney murine leukemia virus reverse transcriptase), primer Random-6, deoxynucleotide triphosphates (dNTP), and RNase inhibitor.
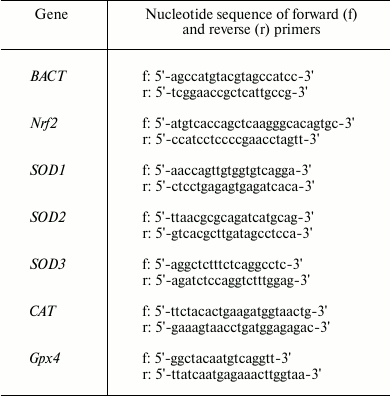
The gene expression patterns of transcription factor Nrf2 (Nrf2), superoxide dismutase isoforms Cu,Zn-SOD (SOD1), Mn-SOD (SOD2), extracellular SOD or ECSOD (SOD3), catalase (CAT), and glutathione peroxidase 4 (GPx4) were analyzed by real-time PCR (RT-PCR) using intercalating dye EvaGreen (Molecular Probes, USA) and the RT-PCR kit (Syntol). RT-PCR was performed using the iQ5 real-time PCR detection system (Bio-Rad, USA). BACT (β-actin) served as an internal control gene. Primer BLAST and Primer 3 were used to design target-specific primers. The primer sequences are presented in Table 1.
Table 1. Primers for RT-PCR
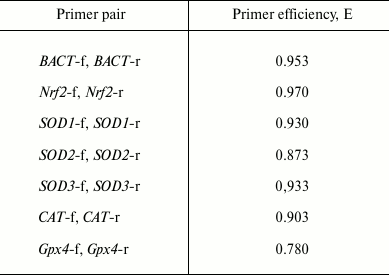
To evaluate the efficiency (E) of the primer pairs, PCR was performed with serial dilutions of the cDNA (1 : 1, 1 : 2, 1 : 4, 1 : 8), and then average ΔCt values were calculated. The efficiencies of the primer pairs are presented in Table 2.
Table 2. Efficiency of the primer pairs
RT-PCR was conducted under the following conditions: the first step at 95°C for 5 min, then 40 cycles of 58(60)°C for 50 sec (fluorescence detection) and 95°C for 15 sec. Then fluorescence intensity was plotted as a function of time. The melting curve was analyzed to determine the specificity of the amplification.
The software package Bio-Rad IQ5 Optical System Software v.2.0 was used to analyze the PCR amplification plots. The RT-PCR data were analyzed using iCycler IQ5 software (BioRad). Relative quantification of gene expression level was performed by the ΔCt method using the amount of cDNA of the comparison gene.
The lymphocyte suspension was separated by centrifugation of the whole blood in a Ficoll–Verografin density gradient (ρ = 1.077) according to the method of Boyum [11] and was used for the following biochemical analyses. The lymphocytes were washed three times and were suspended in Tris-buffered saline (pH 7.4).
The superoxide dismutase (SOD) activity evaluation was based on the ability of SOD to inhibit nitroblue tetrazolium reduction by superoxide generated during autoxidation of adrenaline [12]. The catalase activity was measured using the reaction of hydroperoxide with ammonium molybdate [13]. The glutathione peroxidase (GPx) activity assay was based on the intensity of oxidation of reduced glutathione triggered by tert-butyl hydroperoxide [14], and the glutathione-S-transferase (GST) activity was evaluated using the reaction between reduced glutathione and 1-chloro-2,4-dinitrobenzene [15].
All statistical calculations were carried out with Biostat software (version 2009 Professional – 5.8.4.3 Analyst Soft). The Kolmogorov–Smirnov and Lilliefors test was used to assess the normality of distribution. If the data were not normally distributed, nonparametric Mann–Whitney U-test was applied for comparing two samples. If the distribution was normal, Student’s t-test for small sample sizes was applied. All p-values of less than 0.05 were regarded as significant. A tendency towards statistical significance was postulated in cases when 0.05 < p < 0.1.
RESULTS
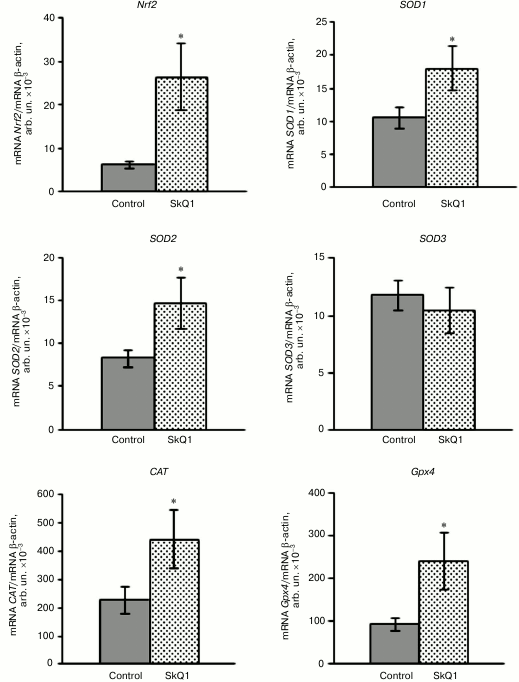
The research demonstrated that SkQ1 supplementation (50 nmol/kg for 5 days) increased the Nrf2 gene expression level in the rat blood leukocytes by 318% compared to the control (Fig. 1). The relative mRNA amount of the Nrf2 transcription factor was confirmed to increase respectively compared to normal values. Concurrently, the effects of SkQ1 supplementation on expression patterns of Nrf2-induced genes encoding antioxidant enzymes SOD isoforms (SOD1-3), catalase (CAT), and glutathione peroxidase 4 (GPx4) in the rat leukocytes were investigated. The mRNA levels of SOD1 and SOD2 increased by 70 and 77%, respectively, in rat blood leukocytes when mitochondria-targeted antioxidant had been administered. This result is evidence of transcriptional activity enhancement of the genes. There were not statistically significant differences in the mRNA level of SOD3 compared to the control (Fig. 1). It was also shown that SkQ1 pretreatment for 5 days upregulated mRNA expression of the CAT gene by 95% and the GPx4 gene by 156% in rat blood leukocytes. These facts prove the higher expression of these genes under the influence of cationic plastoquinone derivative (Fig. 1).
Fig. 1. Influence of SkQ1 on mRNA level of the Nrf2 gene and genes encoding antioxidant enzymes in rat blood leukocytes (M ± m). The number of animals was 13-26 per group. The statistically significant differences between experimental and control group (p < 0.05) are marked by asterisks.
Based on these results, we conclude that SkQ1 is a positive regulator of transcriptional activity of the Nrf2 gene and the genes encoding antioxidant enzymes. This may contribute to the antioxidant capacity of leukocytes under physiological conditions.
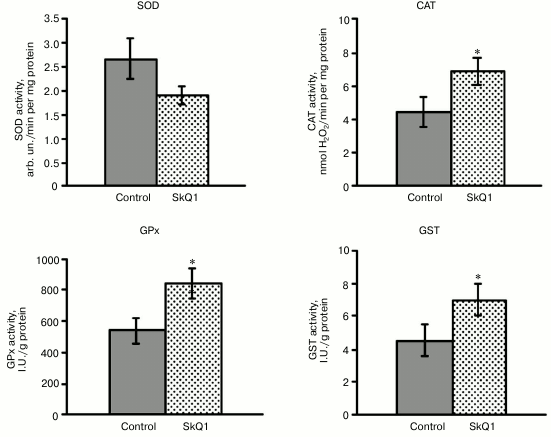
The next step of the research was to study whether SkQ1 affects the activity of antioxidant enzymes in rat blood leukocytes. SkQ1 was found to promote the activation of antioxidant enzymes in rat blood leukocytes, which was confirmed by increased activity of catalase, GPx, and GST by 54-56% compared to the control. It contributes to the antioxidant capacity of blood leukocytes (Fig. 2). At the same time, total SOD activity in the leukocytes remained unchanged under the conditions of SkQ1 supplementation (Fig. 2), in spite of the demonstrated upregulation of transcriptional activity of the SOD1 and SOD2 genes. The unidirectional changes in the expression level of the CAT and Gpx4 genes and catalase and glutathione peroxidase activity were detected in the leukocytes.
Fig. 2. Influence of SkQ1 on the activity of antioxidant enzymes in mononuclear fraction of rat blood (M ± m). The number of animals was 13-26 per group. The statistically significant differences between experimental and control group (p < 0.05) are marked by asterisks.
DISCUSSION
The results of the study showed that administration of cationic plastoquinone derivative SkQ1 upregulated the transcriptional activity of the Nrf2 gene (mRNA level was 4-fold higher). The transcription factor Nrf2 gene is known to express in different cell types [16, 17], including lymphocytes [18]. Nrf2 (NF-E2-related factor 2) is a “cap’n’collar” (CNC) basic-region leucine zipper (bZIP) transcription factor [9].
The activation of the Keap1/Nrf2 signal system is known to occur through dissociation of Nrf2 from inhibitor Keap1, transfer of Nrf2 to the nucleus, dimerization with small Maf protein or transcription factor c-Jun and coactivator proteins, and binding of Nrf2 to cis-acting antioxidant response elements (ARE) located in the promoters of redox-sensitive genes [16, 17]. Many novel inducers activating the Keap1/Nrf2/ARE signal system have been found, including natural and synthetic quinones [18, 19]. One of the main features of quinones – Nrf2 inducers – is the presence of OH-groups at ortho- or para-position [10], which is also characteristic of SkQ1 comprising plastoquinone (1,4-benzoquinone derivative) [1]. Based on the results, SkQ1 is supposed to induce Nrf2 due to its quinone moiety. Importantly, Nrf2 has been shown to possess two ARE-like sequences within its own promoter region, providing a platform for Nrf2 to initiate its own transcription [20]. According to the authors’ opinion [20, 21], this fact proves the presence of a positive feedback loop in the Keap1/Nrf2/ARE signal pathway, enhancing the system sensitivity and adaptive cell defense response.
Our research showed that increased Nrf2 expression level in rat blood leukocytes resulted in enhanced transcriptional activity of target genes encoding antioxidant enzymes SOD1, SOD2, CAT, and GPx4 when SkQ1 had been administered. The genes that code these enzymes possess the antioxidant-response elements (ARE) within the promoters, consequently induction by Nrf2 transcription factor is supposed to be possible [9, 22-24]. Study of Morzadec et al. [25] demonstrated that activation of human Th cells of blood-derived mononuclear fraction strongly increased the levels of Nrf2 protein by increasing Nrf2 gene transcription. The cell activation also enhanced mRNA and protein levels of Nrf2 target genes encoding antioxidant enzymes.
SkQ1 supplementation (250 nmol/kg per day during 1.5 months) did not affect the mRNA levels of the Nrf2 gene and Nrf2-dependent genes encoding glutathione-S-reductase (Gsr) and heme oxygenase 1 (Hmox1) in the retina of senescence-accelerated OXYS rats, whereas thioredoxin reductase 1 (Txrnd1) expression was downregulated [26]. Under the same experimental conditions, the expression levels of all studied genes remained unchanged in the retina of Wistar rats. The discrepancy of SkQ1 effects on gene expression in different rat tissues (retina, peripheral blood leukocytes) is supposed to be linked with genetic traits of the experimental animals and tissue-specific transcriptomes.
According to published studies, SkQ1 has been established to affect the activity of vital antioxidant enzymes in rat blood leukocytes: the activities of catalase, glutathione peroxidase, and glutathione-S-transferase were upregulated, whereas there was no statistically significant difference in SOD activity compared to the control. This might be explained by the fact that total enzyme activity does not directly depend on its transcription level, but can be determined by the translation rate and posttranslational modifications [27]. Indeed, there are several posttranslational modifications of SOD1 and SOD2: nitration, phosphorylation, glutathionylation (glycation for SOD1) that give rise to reduced activity of SODs [28]. In addition, we demonstrated that SkQ1 supplementation upregulated glutathione-S-transferase activity in rat blood leukocytes; this might be a consequence of Nrf2 gene activation, because genes encoding different isoforms of glutathione-S-transferase (GSTA1, GSTM1, GSTP1) are considered to be positively regulated by Nrf2 [9].
Our study concludes that administration of cationic plastoquinone derivative SkQ1 (50 nmol/kg per day for 5 days) results in increase in expression level of Nrf2 transcription factor and ARE-controlled genes in rat blood leukocytes under physiological conditions; it is accompanied by increased activity of catalase, glutathione peroxidase, and glutathione-S-transferase, thus promoting cellular antioxidant capacity.
The authors express their gratitude to V. P. Skulachev for providing the SkQ1 compound for research and help in planning and interpretation of the experiments and their results.
This work was supported by the Ministry of Education and Science of the Russian Federation (grant No. 213.01-11/2014-32).
REFERENCES
1.Skulachev, V. P. (2007) Abiochemical approach to
the problem of aging: “megaproject” on membrane-penetrating
ions. The first results and prospects, Biochemistry (Moscow),
72, 1385-1396.
2.Antonenko, Y. N., Avetisyan, A. V., Bakeeva, L. E.,
Chernyak, B. V., Chertkov, V. A., Domnina, L. V., Ivanova, O. Yu.,
Izyumov, D. S., Khailova, L. S., Klishin, S. S., Korshunova, G. A.,
Lyamzaev, K. G., Muntyan, M. S., Nepryakhina, O. K., Pashkovskaya, A.
A., Pletjushkina, O. Yu., Pustovidko, A. V., Roginsky, V. A.,
Rokitskaya, T. I., Ruuge, E. K., Saprunova, V. B., Severina, I. I.,
Simonyan, R. A., Skulachev, I. V., Skulachev, M. V., Sumbatyan, N. V.,
Sviryaeva, I. V., Tashlitsky, V. N., Vassiliev, J. M., Vyssokikh, M.
Yu., Yaguzhinsky, L. S., Zamyatnin, A. A., Jr., and Skulachev, V. P.
(2008) Mitochondria-targeted plastoquinone derivatives as tools to
interrupt execution of the aging program. 1. Cationic plastoquinone
derivatives: synthesis and in vitro studies, Biochemistry
(Moscow), 73, 1273-1287.
3.Skulachev, M. V., Antonenko, Y. N., Anisimov, V.
N., Chernyak, B. V., Cherepanov, D. A., Chistyakov, V. A., Egorov, M.
V., Kolosova, N. G., Korshunova, G. A., Lyamzaev, K. G., Plotnikov, E.
I., Roginsky, V. A., Savchenko, A. I., Severina, I. I., Severin, F. F.,
Shkurat, T. P., Tashlitsky, V. N., Shidrovsky, K. M., Vyssokikh, M. Y.,
and Zamyatnin, A. A. (2011) Mitochondrial-targeted plastoquinone
derivates. Effect on senescence and acute age-related pathologies,
Curr. Drug. Targets, 12, 800-826.
4.Skulachev, V. P., Antonenko, Yu. N., Cherepanov, D.
A., Chernyak, B. V., Izyumov, D. S., Khailova, L. S., Klishin, S. S.,
Korshunova, G. A., Lyamzaev, K. G., Pletjushkina, O. Yu., Roginsky, V.
A., Rokitskaya, T. I., Severin, F. F., Severina, I. I., Simonyan, R.
A., Skulachev, M. V., Sumbatayan, N. V., Sukhanova, E. I., Tashlitsky,
V. N., Trendeleva, T. A., Vyssokikh, M. Yu., and Zvyagilskaya, R. A.
(2010) Prevention of cardiolipin and fatty acid cycling as two
antioxidant mechanisms of cationic derivatives of plastoquinone (SkQs),
Biochim. Biophys. Acta, 1797, 878-889.
5.Severin, F. F., Severina, I. I., Antonenko, Y. N.,
Rokitskaya, T. I., Cherepanov, D. A., Mokhova, O. V., Yaguzhinsky, L.
S., Korshunova, G. A., Sumbatayan, N. V., Skulachev, M. V., and
Skulachev, V. P. (2010) Penetrating cation/fatty acid anion pair as a
mitochondria-targeted protonophore, Proc. Natl. Acad. Sci. USA,
107, 663-668.
6.Plotnikov, E. Y., Silachev, D. N., Jankauskas, S.
S., Rokitskaya, T. I., Chupyrkina, A. A., Pevzner, I. B., Zorova, L.
D., Isaev, N. K., Antonenko, Y. N., Skulachev, V. P., and Zorov, D. B.
(2012) Mild uncoupling of respiration and phosphorylation as a
mechanism providing nephro- and neuroprotective effects of penetrating
cations from the SkQ family, Biochemistry (Moscow), 77,
1029-1037.
7.Turpaev, K. T. (2013) The Keap1-Nrf2 signaling
pathway. Mechanisms of regulation and role in protection of cells
against toxicity caused by xenobiotics and electrophiles,
Biochemistry (Moscow), 78, 111-126.
8.Jaiswal, A. K. (2004) Nrf2 signaling in coordinated
activation of antioxidant gene expression, Free Radic. Biol.
Med., 36, 1199-1207.
9.Hayes, J. D., and Dinkova-Kostova, A. T. (2014) The
Nrf2 regulatory network provides an interface between redox and
intermediary metabolism, Trends Biochem. Sci., 39,
199-216.
10.Satoh, T., McKercher, S. R., and Lipton, S. A.
(2014) Reprint of: Nrf2/ARE-mediated antioxidant actions of
pro-electrophilic drugs, Free Radic. Biol. Med., 66,
45-57.
11.Boyum, A. (1968) Separation of leukocytes from
blood and bone marrow, Scand. J. Clin. Lab. Invest. (Suppl.),
97, 77-89.
12.Sirota, N. V. (1999) New approach in studies of
adrenalin autoxidation and its use in measurements of superoxide
dismutase activity, Vopr. Med. Khim., No. 3, 14-15.
13.Korolyuk, M. A., Ivanova, L. I., Maiorova, I. G.,
and Tokarev, V. E. (1988) Method for measurements of catalase activity,
Lab. Delo, No. 1, 16-19.
14.Moin, V. M. (1986) Simple and specific approach
for determination of glutathione peroxidase activity in erythrocytes,
Lab. Delo, No. 12, 724-727.
15.Habig, W. H., Pabst, M. J., and Jacoby, W. B.
(1974) Glutathione-S-transferase: the first step in mercapturic acid
formation, J. Biol. Chem., 249, 7130-7139.
16.Kaspar, J. W., Niture, S. K., and Jaiswal, A. K.
(2009) Nrf2: INrf2 (Keap1) signaling in oxidative stress, Free
Radic. Biol. Med., 47, 1304-1309.
17.Ma, Q. (2013) Role of Nrf2 in oxidative stress
and toxicity, Annu. Rev. Pharmacol. Toxicol., 53,
401-426.
18.Forman, H. J., Davies, J. A., and Ursini, F.
(2014) How do nutritional antioxidants really work: nucleophilic tone
and para-hormesis versus free radical scavenging in vivo,
Free Radic. Biol. Med., 66, 24-35.
19.Uruno, A., and Motohashi, H. (2011) The
Keap1-Nrf2 system as an in vivo sensor for electrophiles,
Nitric Oxide, 25, 153-160.
20.Kwak, M. K., Itoh, K., Yamamoto, M., and Kensler,
T. W. (2002) Enhanced expression of the transcription factor Nrf2 by
cancer chemopreventive agents: role of antioxidant response
element-like sequences in the nrf2 promoter, Mol. Cell Biol.,
22, 2883-2892.
21.Bryan, H. K., Olayanju, A., Goldring, C. E., and
Park, B. K. (2013) The Nrf2 cell defensepathway: Keap1-dependent
and -independent mechanisms of regulation, Biochem.
Pharmacol., 85, 705-717.
22.Dhar, S. K., and St. Clair, D. K. (2012)
Manganese superoxide dismutase regulation and cancer, Free Radic.
Biol. Med., 52, 2209-2222.
23.Zenkov, N. K., Men’shikova, E. B., and
Tkachev, V. O. (2009) Some principles and mechanisms of redox
regulation, Kislorod Antioksid., No. 1, 3-64.
24.Miao, L., and St. Clair, D. K. (2009) Regulation
of superoxide dismutase genes: implications in disease, Free Radic.
Biol. Med., 47, 344-356.
25.Morzadec, C., Macoch, M., Sparfel, L.,
Kerdine-Romer, S., Fardel, O., and Vernhet, L. (2014) Nrf2 expression
and activity in human T lymphocytes: stimulation by T cell receptor
activation and priming by inorganic arsenic and
tert-butylhydroquinone, Free Radic. Biol. Med.,
71, 133-145.
26.Perepechaeva, M. L., Grishanova, A. Yu.,
Rudnitskaya, E. A., and Kolosova, A. G. (2014) The
mitochondria-targeted antioxidant SkQ1 downregulates aryl hydrocarbon
receptor-dependent genes in the retina of OXYS rats with AMD-like
retinopathy, J. Ophthalmol., 2014, Article ID530943.
27.Zorov, D. B., Isaev, N. K., Plotnikov, E. Y.,
Silachev, D. N., Zorova, L. D., Pevzner, I. B., Morosanova, M. A.,
Jankauskas, S. S., Zorov, S. D., and Babenko, V. A. (2013) Perspectives
for a mitochondrial medicine, Biochemistry (Moscow), 78,
979-990.
28.Yamakura, F., and Kawasaki, H. (2010)
Post-translational modification of superoxide dismutase, Biochim.
Biophys. Acta, 1804, 318-325.