REVIEW: The Green Alga Chlamydomonas reinhardtii as a Tool for in vivo Study of Site-Directed Mutations in PsbO Protein of Photosystem II
A. V. Pigolev* and V. V. Klimov
Institute of Basic Biological Problems, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; fax: +7 (496) 733-0532; E-mail: alexey-pigolev@rambler.ru* To whom correspondence should be addressed.
Received January 12, 2015; Revision received February 25, 2015
The photosynthetic water oxidation in photosystem II (PS II) takes place in a special water-oxidizing complex (WOC) that consists of a catalytic center, Mn4CaO5 cluster, and also includes a group of extrinsic proteins needed for its stability. The most important of these is PsbO, which binds to the donor side of PS II near the Mn cluster and is directly involved in the regulation of its stability and activity. However, the molecular mechanism of PsbO involvement in photosynthetic water oxidation remains unclear. One of the main approaches to solving this problem is site-directed mutagenesis. Until recently, the effect of mutations in PsbO in vivo has been studied only in cyanobacteria (prokaryotes). In eukaryotic organisms, such studies (site-directed mutagenesis of PsbO) have not been carried out, though it is known that the role of PsbO protein in plants and cyanobacteria may be different. In this review, we consider the possibility of using for this purpose the unicellular green alga Chlamydomonas reinhardtii, a eukaryotic organism with a set of extrinsic proteins of the WOC similar to that of the higher plants. However, in contrast to higher plants, the ΔpsbO mutant of C. reinhardtii is viable. Another reason to use this alga is that the ΔpsbO strain of C. reinhardtii grown in the dark (heterotrophically) is able to build the minimal photochemically active complex of PS II, allowing investigation of the role of individual amino acid substitutions in PsbO in vivo without damaging PS II due to photoinactivation.
KEY WORDS: photosystem II, water-oxidizing complex, PsbO protein, Chlamydomonas reinhardtii, site-directed mutagenesisDOI: 10.1134/S0006297915060036
Abbreviations: DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; ETC, electron transport chain; PS II, photosystem II; Fv/Fm, ratio of variable chlorophyll fluorescence (Fv) to maximum fluorescence level Fm; Fv′ and Fm′, variations in fluorescence yield under continuous light; MSP, manganese-stabilizing protein (PsbO); ΔpsbO, mutant with inactivated gene encoding PsbO protein; RC, reaction center; TMPD, tetramethyl-p-phenylenediamine; WOC, water-oxidizing complex.
Photosystem II (PS II) is the only known in Nature enzyme that can
oxidize water and use it as an electron donor in photosynthesis by
plants and cyanobacteria. To do this, PS II generates the strongest
biological oxidant (the cation radical P680+• of
chlorophyll with redox potential of 1.12-1.26 V) to pull electrons
away from the water molecule (E0′ = 0.81 V)
[1-4]. However, the
P680+• chlorophyll does not oxidize water by itself.
Oxidation occurs in the water-oxidizing complex (WOC) of PS II located
near P680+; the catalytic center of this complex includes
four manganese atoms and one calcium atom
(Mn4CaO5 cluster).
The ligands of the manganese and calcium atoms are proteins D1 and CP43 [5] that, together with proteins D2, CP47, cytochrome b559, and PsbI, form the minimal complex of PS II capable of water oxidation in vitro. However, this complex is unstable and is quickly inactivated. Several more extrinsic proteins of the WOC bound to the donor side of PS II are required for its steady work [6, 7]. In higher plants and green algae, these proteins are PsbO, PsbP, PsbQ, and PsbR (PsbO, PsbU, and PsbV in cyanobacteria), and their main function is probably to stabilize the WOC. The PsbO protein plays the key role in this process directly influencing the Mn cluster stability [7, 8]. The removal of PsbO from PS II preparations leads to an 80% decrease in the rate of photosynthetic oxygen evolution and gradual release of two of the four manganese atoms of the WOC into the medium [9].
Several hypotheses were proposed to explain the function of PsbO. The prevailing hypothesis is that PsbO is necessary for protection of the Mn cluster. According to this, PsbO is bound to the WOC of PS II on the lumenal side of PS II and shields the catalytic center from active chemical compounds in medium (reductants, metals, OH– ions, etc.) [8, 10]. PsbO is also supposed to participate in the organization of proton–water transport between the WOC and the lumen [6-8]. Nevertheless, in spite of the long period of research (since 1979), the functional role of PsbO in photosynthetic water oxidation is still unclear. This is especially so if we wish to indicate the particular amino acids responsible for the supposed function [8, 10, 11].
Site-directed mutagenesis may be a possible solution to this problem. An important stage in this work is to choose the biological model to be studied. Since PsbO is one of the few hydrophilic proteins within PS II, it is most often studied using site-directed mutagenesis in vitro (PS II preparations) [10, 12]. However, under these conditions, it is impossible to study the effect of mutation on PS II biogenesis, what is one of the most interesting trends in the study of the photosynthetic process. One can answer these questions only in living organisms, by investigating the effect of amino acid substitutions in vivo.
Until recently, cyanobacteria (prokaryotes) have been the only object of such manipulations [13, 14]. However, due to some differences between plants and cyanobacteria in their composition of extrinsic proteins of the WOC and because the functions of PsbO in these organisms can be different, it is not quite correct to apply those results to plants. Therefore, work with eukaryotic organisms of similar protein composition is of particular interest.
There are many reasons why it is difficult to work with higher plants. The main problem is that their ΔpsbO mutant (with inactivated gene encoding the PsbO protein) is unviable [15]. In addition, the mutations in PsbO make PS II sensitive to photoinactivation. Therefore, the unicellular green alga Chlamydomonas reinhardtii seems to be more suitable for research, because the protein composition of the WOC in this eukaryotic organism is similar to that in higher plants. In addition, the metabolism of C. reinhardtii is unique to green plants. In contrast to the higher plants, which need light for their growth (obligate phototrophs), C. reinhardtii can grow in the dark (heterotrophically) on acetate and, at the same time, form green active chloroplasts [16, 17]. This feature of C. reinhardtii makes it possible to study the functional state and biogenesis of PS II complex in vivo, excluding damage of WOC due to photoinactivation.
STRUCTURAL AND FUNCTIONAL ORGANIZATION OF PHOTOSYSTEM II
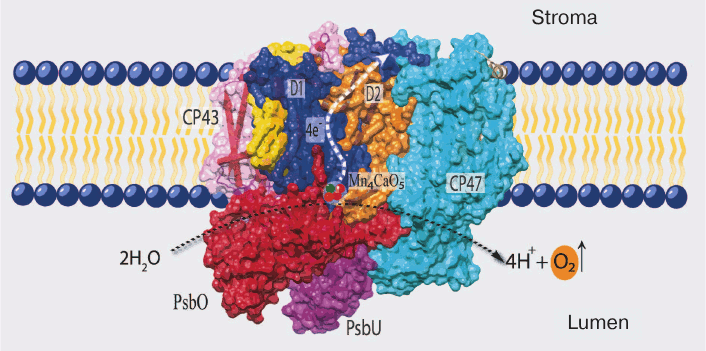
The details of PS II structure are now well established from the data of X-ray structure analysis of cyanobacterial PS II (the last one with resolution up to 1.9 Å in the work of Umena et al.) [5]. According to these data, PS II in the cells of cyanobacteria (and eukaryotes) consists mainly of a dimeric complex with molecular weight of ~700 kDa, and each monomer of this complex consists of 20 proteins and associated molecules of pigments, lipids, and cofactors of electron transfer (Fig. 1). Most of these proteins (17) are incorporated into the membrane and contain one to six transmembrane helices, and only three proteins are peripheral (PsbO, PsbV, and PsbU) [5, 18].
Fig. 1. Positions of the main components of PS II in cyanobacteria (PDB file 3ARC). D1 (PsbA) and D2 (PsbD) are the RC proteins; CP43 and CP47 are light-harvesting complexes tightly bound to RC; PsbO, PsbU, and PsbV (not seen on the picture) are water-soluble proteins of the water-oxidizing complex.
The PS II proteins can be conventionally divided into three groups: (i) proteins of so-called core of PS II and a group of neighboring low molecular weight proteins; (ii) water-soluble proteins of the WOC; and (iii) proteins of the external antenna of this complex.
All of the major electron transfer cofactors are bound to the two key proteins of PS II core – D1 and D2. The cofactors include: (i) six molecules of chlorophyll a (Chl a); two of these are components of the reaction center (RC) and form a dimer with absorption maximum at 680 nm (P680), which is the primary electron donor transferring its electron to a pheophytin (Pheo) molecule upon excitation; (ii) two molecules of Pheo (a magnesium-free Chl derivative), only one of these (bound to D1 protein) being an intermediate electron acceptor; (iii) two quinones – plastoquinone QA (bound to D2 protein) and plastoquinone QB (bound to D1 protein), the secondary and final electron acceptors in PS II located on the stromal side of PS II; and (iv) TyrZ (Tyr161 of D1) and the Mn4CaO5 cluster, forming a redox system for water oxidation (CP43 protein is also a ligand of one atom of manganese) [4, 5, 19].
Together these cofactors form the electron transport chain, which is arranged in the following order from the lumen to the stroma:
H2O → WOC [Mn4CaO5] → Yz/Yz* → P680/P680+ → Pheo a/Pheo a– → QA/QA– → QB/QB.
The functioning of PS II is based on redox reactions: PS II oxidizes water and transfers electrons to the pool of plastoquinones using the energy of absorbed light. The core of this process is the light-induced charge separation in the RC; as a result, the strongest biological oxidizer P680+ with redox potential up to +1.26 V appears on the donor side of PS II [1, 2]. Here, electron deficiency is compensated by electrons received during water oxidation. For this action, PS II has a special WOC with a catalytic center containing manganese ions (the Mn cluster). The Mn cluster is a redox system that successively donates four electrons to the acceptor (TyrZ) and then withdraws them from the donor (2H2O) [20].
The Mn cluster is successively oxidized after each flash of light, passing through five intermediate states known as the Sn states (n = 0-4). As a result, it accumulates four positive charges necessary for the detachment of four electrons from two H2O molecules (2H2O → 4H+ + O2↑ + 4e–) [21]. The side product of this reaction is molecular oxygen; therefore, this type of photosynthesis is called oxygenic.
It should be noted that the redox system of manganese atoms is inherently unstable [22] and, therefore, the extrinsic proteins of the WOC, which are present in all oxygenic organisms, play an important role in optimization of the functioning of the WOC.
EXTRINSIC PROTEINS OF THE WATER-OXIDIZING COMPLEX OF PHOTOSYSTEM
II
The WOC of all known oxygenic organisms has a similar structure: the catalytic center that includes the Mn4CaO5 cluster and also a group of the extrinsic proteins that play an important role in its stabilization. In PS II, these proteins are bound on the lumenal side of the membrane and localized near the catalytic center of the WOC (Fig. 1). The role they play in WOC activity seems to be stabilization of the catalytic center at physiological concentrations of Ca2+ and Cl– ions in the medium [7]. In addition, they can protect the Mn cluster from attack by external reductants present in the lumen [8].
The WOC of different organisms may contain from 1 to 5 proteins [7]. The only WOC protein present in all oxygenic organisms is PsbO. Analysis of the genomes of photosynthetic organisms shows that PsbO might appear simultaneously with the water-oxidizing ability of PS II. It is believed to have been the only WOC protein associated with the Mn cluster in the first protocyanobacteria [6, 23]. There are two strains of the green oxyphotobacterium Prochlorococcus marinus, where PsbO is the only WOC protein encoded in the genome [23, 24]. Then, during evolution, the WOC included other proteins (PsbU and PsbV, as well as CyanoQ and CyanoP of cyanobacteria). In turn, the appearance of eukaryotic photosynthetic organisms (green plants) resulted in the loss of proteins PsbU and PsbV; their place in PS II was occupied by PsbP and PsbQ [23-25].
Thus, the protein composition of the WOC substantially changed during evolution, probably due to adaptation to new environmental conditions [23, 25]. The only constant protein of the WOC was PsbO, which indicates its importance for the interaction with the water oxidation system. This fact is also confirmed by biochemical studies. The extrinsic proteins of the WOC can be successively removed by treating PS II preparations with saline solutions [9, 26, 27]. The removal of PsbP and PsbQ from PS II preparations slightly reduces the oxygen-producing activity and can be compensated for by increasing the content of Ca2+ and Cl– in the medium, whereas removal of PsbO irreversibly reduces the rate of oxygen production (to ~20% of the initial rate) and destabilizes the structure of the manganese cluster [9, 26-29].
PsbO PROTEIN
PsbO is a unique protein of PS II that can be found only in oxygenic photosynthetic organisms: higher plants, algae, and cyanobacteria. The search for homologs in other organisms has given no positive results, and no similar proteins have been found in anoxygenic purple bacteria, the RC of which is a type II photosystem [6, 10, 23]. The conservatism of PsbO throughout algae, plants, and cyanobacteria is very high: ~60-80% among eukaryotes (higher plants and algae) and ~40% between cyanobacteria and plants [10, 23].
The study of PsbO started with the work of Japanese researchers in 1979, when a protein with molecular weight of 33 kDa for the first time was isolated from PS II preparations and characterized [30]. The association between PsbO and the water oxidation function in PS II was shown two years later (1981-1982). Several research teams reported that the treatment of PS II preparations with Tris-HCl (pH ≥ 8.0) resulted in the inhibition of O2 production (the WOC inhibition by 0.8 M Tris buffer had been shown previously, in 1968) [31] and also caused the release from the PS II complex of three proteins with molecular weights of 33, 23, and 17 kDa (PsbO, PsbP, and PsbQ, respectively) [32-34]. The protein with the highest molecular weight, conventionally denoted as 33-kDa protein, is PsbO. This name however does not reflect the true molecular weight of the protein, which is somewhat less (26.5 kDa) [6]. The determination of molecular weight was inaccurate because PsbO is not a typical globular protein, but rather has an elongated spatial structure. Later (in 1984) it was shown that PsbO was necessary for the stability of the Mn cluster: two of four manganese ions are released from the catalytic center upon the removal of PsbO from PS II preparations [9]. Hence, PsbO got its second name: the manganese-stabilizing protein, or MSP.
In PS II, PsbO is the largest of the extrinsic proteins of the WOC, with average molecular weight of 26.5 kDa, and it consists of 231-257 amino acid residues. In cells of higher plants and algae (eukaryotes), PsbO, like other extrinsic proteins, is encoded in the nucleus and synthesized as a precursor protein with two signaling sequences. One of them is responsible for delivery of the protein into the chloroplast, and the other is responsible for delivery into the lumen (Arabidopsis thaliana, UniProt ID P23321) [6, 35].
After getting into the lumen, the signaling sequences of PsbO are cut out; the protein acquires the characteristic tertiary structure and binds to PS II. According to the data of X-ray structure analysis, PsbO in its final native conformation has an elongated structure and consists of two domains (the β-cylinder and the head domain). Both of these domains are involved in the interaction with PS II, and the protein binds to PS II at an angle of 40° to the plane of the membrane (Fig. 1) [5, 6]. It is interesting that there is a pool of PsbO proteins in the lumen that are not bound to PS II [36-38]. The biological sense of this phenomenon is unclear, and it is quite probable that the free proteins may perform some other functions in addition to what is typical of them inside the WOC.
FUNCTIONAL ROLE OF PsbO
Functioning of PS II in the absence of PsbO. The role of PsbO can be determined experimentally by the studying the changes caused by the removal of the protein from PS II. PsbO can be softly removed from subchloroplast preparations of PS II by biochemical methods (incubation in 2 M NaCl, 1 M CaCl2, or 0.2 M NaCl in the presence of 2.6 M urea), with the Mn cluster left intact [9, 39]. In this case, the removal of PsbO leads to a drastic decrease in the rate of oxygen production and gradual dissociation of two of the four manganese atoms from the Mn cluster. However, some part of the oxygen-producing activity of PS II can be preserved (~25%) by increasing the concentrations of Cl– and Ca2+ ions necessary for maintenance of integrity of the Mn cluster in the absence of extrinsic proteins [9]. What changes in the function of the WOC are observed under these conditions?
The kinetics of oxygen production in PS II in response to a series of microsecond flashes shows that the changes affect mostly the higher states of the S-cycle. The removal of PsbO was accompanied by extension of the lifetime of S2 and S3 states [40] and inhibition of the S2→S3 transition [40] and the S3→[S4]→S0 transitions [40, 41]. In addition, the exchange of substrate water bound to the active center in the S3 state slows in the absence of PsbO [42].
ΔpsbO mutations. The loss of PsbO in vivo causes much more serious damage to PS II. In this case, not only the WOC functions are disturbed but also the entire PS II structure is affected until complete disappearance of the PS II complex in mutant cells and the death of the organism. It should be noted that the degree of impairment in PS II in the absence of PsbO is different for plants and cyanobacteria. In plants (Arabidopsis thaliana), the inactivation of PsbO by RNA interference (RNAi) slows plant growth and causes death [15]. In this case, the absence of PsbO is accompanied by the loss of the PS II pigment–protein complex and inability of the plant to grow photoautotrophically. In the ΔpsbO mutant of another model organism, the green alga C. reinhardtii (strain FUD44), the results of mutation were the loss of oxygen-producing activity and the absence of growth under photoautotrophic conditions [43]. When the mutant culture was incubated under mixotrophic conditions (in the light, in the presence of acetate), the formation of a stable PS II complex in the cells of C. reinhardtii was not observed, though the synthesis of other protein components of PS II was not disturbed [36, 43]. However, as our studies have shown, PS II in cells of the ΔpsbO mutant was nevertheless assembled during heterotrophic cultivation of C. reinhardtii in the dark (without the damaging effect of light), though without the ability to evolve oxygen [44].
In contrast to eukaryotes, prokaryotes (cyanobacteria) are able to form PS II and even oxidize water even in the absence of PsbO. For example, the ΔpsbO mutant of Synechocystis sp. PCC 6803 can grow photoautotrophically, though its oxygen evolution is unstable and lower by 60% compared to the wild type [45, 46]. However, it should be noted that for oxygen evolution in this case PS II must have another extrinsic protein of the WOC, PsbV (cytochrome c550), which is absent in the higher plants and green algae [11, 45]. In addition, the ΔpsbO mutant of cyanobacteria is unable to grow in medium with low levels of Cl– and Ca2+ ions. The activity of the WOC in the mutants could be maintained at high (non-physiological) levels of Cl– (≥100 mM) and Ca2+ in the growth medium or at high pH values of the medium (10.0) [45, 47, 48]. These data suggest that the state of the Mn cluster in the absence of PsbO will depend on the chemical composition of the environment.
It can be supposed (and it is in good agreement with the experimental data) that the function of PsbO is to regulate the interactions between the WOC and the medium of the lumenal space of chloroplasts. Now, how can we explain the influence of PsbO on the activity of the WOC of PS II? It is supposedly associated with the location of the protein close to the water oxidation site. According to the data of X-ray crystallography, PsbO is bound on the donor side of PS II, close to manganese atoms, and can perform several major functions.
First, it ensures the structural stabilization of all functional atoms of manganese, as well as chlorine and calcium, in the catalytic center of the WOC of PS II. This may occur due to the interaction between PsbO and proteins of the core complex of PS II. It is known that PsbO per se is not a ligand of manganese and calcium atoms in the Mn4CaO5 cluster. For example, the minimal possible distance between PsbO and manganese atoms in the WOC is 15-20 Å, and this distance is too great for direct interaction [5]. Nevertheless, PsbO can influence the structure and stability of the Mn cluster, because some regions of PsbO contact regions of the D1, D2, and CP43 proteins that are direct ligands of the Mn cluster [5] or necessary for its stability [6].
Second, the binding of PsbO with PS II protects the WOC from inactivation by various compounds due to isolation of the Mn cluster from the lumen. It is believed that the principal cause of withdrawal of Mn ions from the Mn cluster is associated with their reduction [8, 49]. Consequently, the role of PsbO may consist in creating a barrier on the way of these molecules to the catalytic center of water oxidation. The idea about the protective function of PsbO may be supported by the results of the study on immobilization of extrinsic proteins on PS II by means of chemical modification. The fixation of extrinsic proteins on PS II leads to stabilization of the Mn cluster under conditions of thermal, acidic, and saline treatment of PS II preparations [50].
As an extension of this function, the role of PsbO may consist in organization of the transport of necessary molecules between the water oxidation center and the lumen. Thus, PsbO probably regulates the access of chloride, calcium, and bicarbonate ions to the Mn cluster and retention of these molecules close to the Mn cluster. In addition, since the manganese cluster, where water oxidation takes place, is closed by the protein environment from the ambient aqueous medium, water access and proton removal can be regulated [6, 51-54]. The PsbO protein may play an important role in organization of such special transport channels.
Another probable function of PsbO is its involvement in the regulation of dimer-to-monomer transitions of PS II complexes. Thus, the protein may be involved in the biogenesis of PS II. This assumption is based on analysis of the crystal structure of PS II, where PsbO of each PS II monomer interacts with the CP47 protein of the other monomer and, hence, can bind two monomer complexes of PS II together [5, 6]. In addition, the study of ΔpsbO mutation in cyanobacteria showed that the PS II complex in such mutants accumulates in the membranes only as PS II monomers [55].
In conclusion, it should be noted that PsbO might probably have an enzymatic activity, i.e. to function in the cells as GTPase of carboanhydrase. The experimental data obtained by Spetea et al. show that PsbO (in the light) is able to bind and hydrolyze GTP molecules [56]. In previously published works of our and Stemler’s laboratories, it was supposed that PsbO performs the role of carboanhydrase, participating in proton removal from the WOC [57, 58].
The presented above list of possible functions of PsbO is incomplete. Partly, such diversity may be related to the fact that it describes different aspects of the same phenomenon. When binding with PS II, PsbO simultaneously stabilizes the D1, D2, and CP43 protein regions acting as ligands of the Mn cluster and protects the Mn cluster from the lumen. However, on the other hand, such a number of assumptions about the protein function indicate that the exact molecular mechanism of PsbO function in PS II has not yet been established.
Site-directed mutagenesis of PsbO. One of the basic methods to determine at the molecular level how the PsbO protein participates in photosynthetic water oxidation is site-directed mutagenesis, which makes it possible to alter the amino acid sequence of the protein and, consequently, its structure and function. The choice of a model system for study becomes an important factor at this stage.
The PsbO protein is generally studied using mutations in vitro (in PS II preparations), using modified protein to substitute native PsbO in PS II (WOC reconstruction) [10, 12]. The modified PsbO is expressed in E. coli and then in the purified form is added to PS II preparations where the native PsbO was removed. This addition partially restores the function of oxygen evolution. Due to the high homology of PsbO in different organisms, the protein taken from one species can be added to PS II preparations from others to observe the recovery of the rate of oxygen evolution. The rate of oxygen evolution after the reconstruction increased on average to 40-50% of the rate of initial PS II preparations, while without the PsbO it was only 15-20% of the control [59, 60].
The main parameters studied in vitro are the kinetics of protein binding with the membrane and the effect of partial recovery of the rate of O2 evolution [12, 61-63]. In most cases, this system is rapid and convenient; however, there are a number of limitations of this approach. First, in order to remove the native PsbO, the treatment is used that causes the removal of other extrinsic proteins from PS II. Second, oxygen evolution in PS II preparations irreversibly decreases with any method of protein removal. Third, researchers have to perform experiments with the Mn cluster and PS II formed in wild type cells. At the same time, researchers are often concerned with the problem whether the mutation affects exactly the assembly of WOC and PS II components. To answer these questions, they have to study the effect of mutation on PS II functions in vivo.
Until recently, the only object of this research with site-directed mutagenesis of PsbO was the cyanobacterium Synechocystis sp. [13]. Cyanobacteria, being prokaryotes, are a convenient model organism for PsbO studies as they have only one genome and the active complex of PS II can be assembled without the PsbO protein. Cyanobacteria were usually used to investigate the effect of complete inactivation of the psbO gene or in combination with inactivation of the synthesis of other extrinsic proteins of PS II (ΔpsbO:ΔpsbV, ΔpsbO:ΔpsbU, etc.) [14, 45, 47, 48, 64, 65]. There were very few substitutions for individual amino acids in the PsbO protein [10, 13]. As an example, we can mention the work on obtaining Synechocystis strains with cysteine mutations. In Synechocystis sp. PCC6803 mutant constructed for studying the role of disulfide bond in PsbO in vivo, one of the cysteine residues of the protein was replaced by serine (C20S) [13]. This mutant did not accumulate PsbO and was characterized by low rate of oxygen evolution (comparable with the ΔpsbO strain of Synechocystis). The absence of detectable amount of the PsbO in the cell, however, was not associated with lower level of mRNA, which corresponded to that of the wild type [13]. The authors supposed that this substitution caused disturbances in the native conformation of PsbO and in the protein binding with PS II, so the synthesized protein underwent degradation and proteolysis.
When cyanobacteria are considered as a potential object of research, it should be taken into consideration that the subunit composition of WOC proteins strongly varies between cyanobacteria and plants: the PsbO, PsbP, and PsbQ proteins are present in green algae and higher plants, while the PsbO, PsbU, and PsbV proteins are present in cyanobacteria. The PsbV protein, which is absent in higher plants, is very important for the activity of the WOC in prokaryotes; hence, the inactivation of PsbO in cyanobacteria (in contrast to higher plants and algae) does not terminate oxygen evolution and, consequently, it is difficult to estimate the role of individual mutations in PsbO. Under certain conditions, the rate of O2 evolution in cells of the ΔpsbO strain of cyanobacteria can be ≥50% of that in the wild type cells, and oxygen evolution can be completely suppressed only in case of a double mutation – ΔpsbO:ΔpsbV [45, 48].
In this context, it would be particularly interesting to work with eukaryotic organisms of similar protein composition. The studies in higher plants are complicated by the fact that their ΔpsbO mutant in unviable. In addition, protein mutations make PS II highly sensitive to photoinactivation [15]. The problem was also the absence, until recently, of a method that would allow direct gene inactivation in the plant nuclear genome by inserting homologous DNA, as it is possible in yeasts and bacteria and with the chloroplast genome. Now, however, the system for in vivo genome editing (CRISPR/Cas9 system, etc.) can be used for this purpose [66]. In addition, Arabidopsis T-DNA insertion mutants also can be obtained for almost any gene [67, 68]. Currently, gene expression can be also suppressed by the RNAi-silencing technique [69]. The application of these approaches allowed to study the role of two PsbO isoforms in Arabidopsis.
There are two PsbO genes in the cells of Arabidopsis thaliana. The second protein isoform, PsbO-2, appeared approximately 25-50 million years ago as a result of gene duplication during evolution. The amino acid sequences of the two proteins differ in only 11 amino acid residues; however, each of these isoforms is believed to have its own special function, and they cannot be completely substitutable with each other [70-72]. Simultaneous suppression of the expression of two proteins (PsbO-1 and PsbO-2) by RNAi interference resulted in significant disturbances in PS II, so that the plants could not grow under photoautotrophic conditions [15]. The silencing of one of the two genes (a premature stop codon in PsbO-1 [70] or T-DNA insert [72]) also resulted in the development of a certain phenotype with reduced activity of PS II, which supports the idea of specialization of each protein variant. The main protein isoform is PsbO-1 [70-73]. It is the one directly associated with PS II function, while PsbO-2 seems to be specialized in some other functions. In particular, there is an assumption that PsbO-2 is involved in phosphorylation of protein D1 and acts as a GTPase [72].
Researchers have also demonstrated that the levels of PsbO-1 and PsbO-2 expression change differently during cold acclimatization of the plants. The cold treatment of the plants was accompanied by a decrease in the content of the major form, PsbO-1, while the level of PsbO-2 increased [74]. Nevertheless, until quite recently it has been difficult to make amino acid substitutions in the PsbO protein of higher plants using the methods of antisense RNA, in vivo genome editing systems (TALEN, Zinc Finger nucleases, CRISPR/Cas9), and site-directed mutagenesis. But it seems probable that technologies based on the CRISPR/Cas9 method will be the main approach to obtaining such site-directed mutants in vivo in the near future [66]. In addition, as already mentioned, mutations in PsbO make PS II very sensible to photoinactivation, which results in subsequent PS II degradation. The problems described above can be solved by using the green alga C. reinhardtii as the object of research.
Like higher plants, Chlamydomonas is a eukaryotic organism with similar extrinsic protein composition of the WOC. In addition, this alga has some special capabilities [16, 17]. In contrast to higher plants, which need light for their growth (obligate phototrophs), Chlamydomonas is able to grow in the dark (heterotrophically) on acetate and, at the same time, to form green active chloroplasts. Due to this feature, it is possible to study the functional state of the RC and the biogenesis of PS II complex in vivo in algae grown in the dark, avoiding damage to the WOC as a result of photoinactivation. In other words, it becomes possible to study not the consequences of photoinactivation but directly the effects of amino acid substitutions on the efficiency of photosynthetic water oxidation. Hence, it also follows that there will be two model systems at researchers’ disposal simultaneously, which can be compared: the dark and light cultures of C. reinhardtii. Another advantage of C. reinhardtii as a model object is the simplicity of work with it, the relatively short cell cycle (6-8 h), the extent of genetic investigation, and the presence of various mutations in the nuclear, chloroplast, and mitochondrial genes [16, 17, 75].
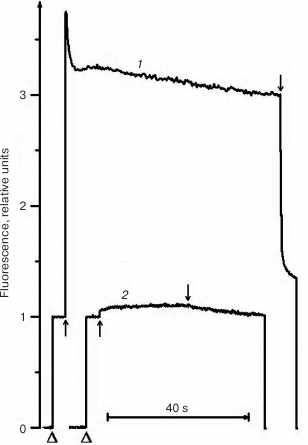
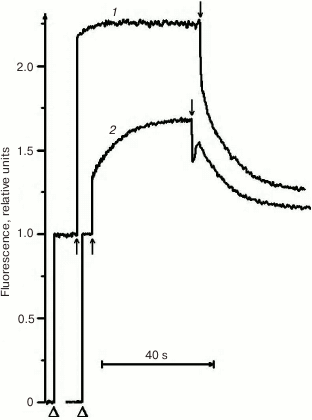
Due to its metabolism, C. reinhardtii is very useful in the study of mutations in PS II. In medium with a carbon source (acetate), the mutants can grow with any PS II damage, up to its complete absence. However, the ΔpsbO strain of C. reinhardtii has not been previously used for site-directed mutagenesis of PsbO until recently. That was partially related to the notions that PsbO in eukaryotes is obligatory for the assembly of a stable PS II complex and that its inactivation leads to almost complete degradation of PS II in cells [43]. For example, the proteins belonging to the core complex of PS II are critically important for PS II assembly, and their loss in C. reinhardtii leads to degradation of incorrectly formed PS II complex by proteases even in the dark [76]. However, as our studies have shown, the degradation of PS II complex in the ΔpsbO mutant of C. reinhardtii is associated largely with the damaging effect of light [44]. The assembly of at least a stable “core” of the PS II complex, capable of light-driven charge separation in RC, took place in cells of the ΔpsbO strain cultivated in the dark. This assembled complex of the PS II was not capable of oxygen evolution but showed some level of photochemical activity in experiments based on the method of variable fluorescence: the Fv/Fm ratio characterizing the efficiency of energy conversion in PS II was 0.37 (Figs. 2 and 3). The kinetics of variable fluorescence was also sensitive to DCMU (an inhibitor of electron transport in PS II), which was evidence of the assembly of functionally active RCs of PS II in this mutant.
Fig. 2. Kinetics of photoinduced changes in chlorophyll fluorescence yield (ΔF) measured in cells of light cultures of the wild type strain (1) and the ΔpsbO strain (2) of C. reinhardtii. Triangles indicate the moments of switching on the measuring light; up and down arrows indicate the moments of switching the actinic light on and off, respectively (1500 µmol⋅m–2·s–1).
Fig. 3. Kinetics of photoinduced changes in chlorophyll fluorescence yield (ΔF) measured in the cells of dark cultures of the wild type strain (1) and the ΔpsbO strain (2) of C. reinhardtii. Triangles indicate the moments of switching on the measuring light; up and down arrows indicate the moments of switching the actinic light on and off, respectively (1500 µmol⋅m–2·s–1).
Chlamydomonas reinhardtii AS A MODEL ORGANISM FOR
SITE-DIRECTED MUTAGENESIS OF PsbO PROTEIN
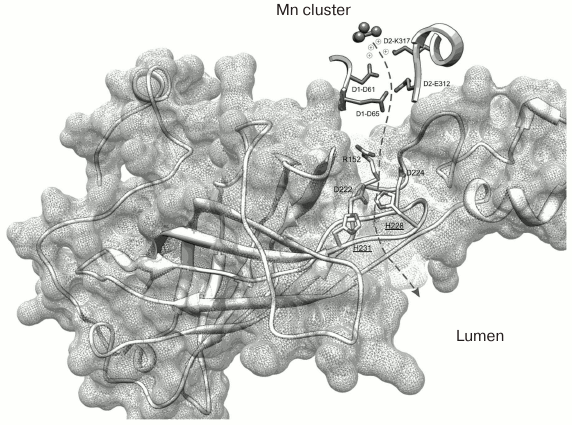
The recently published research of our laboratory can be used as an example of application of C. reinhardtii in the study of amino acid substitutions in the PsbO protein [77]. Two PsbO mutants were obtained using the ΔpsbO strain of C. reinhardtii: there were two lysine substitutions in the PsbO sequence at positions 223 and 226 (K223E and K226E, homologous to His228 and His231 in cyanobacteria). These positively charged amino acids located on the lumenal side of PsbO are highly conserved in the higher plants and green algae. According to some hypotheses (Fig. 4) [6, 78], they are probably involved in formation of a hydrophilic channel connecting the lumen and the WOC.
Fig. 4. Spatial organization of PsbO protein in the PS II complex of the cyanobacterium T. elongatus (file 3BZ1.pdb from PDB). The dashed arrow indicates the supposed proton transport pathway from the Mn cluster into the lumenal space of the thylakoid. The His228 and His231 residues located closer to the lumen (homologous to K223 and K226 in C. reinhardtii) are emphasized.
Investigation of the properties of these mutants has shown that lysine at position 226 is important for the stability of the protein. The substitution of this lysine for oppositely charged glutamine residue resulted in an abrupt decrease in the level of protein in the cells and development of ΔpsbO phenotype (the absence of photoautotrophic growth and oxygen evolution in PS II) [77]. The analysis of protein structure may suggest the causes of such disturbances. It may be because the lysine at position 226, Lys226 (His231 in cyanobacteria), is located immediately before the C-terminal β-strand (K*DIKVTGLWYAQLK) which, as has been shown previously, is necessary to bind to PS II complex [79]. In addition, this C-terminal region is an important structural element of the major domain of PsbO: the β-cylinder responsible for stability of the protein. Therefore, substitution in this region may interfere with normal protein folding, leading to the impairment of binding with PS II and subsequent degradation of the protein.
Investigation of the properties of the other mutant (K223E) showed that the content of PsbO in its cells was maintained within normal limits, so that it was capable of photoautotrophic growth and photosynthetic oxygen evolution. However, the rate of oxygen evolution and the Fv/Fm ratio were lower by 15-20% compared to the control (pseudo wild type (pWT)). The dark relaxation time of Fv in the presence of DCMU was also shown to increase for the K223E mutant, which may indicate impaired function of the water-oxidizing complex.
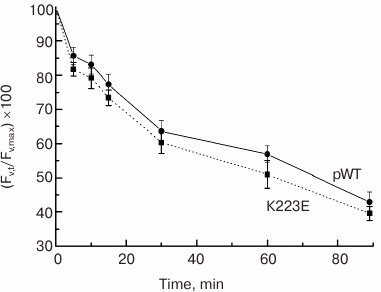
What could the observed changes be associated with? According to one hypothesis, Lys223 may be important for proton removal from the Mn cluster into the lumen, forming part of a channel for their transfer from the WOC to the lumen. One of the methods to test this hypothesis is to assess the sensitivity of PS II to photoinactivation. Strong light must cause an increase in RC and WOC activity and lead to acidification of the lumen and impairment of the WOC. Therefore, if proton removal is impeded, e.g. due to a mutation, it may enhance photodamage to PS II and reduce its activity. However, the testing of K223E cells for sensitivity to photoinactivation showed no significant differences between the mutant strain and the control pWT (Fig. 5).
Fig. 5. Sensitivity to photoinactivation of cells in light cultures of pWT-type and K223E mutant during 90-min illumination at light intensity of 2500 µmol·m–2·s–1 measured by the decrease in the level of Fv′ = (Fm′ – F0)/Fm′.
The disturbances in PS II activity can be caused also by the effect of structural changes in the protein on its protective function. It is known that many molecules – electron donors – are able to inhibit the function of the WOC of PS II [49, 80]. During the interaction with the Mn cluster, reductants can reduce manganese to the degree of Mn2+ oxidation, which can lead to dissociation of Mn from the WOC and inhibition of oxygen evolution. The rate of inactivation will largely depend on accessibility of the Mn cluster for these electron donors, which is determined by the size of the electron donor molecule and by the structure of the protein environment of the WOC. Partial removal or modification of extrinsic proteins of the WOC open the Mn cluster to reductants and make it more available for inhibition [49, 80-82].
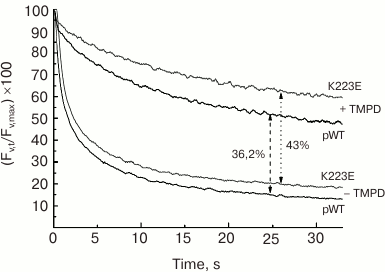
The study of subchloroplast preparations of PS II has shown that the impaired function of the WOC in the K223E mutant of C. reinhardtii may be associated with change in the protein structure resulting in enhanced availability of the Mn cluster for inactivation by electron donors. This may be indicated by the enhanced sensitivity of PS II preparations isolated from the K223E mutant to inactivation by TMPD (tetramethyl-p-phenylene diamine), an artificial electron donor for PS II, compared to the control (Fig. 6). It seems that this residue may be important for regulation of the interaction between the WOC and the lumen.
Fig. 6. Kinetics of dark relaxation of the variable fluorescence (Fv) in PS II preparations of C. reinhardtii strains pWT and K223E after a 1-s flash of actinic light at intensity of 4500 µmol·m–2·s–1. The measurements were made in the absence (a) and presence (b) of TMPD (500 µM).
The data presented above indicate that C. reinhardtii is a convenient model for studying mutations in the PsbO protein in vivo (especially because in vivo it is possible to study the effects of mutations in PsbO on the biogenesis of PS II, in particular, the role of PsbO in the formation of PS II dimer). According to the structural data, several regions of the PsbO protein (E55-R60, V66-K69, and M110-G113) are in close contact with the CP47 protein of another PS II monomer [6]. In this context, it is interesting what effect the mutations in this part of the protein will have on the formation of PS II dimeric complex in the cells. In addition, it is possible to distinguish the regions of PsbO sequence contacting other extrinsic proteins of the WOC (proteins PsbP and PsbQ in Chlamydomonas), where the substitutions will help to reveal regularities in formation of the WOC protein complex.
This work was supported by the Russian Science Foundation (project No. 14-14-00535).
REFERENCES
1.Rappaport, F., Guergova-Kuras, M., Nixon, P. J.,
Diner, B. A., and Lavergne, J. (2002) Kinetics and pathways of charge
recombination in photosystem II, Biochemistry, 41,
8518-8527.
2.Klimov, V. V., Allakhverdiev, S. I., Demeter, Sh.,
and Krasnovsky, A. A. (1979) Photoreduction of pheophytin in
photosystem II of chloroplasts depending on the redox potential of the
medium, Dokl. Acad. Nauk SSSR, 249, 227-230.
3.Allakhverdiev, S. I., Tomo, T., Shimada, Y., Kindo,
H., Nagao, R., Klimov, V. V., and Mimuro, M. (2010) Redox potential of
pheophytin a in photosystem II of two cyanobacteria having the
different special pair chlorophylls, Proc. Natl. Acad. Sci. USA,
107, 3924-3929.
4.Renger, G. (2011) Light induced oxidative water
splitting in photosynthesis: energetics, kinetics and mechanism, J.
Photochem. Photobiol. B, 104, 35-43.
5.Umena, Y., Kawakami, K., Shen, J. R., and Kamiya,
N. (2011) Crystal structure of oxygen-evolving photosystem II at a
resolution of 1.9 Å, Nature, 473, 55-60.
6.De Las Rivas, J., and Barber, J. (2004) Analysis of
the structure of the PsbO protein and its implications, Photosynth.
Res., 81, 329-343.
7.Bricker, T. M., Roose, J. L., Fagerlund, R. D.,
Frankel, L. K., and Eaton-Rye, J. J. (2012) The extrinsic proteins of
photosystem II, Biochim. Biophys. Acta, 1817,
121-142.
8.Popelkova, H., and Yocum, C. F. (2011) PsbO, the
manganese-stabilizing protein: analysis of the structure-function
relations that provide insights into its role in photosystem II, J.
Photochem. Photobiol. B, 104, 179-190.
9.Miyao, M., and Murata, N. (1984) Role of the 33-kDa
polypeptide in preserving Mn in the photosynthetic oxygen-evolution
system and its replacement by chloride ions, FEBS Lett.,
170, 350-354.
10.Williamson, A. K. (2008) Structural and
functional aspects of the MSP (PsbO) and study of its differences in
thermophilic versus mesophilic organisms, Photosynth. Res.,
98, 365-389.
11.Bricker, T. M., and Frankel, L. K. (2011)
Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant
photosystem II: a critical analysis, J. Photochem. Photobiol. B,
104, 165-178.
12.Motoki, A., Usui, M., Shimazu, T., Hirano, M.,
and Katoh, S. (2002) A domain of the manganese-stabilizing protein from
Synechococcus elongatus involved in functional binding to
photosystem II, J. Biol. Chem., 17, 14747-14756.
13.Burnap, R. L., Qian, M., Shen, J. R., Inoue, Y.,
and Sherman, L. A. (1994) Role of disulfide linkage and putative
intermolecular binding residues in the stability and binding of the
extrinsic manganese-stabilizing protein to the photosystem II reaction
center, Biochemistry, 33, 13712-13718.
14.Eaton-Rye, J. J. (2005) Requirements for
different combinations of the extrinsic proteins in specific
cyanobacterial photosystem II mutants, Photosynth. Res.,
84, 275-281.
15.Yi, X., McChargue, M., Laborde, S., Frankel, L.
K., and Bricker, T. M. (2005) The manganese-stabilizing protein is
required for photosystem II assembly/stability and photoautotrophy in
higher plants, J. Biol. Chem., 280, 16170-16174.
16.Harris, E. H. (2001) Chlamydomonas as a
model organism, Annu. Rev. Plant Physiol. Plant Mol. Biol.,
52, 363-406.
17.Grossman, A. R., Lohr, M., and Im, C. S. (2004)
Chlamydomonas reinhardtii in the landscape of pigments, Annu.
Rev. Genet., 38, 119-173.
18.Pagliano, C., Saracco, G., and Barber, J. (2013)
Structural, functional and auxiliary proteins of photosystem II,
Photosynth. Res., 116, 167-188.
19.Cardona, T., Sedoud, A., Cox, N., and Rutherford,
A. W. (2012) Charge separation in photosystem II: a comparative and
evolutionary overview, BBA, 1817, 26-43.
20.Debus, R. J. (2006) in Photosystem II: The
Light-Driven Water:Plastoquinone Oxidoreductase (Wydrzynski, T.,
and Satoh, K., eds.) Springer, Netherlands, pp. 261-284.
21.Joliot, P., Joliot, A., Boughes, B., and
Barbieri, G. (1971) Studies of system II photocenters by comparative
measurements of luminescence, fluorescence and oxygen emission,
Photochem. Photobiol., 14, 287-305.
22.Tyystjarvi, E. (2013) Photoinhibition of
photosystem II, Int. Rev. Cell Mol. Biol., 300,
243-303.
23.De Las Rivas, J., Balsera, M., and Barber, J.
(2004) Evolution of oxygenic photosynthesis: genome-wide analysis of
the OEC extrinsic proteins, Trends Plant Sci., 9,
18-25.
24.Partensky, F., Hess, W. R., and Vaulot, D. (1999)
Prochlorococcus, a marine photosynthetic prokaryote of global
significance, Microbiol. Mol. Biol. Rev., 63,
106-127.
25.Thornton, L., Ohkawa, H., Roose, J., Kashino, Y.,
Keren, N., and Pakrasi, H. (2004) Homologs of plant PsbP and PsbQ
proteins are necessary for regulation of photosystem II activity in the
cyanobacterium Synechocystis 6803, Plant Cell, 16,
2164-2175.
26.Ghanotakis, D. F., Babcock, G. T., and Yocum, C.
F. (1984) Calcium reconstitutes high-rates of oxygen evolution in
polypeptide depleted photosystem II preparations, FEBS Lett.,
167, 127-130.
27.Ono, T., and Inoue, Y. (1983) Mn-preserving
extraction of 33-, 24- and 16-kDa proteins from O2-evolving
PS II particles by divalent salt-washing, FEBS Lett.,
164, 255-260.
28.Bricker, T. M. (1992) Oxygen evolution in the
absence of the 33-kDa manganese-stabilizing protein,
Biochemistry, 31, 4623-4628.
29.Semin, B. K., Davletshina, L. N., Ivanov, I. I.,
Seibert, M., and Rubin, A. B. (2012) Rapid degradation of the
tetrameric Mn cluster in illuminated, PsbO-depleted photosystem II
preparations, Biochemistry (Moscow), 77, 152-156.
30.Kuwabara, T., and Murata, N. (1979) Purification
and characterization of 33 kDa protein of spinach chloroplasts,
Biochim. Biophys. Acta, 581, 228-236.
31.Yamashita, T., and Butler, W. L. (1968)
Photoreduction and photophosphorylation with Tris-washed chloroplasts,
Plant Physiol., 43, 1978-1986.
32.Akerlund, H.-E., and Jansson, C. (1981)
Localization of a 34,000 and 23,000 MW polypeptide to the lumenal
side of the thylakoid membrane, FEBS Lett., 124,
229-232.
33.Yamamoto, Y., Doi, M., Tamura, N., and Nishimura,
M. (1981) Release of polypeptides from highly active
O2-evolving photosystem-2 preparation by Tris-treatment,
FEBS Lett., 133, 265-268.
34.Kuwabara, T., and Murata, N. (1982) Inactivation
of photosynthetic oxygen evolution and concomitant release of three
polypeptides in the photosystem II particles of spinach chloroplasts,
Plant Cell Physiol., 23, 533-539.
35.Hashimoto, A., Ettinger, W. F., Yamamoto, Y., and
Theg, S. M. (1997) Assembly of newly imported oxygen-evolving complex
subunits in isolated chloroplasts: sites of assembly and mechanism
binding, Plant Cell, 9, 441-452.
36.De Vitry, C., Olive, J., Drapier, D., Recouvreur,
M., and Wollman, F. A. (1989) Posttranslational events leading to the
assembly of photosystem II protein complex: a study using
photosynthesis mutants from Chlamydomonas reinhardtii, J.
Cell Biol., 109, 991-1006.
37.Mayfield, S. P., Schirmer-Rahire, M., Frank, G.,
Zuber, H., and Rochaix, J.-D. (1989) Analysis of the genes of the OEE1
and OEE3 proteins of the photosystem II complex from Chlamydomonas
reinhardtii, Plant Mol. Biol., 12, 683-693.
38.Ettinger, W. F., and Theg, S. M. (1991)
Physiologically active chloroplasts contain pools of unassembled
extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex
in the thylakoid lumen, J. Cell Biol., 115, 321-328.
39.Mavankal, G., McCain, D. C., and Bricker, T. M.
(1986) Effects of chloride on paramagnetic coupling of manganese in
calcium chloride-washed photosystem II preparations, FEBS Lett.,
202, 235-239.
40.Miyao, M., Murata, M., Lavorel, J., Maison-Petri,
B., Boussac, A., and Etienne, A. L. (1987) Effects of the 33 kDa
protein on the S-state transition in photosynthetic oxygen evolution,
Biochim. Biophys. Acta, 890, 151-159.
41.Ono, N., and Inoue, Y. (1985) S-state turnover in
the O2-evolving system of the CaCl2-washed
photosystem II particles depleted of three peripheral proteins as
measured by thermoluminescence. Removal of 33 kDa protein inhibits
S3 to S4 transition, Biochim. Biophys.
Acta, 806, 331-340.
42.Hillier, W., Hendry, R. L., Burnap, T., and
Wydrzynski, T. (2001) Substrate water exchange in photosystem II
depends on the peripheral proteins, J. Biol. Chem., 276,
46917-46924.
43.Mayfield, S. P., Bennoun, P., and Rochaix, J. D.
(1987) Expression of the nuclear encoded OEE1 protein is required for
oxygen evolution and stability of photosystem II particles in
Chlamydomonas reinhardtii, EMBO J., 6,
313-318.
44.Pigolev, A. V., Zharmukhamedov, S. K., and
Klimov, V. V. (2009) The ΔpsbO mutant of Chlamydomonas
reinhardtii is capable of assembling stable, photochemically active
reaction center of photosystem II, Biochemistry (Moscow) Suppl.
Ser. A: Membr. Cell Biol., 1, 31-40.
45.Burnap, R. L., and Sherman, L. A. (1991) Deletion
mutagenesis in Synechocystis sp. PCC6803 indicates that the
Mn-stabilizing protein of photosystem II is not essential for
O2 evolution, Biochemistry, 30, 440-446.
46.Komenda, J., and Barber, J. (1995) Comparison of
psbO and psbH deletion mutants of Synechocystis
PCC 6803 indicates that degradation of D1 protein is regulated by the
QB site and dependent on protein synthesis, Biochemistry,
34, 9625-9631.
47.Philbrick, J. B., Diner, B. A., and Zilinskas, B.
A. (1991) Construction and characterization of cyanobacterial mutants
lacking the manganese-stabilizing protein of photosystem II, J.
Biol. Chem., 266, 13370-13376.
48.Eaton-Rye, J. J., Shand, J. A., and Nicoll, W. S.
(2003) pH-dependent photoautotrophic growth of specific photosystem II
mutants lacking lumenal extrinsic polypeptides in Synechocystis
PCC 6803, FEBS Lett., 543, 148-153.
49.Kuntzleman, T., and Yocum, C. F. (2005)
Reduction-induced inhibition and Mn(II) release from photosystem II
oxygen-evolving complex by hydroquinone or NH2OH are
consistent with a Mn(III)/Mn(III)/Mn(IV)/Mn(IV) oxidation state for
darkadapted enzyme, Biochemistry, 44, 2129-2142.
50.Enami, I., Kamo, M., Ohta, H., Takahashi, S.,
Miura, T., Kusayanagi, M., Tanabe, S., Kamei, A., Motoki, A., Hirano,
M., Tomo, T., and Satoh, K. (1998) Intramolecular cross-linking of the
extrinsic 33-kDa protein leads to loss of oxygen evolution but not its
ability of binding to photosystem II and stabilization of the manganese
cluster, J. Biol. Chem., 273, 4629-4634.
51.Guskov, A., Kern, J., Gabdulkhakov, A., Broser,
M., Zouni, A., and Saenger, W. (2009) Cyanobacterial photosystem II at
2.9-Å resolution and the role of quinones, lipids, channels and
chloride, Nat. Struct. Mol. Biol., 3, 334-342.
52.Ho, F. M., and Styring, S. (2008) Access channels
and methanol binding site to the CaMn4 cluster in
photosystem II based on solvent accessibility simulations, with
implications for substrate water access, Biochim. Biophys. Acta,
1777, 140-153.
53.Shutova, T., Irrgang, K. D., Shubin, V., Klimov,
V. V., and Renger, G. (1997) Analysis of pH-induced structural changes
of the isolated extrinsic 33 kDa protein of photosystem II,
Biochemistry, 36, 6350-6358.
54.Shutova, T., Klimov, V. V., Andersson, B., and
Samuelson, G. (2007) A cluster of carboxylic groups in PsbO protein is
involved in proton transfer from the water oxidizing complex of
photosystem II, BBA Bioenerg., 1767, 434-440.
55.Bentley, F. K., and Eaton-Rye, J. (2008) in
Photosynthesis. Energy from the Sun: 14th Int. Congr. on
Photosynthesis (Allen, J. F., Gantt, E., Golbeck, J. H., and
Osmond, B., eds.) Dordrecht, pp. 715-717.
56.Spetea, C., Hundal, T., Lundin, B., Heddad, M.,
Adamska, I., and Andersson, B. (2004) Multiple evidence for nucleotide
metabolism in the chloroplast thylakoid lumen, Proc. Natl. Acad.
Sci. USA, 101, 1409-1414.
57.Lu, Y. K., Theg, S. M., and Stemler, A. J. (2005)
Carbonic anhydrase activity of the photosystem II OEC33 protein from
pea, Plant Cell Physiol., 46, 1944-1953.
58.Shitov, A. V., Pobeguts, A. V., Smolova, T. N.,
Allakhverdiev, S. I., and Klimov, V. V. (2009) Manganese-dependent
carboanhydrase activity of photosystem II proteins, Biochemistry
(Moscow), 74, 509-517.
59.Betts, S. D., Hachigian, T. M., Pichersky, E.,
and Yocum, C. F. (1994) Reconstitution of the spinach oxygen-evolving
complex with recombinant Arabidopsis manganese-stabilizing
protein, Plant Mol. Biol., 26, 117-130.
60.Enami, I., Yoshihara, S., Tohri, A., Okumura, A.,
Ohta, H., and Shen, J. R. (2000) Cross-reconstitution of various
extrinsic proteins and photosystem II complexes from cyanobacteria, red
algae and higher plants, Plant Cell Physiol., 41,
1354-1364.
61.Seidler, A. (1996) Intermolecular and
intramolecular interactions of the 33-kDa protein in photosystem II,
Eur. J. Biochem., 242, 485-490.
62.Popelkova, H., Im, M. M., D’Auria, J.,
Betts, S. D., Lydakis-Simantiris, N., and Yocum, C. F. (2002)
N-terminus of the photosystem II manganese stabilizing protein: effects
of sequence elongation and truncation, Biochemistry, 41,
2702-2711.
63.Popelkova, H., Im, M. M., and Yocum, C. F. (2003)
Binding of manganese stabilizing protein to photosystem II:
identification of essential N-terminal threonine residues and domains
that prevent nonspecific binding, Biochemistry, 42,
6193-6200.
64.Burnap, R. L., Shen, J. R., Jursinic, P. A.,
Inoue, Y., and Sherman, L. A. (1992) Oxygen yield and
thermoluminescence characteristics of a cyanobacterium lacking the
manganese-stabilizing protein of photosystem-II, Biochemistry,
31, 7404-7410.
65.Summerfield, T. C., Crawford, T. S., Young, R.
D., Chua, J. P., Macdonald, R. L., Sherman, L. A., and Eaton-Rye, J. J.
(2013) Environmental pH affects photoautotrophic growth of
Synechocystis sp. PCC 6803 strains carrying mutations in the
lumenal proteins of PSII, Plant Cell Physiol., 54,
859-874.
66.Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and
Nekrasov, V. (2013) Plant genome editing made easy: targeted
mutagenesis in model and crop plants using the CRISPR/Cas system,
Plant Methods, 9, 39.
67.O’Malley, R. C., and Ecker, J. R. (2010)
Linking genotype to phenotype using the Arabidopsis unimutant
collection, Plant J., 61, 928-940.
68.Alonso, J. M., Stepanova, A. N., Leisse, T. J.,
Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J.,
Barajas, P., Cheuk, R., Gadrinab, C., Heller, C., Jeske, A., Koesema,
E., Meyers, C. C., Parker, H., Prednis, L., Ansari, Y., Choy, N., Deen,
H., Geralt, M., Hazari, N., Hom, E., Karnes, M., Mulholland, C.,
Ndubaku, R., Schmidt, I., Guzman, P., Aguilar-Henonin, L., Schmid, M.,
Weigel, D., Carter, D. E., Marchand, T., Risseeuw, E., Brogden, D.,
Zeko, A., Crosby, W. L., Berry, C. C., and Ecker, J. R. (2003)
Genome-wide insertional mutagenesis of Arabidopsis thaliana,
Science, 301, 653-657.
69.Cerutti, H., Ma, X., Msanne, J., and Repas, T.
(2011) RNA-mediated silencing in algae: biological roles and tools for
analysis of gene function, Eukaryot. Cell, 10,
1164-1172.
70.Murakami, R., Ifuku, K., Takabayashi, A.,
Shikanai, T., Endo, T., and Sato, F. (2002) Characterization of an
Arabidopsis thaliana mutant with impaired psbO, one of two genes
encoding extrinsic 33-kDa proteins in photosystem II, FEBS
Lett., 523, 138-142.
71.Murakami, R., Ifuku, K., Takabayashi, A.,
Shikanai, T., Endo, T., and Sato, F. (2005) Functional dissection of
two Arabidopsis PsbO proteins: PsbO1 and PsbO2, FEBS J.,
272, 2165-2175.
72.Lundin, B., Hansson, M., Schoefs, B., Vener, A.
V., and Spetea, C. (2007) The Arabidopsis PsbO2 protein
regulates dephosphorylation and turnover of the photosystem II reaction
centre D1 protein, Plant J., 49, 528-539.
73.Bricker, T. M., and Frankel, L. K. (2008) The
psbo1 mutant of Arabidopsis cannot efficiently use
calcium in support of oxygen evolution by photosystem II, J. Biol.
Chem., 283, 29022-29027.
74.Goulas, E., Schubert, M., Kieselbach, T.,
Kleczkowski, L. A., Gardestrom, P., Schroder, W. P., and Hurry, V.
(2006) The chloroplast lumen and stromal proteomes of Arabidopsis
thaliana show differential sensitivity to short- and long-term
exposure to low temperature, Plant J., 47, 720-734.
75.Rochaix, J. D. (1995) Chlamydomonas
reinhardtii as the photosynthetic yeast, Annu. Rev. Genet.,
29, 209-230.
76.Morais, F., Barber, J., and Nixon, P. (1998) The
chloroplast-encoded alpha subunit of cytochrome b-559 is
required for assembly of the photosystem two complex in both the light
and the dark in Chlamydomonas reinhardtii, J. Biol.
Chem., 273, 29315-29320.
77.Pigolev, A. V., Timoshevsky, D. S., and Klimov,
V. V. (2012) Effect of K223E and K226E amino acid substitutions in PsbO
protein of photosystem 2 on stability and functional activity of the
water-oxidizing complex in Chlamydomonas reinhardtii,
Biochemistry (Moscow), 77, 71-77.
78.Murray, J. W., and Barber, J. (2006)
Identification of a calcium-binding site in the PsbO protein of
photosystem II, Biochemistry, 45, 4128-4141.
79.Betts, S. D., Lydakis-Simantiris, N., Ross, J.
R., and Yocum, C. F. (1998) The carboxyl-terminal tripeptide of the
manganese-stabilizing protein is required for quantitative assembly
into photosystem II and for high rates of oxygen evolution activity,
Biochemistry, 40, 14230-14236.
80.Mei, R., and Yocum, C. (1993) Characterization of
inhibitory effects of NH2OH and its N-methyl derivatives on
the O2-evolving complex of photosystem II, Photosynth.
Res., 38, 449-453.
81.Ghanotakis, D. F., Topper, J. N., and Yocum, C.
F. (1984) Structural organization of the oxidizing side of photosystem
II. Exogenous reductants reduce and destroy the Mn-complex in
photosystem II membranes depleted of the 17 and 23 kDa
polypeptides, BBA Bioenerg., 767, 524-531.
82.Kuntzleman, T., McCarrick, R., Penner-Hahn, J.
E., and Yocum, C. F. (2004) Probing reactive sites within the
photosystem II manganese cluster: evidence for separate populations of
manganese that differ in redox potential, Phys. Chem. Chem.
Phys., 6, 4897-4904.