Construction of a Single Chain Variable Fragment Antibody (scFv) against Carbaryl and Its Interaction with Carbaryl
Zhang Xiuyuan1*, Huang Zhihong1, Wang Lixia1, and Liu Xiaonan2
1Hebei North University, Zhangjiakou 075000, Hebei, China; E-mail: zhangxiuyuan917@163.com2Tianjin University, Tianjin 300457, China
* To whom correspondence should be addressed.
Received November 20, 2014; Revision received March 12, 2015
Carbaryl is a low molecular weight insecticide that inhibits cholinesterase. Residues of carbaryl in food and the environment have damaged human health. A high-specificity scFv that can identify carbaryl is still lacking. In the present study, an anti-carbaryl scFv gene was prepared by cloning VL and VH genes from hybridoma cells secreting monoclonal antibody, then VH and VL were fused together using splicing by overlap extension (SOE) PCR with a flexible polypeptide linker connector (Gly4Ser)3, and then the scFv-pET-26b recombinant plasmid was constructed and transformed into E. coli BL21 for expression using IPTG as an inducer. The expressed recombinant protein was identified by SDS-PAGE and ELISA. The three-dimensional structure of the anti-carbaryl scFv was constructed by computer modeling, and carbaryl was docked to the scFv model to obtain the structure of the binding complex. The binding site was composed of Ala51, Ser52, Ile51, Gly54, Ser56, Arg98, and Gly100. This helps to understand the mechanism of interaction between anti-carbaryl antibody and antigen. Furthermore, it provides guidance for in vitro affinity maturation of anti-carbaryl antibody.
KEY WORDS: anti-carbaryl scFv, ELISA, construction, binding complexDOI: 10.1134/S0006297915050181
Abbreviations: IPTG, isopropyl β-D-1-thiogalactopyranoside; TMB, 3,3′,5,5′-tetramethylbenzidine.
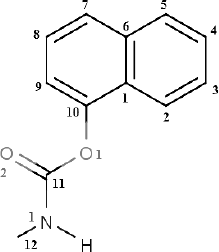
Carbaryl (C12H11NO2,
1-naphthyl-N-methyl carbamate) (Fig. 1), a
carbamate insecticide, is used as a broad-spectrum contact and
intestinal insecticide with some systemic properties used on crops and
trees. The mode of action of carbaryl, like the organophosphates, is
reversible inhibition of cholinesterase [1], and
the signs and symptoms are typically cholinergic with lacrimation,
salivation, miosis, convulsions, and death [2].
Residues of carbaryl on crops and in the environment pose a potential
hazard to humans.
Fig. 1. Structure of carbaryl.
Pesticide residue monitoring has attracted enormous attention to carbaryl residues in recent years. An official document of the European Union (Regulation (EC) No. 396/2005 of the European Parliament and of the Council) established maximum residue level of carbaryl in food as 0.05 mg/kg. This has resulted in a growing demand for rapid and simple testing procedures for determining carbaryl. Earlier, routine analysis of carbaryl content was based on chromatographic methods after derivatization or spectrophotometry and colorimetry, thin layer chromatography, and high performance liquid chromatography [3, 4]. These methods are laborious and time intensive for sample pretreatment. Enzyme linked immunosorbent assay (ELISA) is becoming increasingly popular as a screening methodology to meet the testing demand. It is suitable for surveying larger numbers of samples than could be accomplished by conventional analysis, and it exhibit high sensitivity and low detection limits [5].
The single-chain variable fragment (scFv) antibody is a third generation antibody. In recent years, with molecular biological techniques it is possible to produce a single-chain variable fragment (scFv) antibody in bacterial culture by recombinant DNA technology. Compared to monoclonal and polyclonal antibody, the cost of scFv production is very low; in addition, scFv can be produced in batch and fused with a marker molecule for effective immunological detection [6]. Therefore, one of objectives of this research was to construct a high-affinity scFv against carbaryl that might provide reliable carbaryl detection.
The scFv usually suffers from low binding affinity compared to those for macromolecular antigens. The study of the binding mechanism of scFv with antigen can help to improve their binding affinities. But the antigen–antibody reaction mechanism is poorly understood, this leading to hapten design based on trial and error and difficulty in antibody transformation. The usual approach for studying protein–ligand binding mechanism is by crystallization, which is time-consuming and difficult. Therefore, the other objective of the present study was to use construct recombinant antibody (scFv) against carbaryl and elucidate its recognition mechanism with carbaryl by computer bioinformatics technology, which lays a foundation for enhancing carbaryl antibody affinity in the future.
MATERIALS AND METHODS
Materials. PGEM-T-Easy plasmid was purchased from Promega (USA). The pET-26b plasmid was purchased from Beijing Bao Covey Food Safety Biological Technology Co. (China). Escherichia coli strain JM109 was purchased from TaKaRa Biotechnology Co. Ltd (China). The oligonucleotide primers [7] (Table 1) were synthesized by Invitrogen (USA) and used for polymerase chain reaction (PCR) cloning of the variable genes. Restriction enzymes EcoRI and XhoI were purchased from TaKaRa. Pfu DNA polymerase and T4 DNA ligase were supplied by TransGen Biotechnology Co. Ltd. (China). The GoScript reverse transcription system was purchased from Promega. DNA gel extraction kit, RNeasy Midi kit, Oligotex mRNA Mini Kit, and QIA quick PCR Purification Kit were purchased from Qiagen (USA). A hybridoma cell line that secretes mAb against carbaryl was established in our laboratory previously. Other reagents were of analytical quality. PCR was performed in a DNA Engine thermal cycler, and gels and film were imaged and analyzed in the Gel Doc XR system, which were all obtained from Bio-Rad (USA).
Table 1. Primers used for cloning
variable-region genes and generating the synthetic gene encoding
scFv
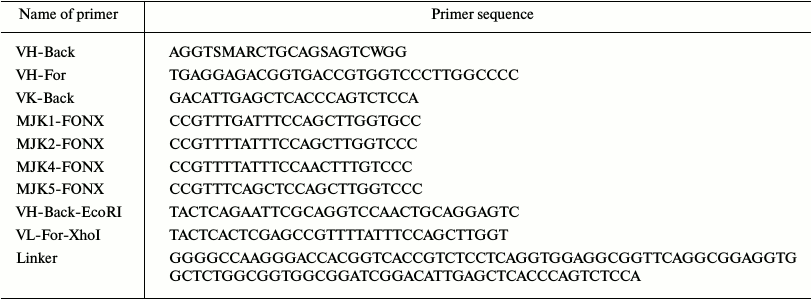
Notes: S, M, R, and W are degenerate bases; they denote (G, C), (A, C), (A, G), and (A, T), respectively.
Total RNA extraction. Total RNA was extracted from the carbaryl hybridoma cell line (about 106-107 cells) using the RNeasy Midi kit according to the manufacturer’s instructions.
Cloning of VL and VH genes. The first-strand cDNA was synthesized from RNA using a reverse transcription PCR kit, then cDNA as template to amplify heavy chain and light chain of antibody variable region using primers VL-back, VL-forward, VH-back, and VH-forward. The PCR protocol involved an initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min, with final extension at 72°C for 10 min. The amplification products were purified using an agarose gel/DNA purification kit.
Construction of ScFv by SOE (splicing by overlap extension). VH and VL DNA fragments were connected by SOE to obtain scFv. The VL gene fragment was used as a template for amplification of the VL fragment containing the XhoI site and a portion of the linker in the absence of VL-For-XhoI and VL-back-linker. The VH fragment was used as a gene template to amplify the VH fragment containing the EcoRI site and a portion of the linker in the absence of VH-Back-EcoRI and VH-forward-linker. The PCR protocol used was the same as described above. The amplified modified VH and VL fragments were then overlapped to obtain the scFv gene using overlap extension PCR, resulting in the formation of the (Gly4Ser)3 linker from the overlapped gene portions residing between the VH and VL fragments. The PCR protocol for the overlap extension step involved an initial denaturation at 94°C for 5 min, followed by 10 cycles at 94°C for 45 s, 50°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. Finally, the gene for the scFv fragment containing the EcoRI and XhoI sites was amplified using the primers VL-For-XhoI and VH-Back-EcoRI with the same PCR protocol as used for cloning of the VL and VH genes. The amplification products were purified with an agarose gel/DNA purification kit [8, 9].
Construction of recombinant plasmid pET-26b-scFv. The resulting ScFv was recovered using an agarose gel/DNA purification kit and then digested by EcoRI and XhoI and inserted into plasmid pET-26b. The recombinants were transformed into E. coli JM109 competent cells. The colonies were identified by colony PCR, and then positive colonies were used for both restriction enzyme analysis and sequence analysis. The sequences were determined by a commercial facility (Genewiz Biological Technology Co. Ltd, China).
Expression and characterization of anti-carbaryl protein. To analyze the activity of soluble anti-carbaryl scFv. A positive colony was selected, and the recombinant plasmid was extracted and transformed into E. coli BL21 competent cells. The colony was cultured in 2× YT medium containing 100 mg/ml ampicillin, and IPTG (1 mM) was added to the medium when the OD600 reached 0.8-1.0 to induce the production of the soluble scFv. The medium was then incubated at 25°C with shaking at 200 rpm overnight followed by centrifugation at 5000g for 10 min, and the anti-carbaryl scFv existing in periplasm was extracted using cold osmotic shock. The pellet was resuspended with 1 ml of ice-cold 1× TES (0.2 M Tris-HCl, pH 8.0, 0.5 mM EDTA, 0.5 mM sucrose), supplemented with 0.2× TES, and incubated on ice for 30 min, followed by centrifugation at 12,000 rpm for 10 min at 4°C. The supernatant was analyzed for the presence of soluble scFv by SDS-PAGE.
Specificity analysis of anti-carbaryl scFv. To determine the specificity of the anti-carbaryl scFv, an indirect competition ELISA (ic-ELISA) method was used to construct a standard curve. The ic-ELISA approach can be described as follows: carbaryl-OVA was coated in 96-wells plate (1 µg/ml, 100 µl/well) at 4°C overnight and then blocked with 0.5% skim milk. The scFv was added to the reaction wells and incubated at 37°C for 1 h, and 50 µl of goat pAb to His-tag (HPR) solution (1 : 5000 in PBS) was added to the wells. Then the enzymatic reaction was performed with TMB as the substrate. After incubation for 20 min at room temperature, the absorbance was measured at 450 nm.
To further identify the specificity of anti-carbaryl scFv, cross-reactivity was evaluated based on the scFv using some carbaryl analogs. We determined the average analyte concentration required for 50% inhibition (IC50) and compared the values to the value from a standard curve for carbaryl running on the same plate. Cross-reactivity was calculated as follows:
Cross-reactivity (CR), % = IC50(carbaryl)/IC50(competitor) × 100%.
Homology modeling. SWISS-MODEL Workspace [10] was used to identify a suitable template for determining the 3D structure of scFv by homology modeling. Three different 3D structural models for anti-carbaryl scFv were then generated using Modeller9 v2 [11] based on the 3D structures of the templates. These models were optimized by adding hydrogen atoms to each of them at pH 7.0 and then using CHARMM all-atom force-field minimization with the implicit solvent model set to none. Energy was minimized for at least 1000 steps until the gradient converged to 0.5 kcal/mol using the steepest descent protocol available in Discovery Studio to remove any steric clashes and to stabilize the models. Ramachandran values were determined for subsequent optimization of all five models using the SAVES [12] web server, and the model exhibiting the best Ramachandran score was retained for further studies [13].
Molecular docking. Docking of carbaryl antigen to scFv was carried out using Discovery Studio software, which is a fast automated docking program that is considered one of the most reliable bioinformatics tools. Structure of carbaryl was drawn by ChemDraw3D software. All water molecules from the scFv model structure were removed, and hydrogen atoms were added. The active site for docking was defined as all atoms within 6.5 Å radius of the co-crystallized ligand; the default parameters in the Discovery Studio Program module were used [14].
RESULTS AND DISCUSSION
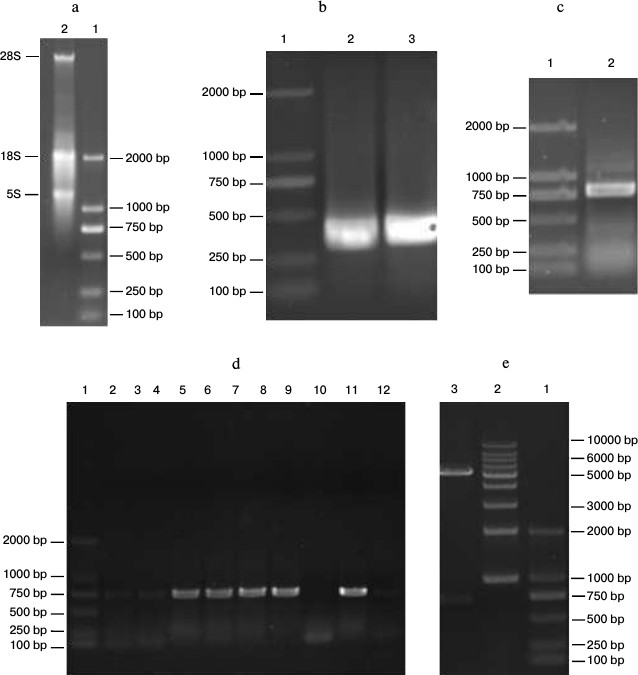
Construction of anti-carbaryl pET-26b-scFv recombinant plasmid. Total RNA extraction. Total RNA of carbaryl hybridoma cell line was extracted using the RNeasy Midi kit according to the manufacturer’s instructions. The results are shown in Fig. 2a. The 28S, 18S, and 5S bands were clearly formed, which indicated total RNA integrity was successful.
Amplification of VH and VL gene. By the primers for different subgroups, nest PCR successfully amplified the different VH and VL DNA fragments. VH was about 340 bp, and VL was about 320 bp (Fig. 2b).
Construction of ScFv by SOE. The VH and VL fragments containing linker overhangs were assembled by splicing overlap extension PCR, and the electrophoresis result showed that obtained scFv was about 750 bp (Fig. 2c), which was as anticipated.
Positive clone selection, identification, and sequencing. The scFv products and vector pET-26b were both digested with EcoRI and XhoI restriction enzymes. The digested products were purified with a PCR product purification kit and then ligated using the T4 DNA ligation enzyme. The ligation products were transformed into E. coli strain JM109 competent cells. Positive clone identification and sequencing are shown in Figs. 2 (d and e) and 3.
Fig. 2. Construction of anti-carbaryl pET-26b-scFv recombinant plasmid. a) Total RNA from hybridoma. Lanes: 1) marker 2000; 2) total RNA from carbaryl hybridoma. b) PCR amplification of VH and VL. Lanes: 1) marker 2000; 2) VL gene; 3) VH gene. c) PCR amplification of scFv gene. Lanes: 1) marker 2000; 2) scFv fragment of carbaryl. d) PCR detection of scFv-pET-26b positive clones. Lanes: 1) marker 2000; 2-12) colony products. e) Double enzyme validation.
Fig. 3. Sequence of anti-carbaryl scFv and partition.
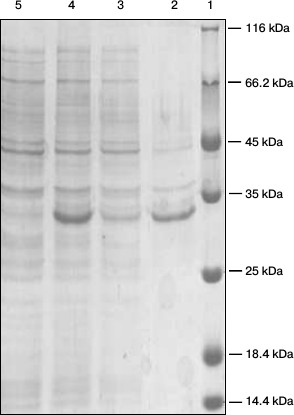
Expression and identification of anti-carbaryl scFv. The positive recombinant plasmid was used to transform E. coli BL21 competent cells. When the OD600 reached 0.8-1.0, expression of protein was induced using IPTG for overnight at 25°C. The periplasmic protein was extracted by cold osmotic shock. As shown in Fig. 4, a soluble protein of about 31 kDa was expressed as determined by SDS-PAGE.
Fig. 4. SDS-PAGE analysis of the expressed products. Lanes: 1) markers; 2) precipitate; 3) supernatant; 4) after induction; 5) before induction.
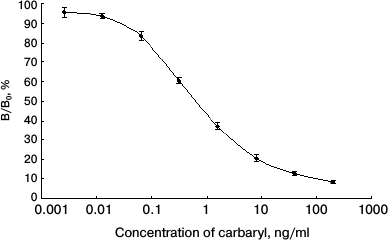
Specificity of anti-carbaryl scFv. To identify the specificity of anti-carbaryl scFv, indirect ELISA was used, and each experiment was performed in triplicate. A standard curve of ic-ELISA is shown in Fig. 5. The carbaryl antigen was strongly recognized by the scFv, and the binding activity was dose-dependent. The limit of detection (LOD) of dc-ELISA (IC15) was 0.04 ± 0.01 ng/ml. The sensitivity of dc-ELISA (IC50) was 0.73 ± 0.06 ng/ml. Compared with earlier known antibodies, this is lower than that of its original monoclonal antibody and reported ELISA by polyclonal antibody [15], and a somewhat higher than reported ELISA by monoclonal antibody [16]. But the approach using scFv to prepare immunoassay is simple and reproducible (with the ability to store bacterial cells indefinitely). The production of a soluble protein can be adapted to industrial scale and be rapidly purified in one step.
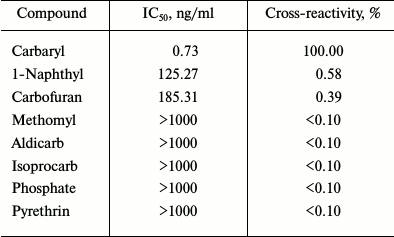
The cross-reactivities of the scFv were determined using optimized ic-ELISA (Table 2). The cross-reactivity values of scFv with other pesticides were below 0.1% except for 1-naphthyl (0.58%) and carbofuran (0.39%). All these indicated that expression of anti-carbaryl scFv had high specificity.
Fig. 5. Normalized standard curve obtained by ic-ELISA.
Table 2. Cross-reactivity of the scF
with other pesticides in ic-ELISA
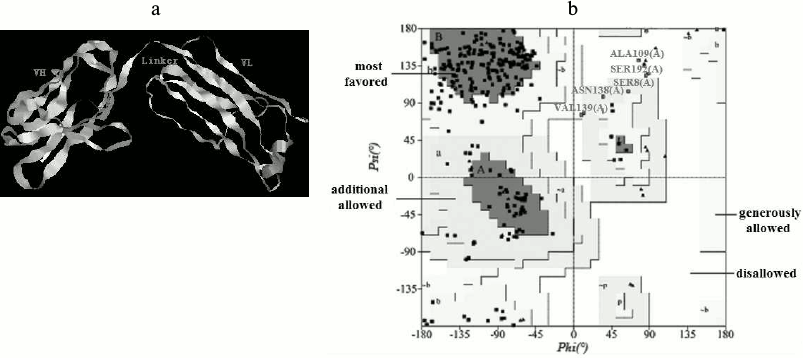
Homology modeling. The 3D structure of the anti-carbaryl scFv was built by homology modeling based on the template crystal structure (PDB code: 1IGY). Models generated using Modeller9 v2 were optimized and evaluated. The best model of anti-carbaryl scFv and its Ramachandran plot is shown in Fig. 6. The scFv is composed of VH, VL, and linker, which is in accord with typical structure of single chain antibody. About 89.7% of the amino acid residues are in most favored regions and 2.5% in disallowed regions by the PROCHECK program. The secondary structure of modeling is composed of 16 α-helices, 93 extended strands, 23 β-turns, and 90 random coils, which was determined by analysis using the Swiss Model website (http://swissmodel.expasy.org/).
Fig. 6. Three-dimensional model of scFv and Ramachandran plot. a) Three-dimensional model of anti-carbaryl scFv. b) Ramachandran plot of the anti-carbaryl scFv model.
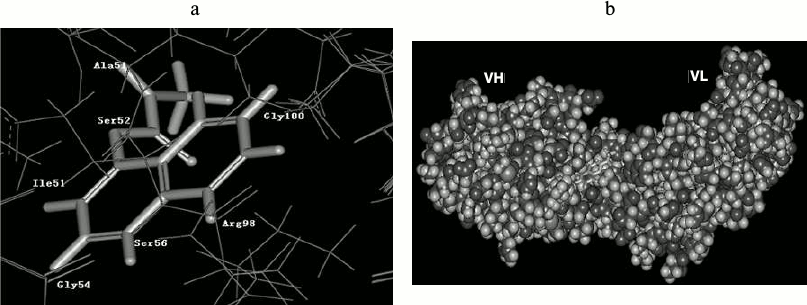
Molecular docking. The resulting structure of the scFv (receptor) and the docked carbaryl molecule, centered on the docking active site, is shown in Fig. 7. Accordingly, the carbaryl moiety is in the hydrophobic pocket of the active site, with hydrophobic conditions imposed on the carbaryl molecule. The binding domain is mainly formed by Ser52, Ile51, Gly54, Ser56, Arg98, Gly100, etc. Arg98 and Gly100 belong to VH-CDR2, Ile51, Gly54, and Ser56 belong to VH-CDR3, Ala51 and Ser52 belong to VL-CDR2. The naphthyl end of carbaryl interacts with VH, the left benzene ring is buried in VH, and Arg98 and Gly100 interact with it. Arg98 is a long side chain residue, and its C–C and C–N bonds can freely rotate to change conformation; these changes affect affinity of scFv. The right benzene ring lies in a groove; Ile51, Gly54, and Ser56 interact with it. The methyl carbamate end of carbaryl interacts with VL, N1, and Ala51, C11, and Ser52 interact respectively. C12 and O2 are warped and naked out of the groove.
Fig. 7. Anti-carbaryl scFv interaction with ligand. a) Structure of binding domain. b) Complex structure of carbaryl and scFv.
The VL and VH genes of an anti-carbaryl antibody were cloned and assembled using a linker. The anti-carbaryl scFv recombinant fusion protein was expressed in E. coli strain BL21 by IPTG induction. Anti-carbaryl scFv demonstrated high specificity with ic-ELISA, and cross-reactivity was determined.
To date, the 3D structure of anti-carbaryl antibody has not been solved, and for the first time, to the best of our knowledge, the 3D structure of anti-carbaryl scFv was constructed based on the known crystal structure of phenobarbital antibody (PDB ID: 1IGY). The docking results revealed that residues form a hydrophobic groove from H-CDR2, H-CDR3, and L-CDR2, and the carbaryl molecule and is firmly retained in it. The active binding site is composed of residues of Ser52, Ile51, Gly54, Ser56, Arg98, Gly100, etc. In conclusion, this study will be helpful for further experimental analysis of anti-carbaryl antibody and its binding affinity with ligands.
This work was supported by the National Natural Science Foundation of China (grant No. 20905058) and China Zhangjiakou Science and Technology Project (grant No. 1311018C-3). We are grateful to Tianjin International Joint Research Institute of Pharmaceutical Biology for use of the Discovery Studio software.
REFERENCES
1.O’Malley, M. (1997) Clinical evaluation of
pesticide exposure and poisonings, Lancet, 349,
1161-1166.
2.Casida, J. E., and Augustinsson, K. B. (1959)
Reaction of plasma albumin with l-naphthyl-N-methyl-carbamate and
certain other esters, Biochim. Biophys. Acta, 36,
411-426.
3.Raminderjit, S. B., and Kousik, M. (2012) Direct
estimation of carbaryl by gas liquid chromatography with nitrogen
phosphorus detection, Bull. Environ. Contam. Toxicol.,
89, 15-20.
4.Hodgson, E. (1991) Pesticides – past, present
and future: review, Pest. Toxicol., 1, 3-12.
5.Jeanne, A. I., Eric, G. O., and James, R. F. (1993)
Validation of a paramagnetic particle-based ELISA for the quantitative
determination of carbaryl in water, Bull. Environ. Contam.
Toxicol., 51, 260-267.
6.Frame, K. K., and Hu, W. S. (1990) The loss of
antibody productivity in continuous culture of hybridoma cells,
Biotechnol. Bioeng., 35, 469-476.
7.Wang, S. H., Zhang, J. B., and Zhang, Z. P. (2006)
Construction of single chain variable fragment (ScFv) and
bis-cFv-alkaline phosphatase fusion protein for detection of
Bacillus anthracis, Anal. Chem., 78, 997-1004.
8.Dong, J. X., Li, Z. F., and Lei, H. T. (2012)
Development of a single-chain variable fragment-alkaline phosphatase
fusion protein and a sensitive direct competitive chemiluminescent
enzyme immunoassay for detection of ractopamine in pork, Analyt.
Chim. Acta, 736, 85-91.
9.Xu, Z. L., Dong, J. X., and Wang, H. (2012)
Production and characterization of a single-chain variable fragment
linked alkaline phosphatase fusion protein for detection of
o,o-diethyl organophosphorus pesticides in a one-step
enzyme-linked immunosorbent assay, J. Agric. Food Chem.,
60, 5076-5083.
10.Arnold, K., Bordoli, L., and Kopp, J. (2006) The
SWISS-MODEL workspace: a web-based environment for protein structure
homology modeling, Bioinformatics, 22, 195-201.
11.Sali, A., and Blundell, T. L. (1993) Comparative
protein modelling by satisfaction of spatial restraints, J. Mol.
Biol., 234, 779-815.
12.NIH MBI Laboratory for Structural Genomics and
Proteomics SAVES (Structural Analysis and Verification Server; http://nihserver.mbi.ucla.edu/SAVES/).
13.Adeel, M., Ahmad, F., and Vivekanand, J. (2010)
Modeling the three-dimensional structures of an unbound single-chain
variable fragment (scFv) and its hypothetical complex with a
Corynespora cassiicola toxin, cassiicolin, J. Mol.
Model., 16, 1883-1893.
14.Tu, Z., Xu, Y., and Fu, J. H. (2011) Sequence
analysis, 3D modeling and molecular docking of anti-deoxynivalenol
single-domain heavy chain antibody, Jiangsu J. Agric. Sci.,
27, 893-898.
15.Sun, J. W., Dong, T. T., Zhang, Y., and Wang, S.
(2010) Development of enzyme linked immunoassay for the simultaneous
detection of carbaryl and metolcarb in different agricultural products,
Analyt. Chim. Acta, 666, 76-82.
16.Abad, A., and Montoya, A. (1995) Application of a
monoclonal antibody-based ELISA to the determination of carbaryl in
apple and grape juices, Analyt. Biochem., 311,
365-370.