Parallel G-Quadruplexes Formed by Guanine-Rich Microsatellite Repeats Inhibit Human Topoisomerase I
A. M. Ogloblina1, V. A. Bannikova2, A. N. Khristich2, T. S. Oretskaya2, M. G. Yakubovskaya1, and N. G. Dolinnaya2*
1Blokhin Russian Cancer Research Center, 115478 Moscow, Russia; fax: (495) 324-1205; E-mail: globbi@mail.ru2Faculty of Chemistry, Lomonosov Moscow State University, 119991 Moscow; Russia; fax: (495) 939-3181; E-mail: dolinnaya@hotmail.com
* To whom correspondence should be addressed.
Received December 15, 2014; Revision received February 16, 2015
Using UV and CD spectroscopy, we studied the thermodynamic stability and folding topology of G-quadruplexes (G4), formed by G-rich fragments in human microsatellites that differ in the number of guanosines within the repeating unit. The oligonucleotides d(GGGT)4 and d(GGT)4 were shown to form propeller-type parallel-stranded intramolecular G-quadruplexes. The G4 melting temperature is dramatically decreased (by more than 45°C) in the transition from the tri-G-tetrad to the bi-G-tetrad structure. d(GT)n-repeats do not form perfect G-quadruplexes (one-G-tetrad); folded G4-like conformation is not stable at room temperature and is not stabilized by monovalent metal ions. The minimum concentration of K+ that promotes quadruplex folding of d(GGT)4 was found to depend on the supporting Na+ concentration. It was demonstrated for the first time that the complementary regions flanking G4-motifs (as in d(CACTGG-CC-(GGGT)4-TA-CCAGTG)) cannot form a double helix in the case of a parallel G4 due to the steric remoteness, but instead destabilize the structure. Additionally, we investigated the effect of the described oligonucleotides on the activity of topoisomerase I, one of the key cell enzymes, with a focus on the relationship between the stability of the formed quadruplexes and the inhibition degree of the enzyme. The most active inhibitor with IC50 = 0.08 µM was the oligonucleotide d(CACTGG-CC-(GGGT)4-TA-CCAGTG), whose flanking G4-motif sequences reduced the extreme stability of G-quadruplex formed by d(GGGT)4.
KEY WORDS: G-quadruplexes, microsatellite repeats, stability and structure of G-quadruplexes, topoisomerase I inhibitorsDOI: 10.1134/S0006297915080088
Microsatellite DNA repeats ranging in size from two to six nucleotides per repeated unit are widespread within the eukaryotic genome. These noncoding DNA sequences were for a long time believed to be idle. Only two decades ago, data suggesting their special role in DNA metabolism and genome functioning appeared. It was established that the microsatellite repeats are hot points for homologous recombination, and they are regions of elevated genome instability [1]. These processes play an important role in multistep carcinogenesis and other pathological conditions [2]. One of the reasons for unusual properties of the microsatellite DNAs could be that they adopt noncanonical conformations. Uniform repeated nonrandom sequence is able to form a new structural motif due to synchronous local changes in DNA secondary structure, which is determined by special geometry of the stacking contacts. Indeed, all the known noncanonical forms of nucleic acids are made of sequences that possess symmetrical elements, repeats and other deviations from random distribution of the nucleotide residues. Our study is focused on several guanine-containing sequences – d(GGGT)4, d(GGT)4, and d(GT)n that mimic one chain of the microsatellite repeats and may potentially form G-quadruplex, one of the most amazing, and extensively studied alternative forms of DNA [3-5]. G-Quadruplexes (G4) are formed via intra- or intermolecular interactions in DNA or RNA molecules containing oligoG sequences (so-called G-tracts). The core of the quadruplex consists of two or more G-tetrads, in which four guanine residues from different G-tracts are combined by a system of Hoogsteen hydrogen bonds (Fig. 1). Stacking interactions of the G-tetrad planes form specific structure of G4. DNA and RNA quadruplexes are the only noncanonical structures the formation of which in the living cell was strictly proved [6-8]. Being structural elements of the genome, G-quadruplexes are recognized by numerous proteins and enzymes, and they affect the most important biological processes such as replication, chromosome end protection, transcription, translation, mutagenesis, and DNA recombination [9-13]. Besides, G-quadruplexes are considered as targets for low molecular weight ligands (potential antitumor agents) that influence gene expression [14, 15]. Moreover, oligonucleotide quadruplexes (DNA aptamers) introduced into the cell are potential medications as such [16, 17].
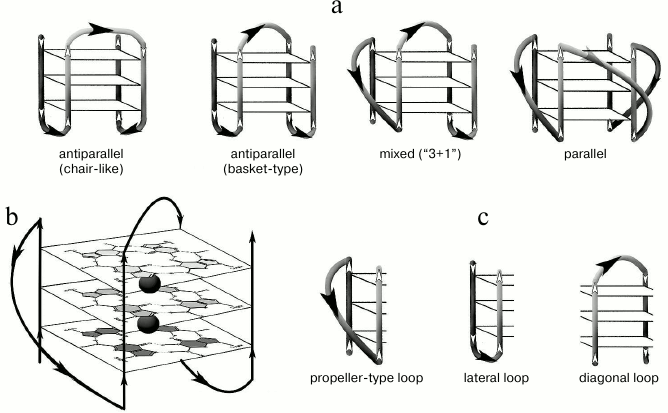
Fig. 1. Schematic view of structurally different quadruplexes. a) Intramolecular G4 that differ in strand orientation within the quadruplex core. b) Parallel G4, for which G-tetrad structure and potassium ion binding regions (black circles) are shown. c) Types of loops that join G-tracts in the quadruplex.
Structures of G-quadruplexes are extremely diverse. This is explained by such variables as quantity of the quadruplex-forming molecules (1, 2, or 4), G-tract length, strand mutual orientation [22], nucleotide sequence (composition) and size of the loops joining the G-tracts [23-25] (Fig. 1). All this hinders our understanding of functioning mechanisms of the G-quadruplexes in the genome and makes it necessary to study secondary structures formed by certain G4 motifs in detail.
In the present work, we described the structure and stability (in different experimental conditions) of G-quadruplexes formed by oligonucleotide models of microsatellites with tetra-, tri-, and dinucleotide repeats d(GGGT)4, d(GGT)4, and d(GT)n that differ by number of guanosine residues within the G-tract and number of repeated units. In addition, studies were made of the effect of characterized oligonucleotides on the activity of topoisomerase I, one of the key DNA metabolism enzymes. Because in the genome microsatellite repeats are flanked with irregular sequences, we also used oligonucleotides having two motifs: the repeated sequence to be analyzed and flanking duplex-forming fragments. UV and CD spectroscopies were used to dissect conformation potential of the oligonucleotides under study and to evaluate the thermodynamic stability of secondary structures formed by them. Ability of the oligonucleotides to inhibit conversion of plasmid DNA into the relaxed form by topoisomerase I was studied using electrophoresis techniques.
MATERIALS AND METHODS
Oligonucleotides. PAGE-purified oligodeoxyribonucleotides made by Syntol (Russia) were used in this work: d(GGGT)4 (1), d(CACTGG-CC-(GGGT)4-TA-CCAGTG) (2), d(GGT)4 (3), d(GT)15 (4), d(GT)16 (5), d(GT)31 (6), d(CACTGG-T-(GT)11-T-CCAGTG) (7), d(GCACTGG-T-(GT)12-T-CCAGTGC) (8), d(GCACTGG-T-(GT)13-T-CCAGTGC) (9), d(GCACTGG-T-(GT)14-T-CCAGTGC) (10), d(CACTGG-T-(GT)15-T-CCAGTG) (11), d(GCACTGG-T-(GT)15-T-CCAGTGC) (12), d(GCACTGG-T-(GT)16-T-CCAGTGC) (13), d(GCACTGG-CC-(GT)16-TA-CCAGTGC) (14), d(TAGTCTGGTACTGCATGCTATGCACG) (15). Concentrations of the oligonucleotides were measured spectrophotometrically using molar extinction coefficients calculated by the nearest neighbor algorithm. Oligonucleotides were dissolved in buffer A (20 mM HEPES-KOH, 140 mM NaCl, 5 mM KCl, pH 7.3). The solutions obtained were put into a water bath preheated to 95°C and slowly (during several hours) chilled down to the room temperature (annealing).
To assess the influence of monovalent metals ions on stability of the secondary structure formed by oligonucleotide d(GGT)4, three sets of solutions were made. Sample 1 of the first set was prepared by dissolving d(GGT)4 in buffer B (10 mM Tris-HCl, pH 7.2) at a final concentration 9.4·10–6 M in 400 µl of solution. This sample was used as a control containing no potassium ions. To make other solutions of this set, KCl (concentrated solution or dry powder) was added to the sample 1 to the final salt concentration 5-750 µM. A second set of solutions was prepared similarly in buffer C (10 mM Tris-HCl, pH 7.2, 20 mM NaCl). KCl concentration range was 4-546 mM. For the third set of d(GGT)4 solutions made in buffer D (20 mM HEPES, 120 mM NaCl, pH 7.3), KCl concentration range was 14-307 mM. In the latter two sets, NaCl concentrations were constant (20 and 120 mM for the second and the third sets, respectively) while KCl concentrations varied. Samples were annealed as described above.
UV spectroscopy. Melting curves of the oligonucleotide samples were registered in thermostatted quartz cuvettes (Hellma, Germany) with optical pathlength 10 or 1 mm using a double-beam Hitachi U-2900 UV/Visible spectrophotometer (Hitachi, Japan) equipped with an SPR-10 thermoelectric controller. Conformation changes were registered between 10 and 90°C at 295 or 260 nm. Heating rate was 0.5°C/min. Oligonucleotide concentrations varied in the range of 5.3-70 µM.
Circular dichroism. CD spectra were registered in 10 mm quartz cuvettes (Hellma) using a Chirascan spectrometer (Applied Photophysics Ltd, UK) equipped with a thermoelectric controller, between 7 and 85°C. Temperature was increased uniformly at 0.5°C/min rate. Measurements were taken every 3-8°C in the wavelength range 230-330 nm. Scanning speed was 1 nm/sec. To avoid water condensation on the cuvette surface, the cuvette chamber was blown with nitrogen. Spectrum of buffer A without oligonucleotide was taken as a baseline that was subtracted from the sample spectrum. CD values were presented as molecular dichroism Δε (cm–1·M–1) counting per oligonucleotide. The spectra were processed with the Origin 8.0 software using the Savitsky–Golay filter.
Studying the influence of guanine-rich oligonucleotides on topoisomerase I activity. Nuclear extract from the HeLa cells that contains topoisomerase I was used in the work. To prepare the extract, cells were double washed with PBS. Then, 3 ml of buffer E (10 mM HEPES, pH 7.6, 60 mM KCl, 1 mM EDTA, 1 mM DTT) and 0.1% NP-40 were added. The mixture was centrifuged for 5 min at 200g and 4°C. The sediment (cell nuclei) was suspended in 100 µl of buffer E and centrifuged under the same conditions. The sediment was mixed with double volume of buffer F (20 mM Tris-HCl, pH 8.0, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol) and 1/6.25 volume of 2.5 M NaCl. The mixture was thoroughly mixed and left on ice for 10 min. Then, the mixture was centrifuged at 16,000g and 4°C for 10 min. The supernatant (nuclear extract) was stored at –20°C. One activity unit of topoisomerase I was defined as nuclear extract concentration that converted 250 ng of supercoiled pUC19 plasmid to relaxed form during 30 min at 37°C [26]. The plasmid relaxation was monitored by electrophoresis in 1% agarose gel. Plasmid pUC19 was isolated from E. coli (strain XL1-Blue) using GeneJET plasmid miniprep kit (Fermentas, USA), according to the manufacturer’s manual. Cells were cultivated in the LB medium supplemented with 0.1 mg/ml ampicillin at 37°C with shaking (300 rpm) for 10 h.
To assess ability of oligonucleotides to affect activity of the topoisomerase I, 0.15 µg of supercoiled pUC19 plasmid DNA was mixed in buffer G (10 mM Tris-HCl, pH 7.9, 1 mM EDTA, 0.15 M NaCl, 0.1% BSA, 0.1 mM spermidine, 5% glycerol) with 1 unit of nuclear extract of HeLa cells and the studied oligonucleotides at different concentrations. Reaction was performed at 37°C for 30 min, then it was stopped by adding SDS to final concentration 1% and treating with proteinase K (50 µg/ml) for 30-60 min at 55°C. Reaction products were separated by electrophoresis in 1% agarose gel at electric field strength 2 V/cm in standard Tris-borate buffer at room temperature. The gels were stained in water solution of ethidium bromide (0.5 µg/ml). DNA quantity in the bands was measured by fluorescence in the transmitted UV light with wavelength 240-360 nm. The enzyme inhibition extent was calculated using the equation:
(S – S0)/(Scontrol – S0) × 100%, (1)
where S was an amount of supercoiled plasmid after treatment with topoisomerase I in the presence of oligonucleotide, Scontrol was an amount of supercoiled plasmid without enzyme treatment, S0 was an amount of supercoiled plasmid after topoisomerase I treatment in the absence of oligonucleotide inhibitor.
Specific topoisomerase I inhibitor camptothecin at concentration range 0.1-10 µM was used as a positive control.
RESULTS
Analysis of the G-quadruplexes formed by oligonucleotide d(GGGT)4 and its derivative, in which G4-motif is flanked by mutually complementary sequences. To characterize secondary structures formed by a set of the oligonucleotide models, the combination of UV and CD spectroscopies was utilized. UV-spectroscopy allows independent monitoring of denaturation of G-quadruplexes and DNA duplexes, while CD gives an opportunity to determine the quadruplex topology [27, 28]. The pattern of temperature dependence of UV absorbance at 295-297 nm is a marker of G-quadruplex structure. In distinction from DNA duplex, whose melting is accompanied by hyperchromic effect (usually at 260 nm), breakdown of G-quadruplex causes a decrease in optical density of the solution, and this is a cooperative process. Furthermore, denaturation of DNA duplex that may coexist with the G4-structure does not contribute to the G-quadruplex melting profile at this wavelength [29].
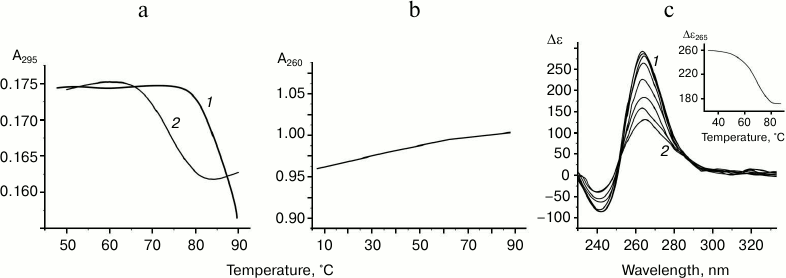
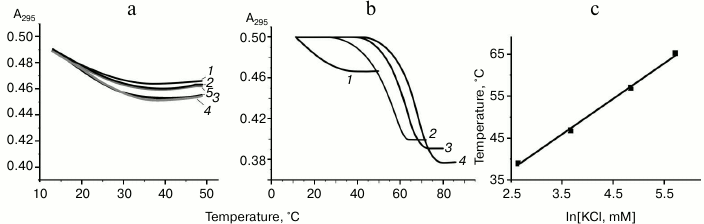
The oligonucleotide d(GGGT)4 UV-melting curve (at 295 nm) in buffer A containing K+ and Na+ shows a cooperative helix-coil transition that corresponds to the G-quadruplex denaturation (Fig. 2a). Hypochromic effect is more than 10%. At the same time, the thermodynamic stability of the structure is so high (Tmelt is over 85°C) that it is not possible to register its full denaturation profile. The melting curve at 295 nm for the d(CACTGG-CC-(GGGT)4-TA-CCAGTG), where mutually complementary hexanucleotide fragments were connected to the 5′- and 3′-ends of (GGGT)-repeat through the short linkers, suggests that this oligonucleotide also forms G4. However, its Tmelt (73°C) is more than 12°C lower than that of the non-flanked analog d(GGGT)4 (Fig. 2a). Moreover, the value of the hypochromic effect that accompanies the G-quadruplex denaturation in this case is only 7%. Thus, it is significantly lower than value shown for the d(GGGT)4 oligonucleotide. This effect is explained by the fact that 16 nucleotide residues (out of 32) in the d(CACTGG-CC-(GGGT)4-TA-CCAGTG) do not contribute to the G4 melting, though they absorb UV light. Destabilizing effect of the oligonucleotide fragments that flank (GGGT)-repeat could be due to that these sequences do not hybridize during the G-quadruplex formation, which is energetically unfavorable. Indeed, d(CACTGG-CC-(GGGT)4-TA-CCAGTG) melting leads to negligible non-cooperative optical density increase at 260 nm suggesting that duplex structure is absent (Fig. 2b).
Fig. 2. Thermodynamic stability and topology of tri-tetrad G4. a) Curves of temperature dependence of UV-absorbance (UV-melting curves) at 295 nm for oligonucleotides d(GGGT)4 (1) and d(CACTGG-CC-(GGGT)4-TA-CCAGTG) (2). b) UV-melting curve of d(CACTGG-CC-(GGGT)4-TA-CCAGTG) at 260 nm. c) CD spectra of d(CACTGG-CC-(GGGT)4-TA-CCAGTG) were measured at different temperatures from 37 (1) to 83°C (2); intermediate temperatures that correspond to CD spectra in the direction of the positive band amplitude decrease were 45, 53, 61, 69, 75, and 79°C. Insert: oligonucleotide melting profile obtained from CD data at 265 nm. All the measurements were taken in buffer A.
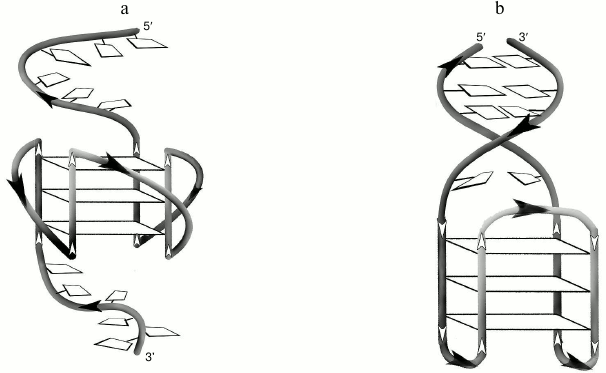
Secondary structure generated during intramolecular folding of the d(CACTGG-CC-(GGGT)4-TA-CCAGTG), which we suggest, is shown in the Fig. 3a. One can see that in case of parallel orientation of the strands in the G-quadruplex structure, the G4-flanking complementary sequences cannot form double helix due to their spatial remoteness. Thus, these sequences only destabilize intramolecular parallel G-quadruplex. For comparison, structure of the antiparallel G4, which coexist with a duplex domain made of G4-flanking sequences, is shown in the Fig. 3b. CD spectral data registered at different temperatures allowed building a melting curve for the d(CACTGG-CC-(GGGT)4-TA-CCAGTG) (Fig. 2c, insert) that corresponds well to the curve of temperature dependence of the UV absorbance at 295 nm. The presence of isodichroic points in a set of CD spectra registered at different temperatures suggests that the secondary structure generated by the (GGGT)4 and d(CACTGG-CC-(GGGT)4-TA-CCAGTG) oligonucleotides breaks up according to the all-or-none model that excludes the presence of intermediate structures.
Fig. 3. a) Putative secondary structure formed during intramolecular folding of d(CACTGG-CC-(GGGT)4-TA-CCAGTG). b) For comparison, structure of the antiparallel G4 that coexists with the duplex domain formed as a result of hybridization of sequences flanking G4-motif is shown.
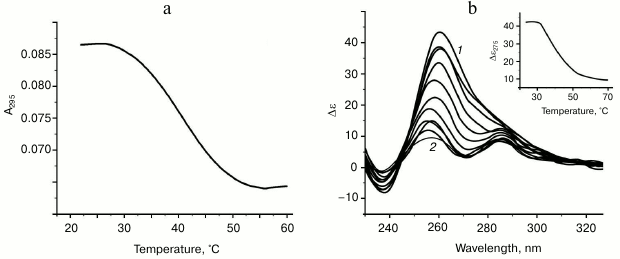
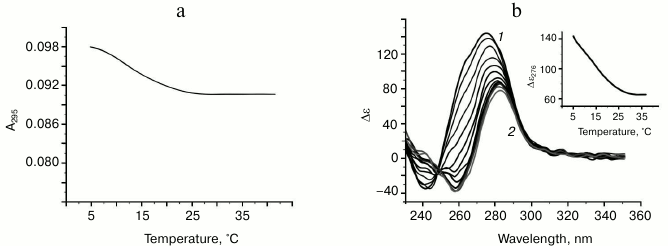
Characteristic of G-quadruplex formed by oligonucleotide d(GGT)4; influence of monovalent metal ions on secondary structure stability. The d(GGT/ACC) microsatellite repeat is the most frequently occurring trinucleotide repeat in the human genome [30]. Nevertheless, experimental studies of the conformation potential of the d(GGT)n have not been performed yet. Our data on the UV-melting of the d(GGT)4 at 295 nm suggest that it forms G-quadruplex with Tmelt 40°C in buffer A (Fig. 4a). Though the melting curve corresponded to cooperative structural transition, value of the hypochromic effect was largely dependent on the sample preparation conditions (annealing time, sample incubation time at low temperature, concentration of the oligonucleotide) varying between 11 and 22%. This indicates the presence of several quadruplex conformations that slowly turn to each other. This suggestion is supported by the CD data. According to the spectrum, parallel G4-topology prevails, but the shoulder at 287 nm evidences possible presence of the other quadruplex conformations (Fig. 4b). The absence of distinct isodichroic points in a set of the CD spectra registered at different temperatures confirms that the d(GGT)4 oligonucleotide exists in several structural forms.
Fig. 4. Thermodynamic stability and topology of bi-tetrad G4 formed by oligonucleotide d(GGT)4. a) UV-melting curve at 295 nm. b) CD spectra measured at different temperatures, starting from 24 (1) and finishing at 69°C (2); intermediate temperatures that correspond to CD spectra in the direction of the positive band amplitude decrease were 29, 34, 39, 44, 49, 54, 59, 64°C. Insert: oligonucleotide melting profile obtained from CD data at 275 nm. All the measurements were taken in buffer A.
It is known that parallel G4 tend to stick together by means of stacking interactions between the terminal G-tetrads [31, 32]. However, under conditions of our experiment the quadruplex dimerization did not occur. The absence of dependence of melting temperature on the oligonucleotide concentration in the 5.3-70 µM range speaks in favor of intramolecular complex formation in case of this oligonucleotide.
To study the influence of monovalent metal ions on the formation and stability of parallel G-quadruplexes with only two G-tetrads, we registered the melting curves for the d(GGT)4 oligonucleotide at 295 nm in buffers B, C, and D that have different concentrations of K+ and Na+. The shape of the melting curves obtained in buffer B that contains no monovalent metal ions or only contains potassium ions in the range 5-750 µM suggests that the G-quadruplex is only partially assembled (Fig. 5a). Even if the quadruplex structures appeared, they were unstable at room temperature. It is worth mentioning that in this set Tmelt was estimated approximately, as we did not observe the low temperature plateau; conventional temperature was 18°C, irrespectively to the solution composition, and hypochromic effect did not exceed 6-8%. Unlike buffer B, buffer C solutions contained 20 mM NaCl, and concentration of KCl varied from 4 to 546 mM. It was shown that only at 103 mM K+, significant changes in the shape of the melting curves were registered; hypochromic effect increased to 20% and Tmelt value reached 55°C. Further increase in KCl concentration caused quadruplex stabilization (Fig. 5b). For comparison, superposition of the UV-melting curves for oligonucleotide solutions in buffer C, where potassium ions concentrations varied (before and after the threshold value), is shown in the Fig. 5b. To determine the role of Na+ in the quadruplex structure stabilization, in the next experimental set we increased NaCl concentration to 120 mM, which remained constant for this set (buffer D). Raising NaCl concentration made it possible to form G4 already at 14 mM K+, i.e. seven times lower than in case of buffer C, where NaCl concentration was 20 mM. Further addition of KCl up to 307 mM led to quadruplex structure stabilization. In Fig. 5c, linear dependence Tmelt–ln[K+] for samples in the buffer D containing 120 mM NaCl is shown.
Fig. 5. Influence of the monovalent metal ions on formation of bi-tetrad G4 by the oligonucleotide d(GGT)4. a) Normalized to the equal initial optical density UV-melting curves (at 295 nm) of oligonucleotide solutions in buffer B without sodium ions but containing different K+ concentrations: 0 (1), 5 (2), 30 (3), 150 (4), 750 µM (5). b) Normalized to the equal initial optical density UV-melting curves (at 295 nm) of oligonucleotide solutions in buffer C containing 20 mM Na+ and different concentrations of K+: 36 (1), 103 (2), 258 (3), 546 mM (4). c) Dependence of Tmelt of G-quadruplexes on ln[K+] in buffer D containing 120 mM NaCl.
Complex formation in case of oligonucleotides containing (GT) repeat. A set of synthesized compounds (4-14) contained oligonucleotides with very different number of dinucleotide repeats (from 11 to 31). Some oligonucleotides possessed mutually complementary 6-7-nucleotide sequences connected to the 5′- and 3′-ends of d(GT)n through different mono(di)nucleotide linkers. These systems were designed to determine which factors can affect generation of the secondary structure by repeating (GT)n sequence and, in general, whether it is possible to form these structures in the conditions that, as it was shown above, favor intramolecular assembly of the parallel G-quadruplexes by the (GGT) and (GGGT) repeats. Curves of temperature dependence of the UV absorbance at 295 nm for the d(GT)n and its analogs in buffer A demonstrate a moderate hypochromic effect of 6-7% in case of oligonucleotides that contain no GT-repeat flanking sequences, and 3-4% in case of the ones having such sequences. This fact is of great importance, as it indicates that decrease in the sample optical density during heating is related to the conformational changes in the GT-repeats, while the presence of flanking sequences just decreases this effect. For example, a melting curve for d(GT)16 oligonucleotide is shown in the Fig. 6a. For comparison, heating of the unstructured random sequence oligomer (15) caused marginal non-cooperative increase in optical density at 295 nm due to weakening of base stacking with the temperature increase (data not shown). Corresponding to the weakly-cooperative conformational change, melting curves for d(GT)n are similar to the profiles of the temperature dependence of UV-absorbance at 295 nm for the d(GGT)4 oligonucleotide in buffer B containing no monovalent metal ions or containing K+ at very low concentrations (to 750 µM) (compare Figs. 5a and 6a). One can assume that (GT)n repeats form partially assembled quadruplex structures that are not stabilized by K+ and Na+ present at physiological concentrations, in contrast to the systems with two (d(GGT)4) and three (d(GGGT)4) tetrads at the quadruplex core. Apparently, the presence of thymine residues impedes formation of adequate stacking contacts (at the distance 3.3-3.4 Å) between the G-tetrads and retention of potassium ions that are unable, due to spatial remoteness even if present in the quadruplex core, to form coordinate bonds with oxo groups of guanines of the neighboring G-tetrads and stabilize four-helical structure. Low cooperativity of melting and the absence of dependence of the conventional Tmelt (12°C) on the number of dinucleotide repeats support correctness of the stated supposition. Furthermore, increase of K+ concentration to 100 mM did not increase stability of the secondary structure generated by d(GT)n. Analysis of temperature dependence of UV absorbance at 260 nm for these oligonucleotides revealed that complementary d(GT)n-flanking sequences do not generate duplex structures (S-shaped helix-coil transition is missing). According to the CD spectra registered at different temperatures, strands are arranged in parallel in the putative G-quadruplexes for all the compounds having dinucleotide repeats (Fig. 6b). At the same time, it is worth mentioning that the maximum of the positive band for structured oligomers does not correspond to 260 nm, but is long-wavelength shifted, and bathochromic shift of the CD spectra further increases with temperature. This behavior is unusual for standard G-qudruplexes.
Fig. 6. Analysis of the oligonucleotide d(GT)16 secondary structure. a) UV-melting curve at 295 nm. b) CD spectra measured at different temperatures, starting from 5 (1) and finishing at 38°C (2); intermediate temperatures that correspond to CD spectra in the direction of the positive band amplitude decrease were 9, 11, 14, 17, 20, 23, 26, 29, 32, 35°C. Insert: oligonucleotide melting profile obtained from the CD data at 276 nm. All the measurements were taken in buffer A.
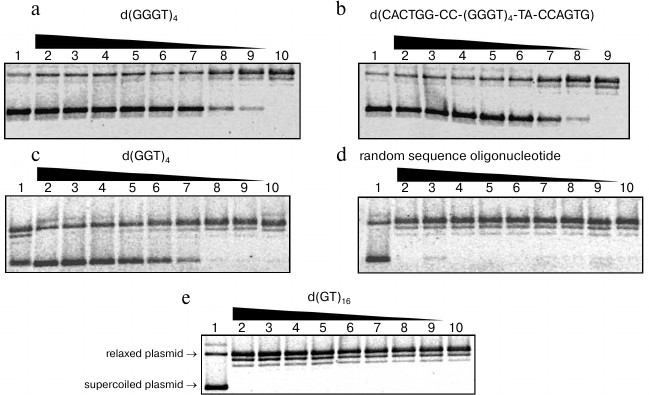
Effect of G-quadruplex-forming oligonucleotides on the topoisomerase I activity. We studied influence of guanine-rich di-, tri-, and tetranucleotide repeats on the ability of topoisomerase I to relax pUC19 plasmid DNA via reversible cleavage of one strand of the double helix. We also analyzed the relation between stability of G-quadruplex formed by repeated sequence and its inhibiting capability. To do so, we added different quantities of the characterized oligonucleotides to the reaction mixtures containing nuclear extract from HeLa cells, and topoisomerase I substrate. Plasmid pUC19 relaxation was monitored using agarose gel electrophoresis, as electrophoretic mobility of the supercoiled topoisomer is significantly higher than that of relaxed or linear forms. At first, the obtained nuclear extract from HeLa cells was checked for topoisomerase activity using highly specific enzyme inhibitor camptothecin [33]. The data confirmed that supercoiled plasmid relaxation is caused by the topoisomerase I and not by some other nuclear protein. The topoisomerase I inhibition rate in the presence of G4-motif-containing oligonucleotides was calculated using Eq. (1). Oligonucleotides d(GGGT)4, d(CACTGG-CC-(GGGT)4-TA-CCAGTG), d(GGT)4, d(GT)16, and other oligomers containing GT-repeats were added to the final concentrations 0.05-12.5 µM. Random sequence oligomer (15) that is unable to form quadruplex structure was used as a control. The d(GGGT)4 and d(CACTGG-CC-(GGGT)4-TA-CCAGTG) oligonucleotides possessed the strongest inhibitory activity. They started to affect topoisomerase I ability to decrease DNA supercoil density already at 0.05 µM (Fig. 7, a and b). The d(GGT)4 oligonucleotide appeared to be less efficient; to reach the same inhibition levels, one had to add 10 times more of this oligonucleotide than d(GGGT)4 (Fig. 7c). Addition of the control oligomer (15) did not affect topoisomerase activity; even at 12.5 µM concentration, it did not inhibit relaxation of supercoiled plasmid DNA (Fig. 7d). Topoisomerase activity was not inhibited by the oligonucleotides containing GT-repeats (Fig. 7e). The number of dinucleotide repeats and the presence of the GT-motif-flanking sequences did not affect the results. The revealed regularities corresponded well to our data regarding stability of secondary structures generated by d(GT)n. Even if G-quadruplex was partially folded during sample chilling, its Tmelt was significantly lower than the temperature at which we studied topoisomerase I activity (37°C).
Fig. 7. Relaxation of supercoiled plasmid pUC19 induced by topoisomerase I in the presence of oligonucleotides: a) d(GGGT)4; b) d(CACTGG-CC-(GGGT)4-TA-CCAGTG); c) d(GGT)4; d) random sequence oligonucleotide; e) d(GT)16. Electrophoregram of enzymatic reaction products in 1% agarose gel; oligonucleotide concentrations (µM) were 12.5 (2), 10 (3), 5 (4), 2.5 (5), 1.0 (6), 0.5 (7), 0.1 (8), 0.05 (9), 0 (10). Lane 1 corresponds to the reaction mixture containing no enzyme. For visualization, gels were stained with ethidium bromide. Positions of supercoiled and relaxed plasmids are indicated on the left side of the panel (e).
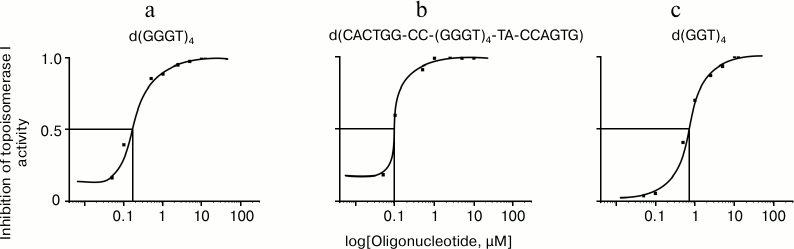
To quantify the inhibitory effect of the G-quadruplex structures, the electrophoregrams were processed using ImageJ software (National Institutes of Health, USA). Based on the curves of dependence of enzyme inhibition activities on logarithm of the oligonucleotide concentration in the reaction mixture, the half-maximal inhibitory concentrations (IC50) for G4-forming oligomers were calculated: 0.63 ± 0.06, 0.12 ± 0.02, and 0.08 ± 0.01 µM for d(GGT)4, d(GGGT)4, and d(CACTGG-CC-(GGGT)4-TA-CCAGTG), respectively (Fig. 8). It is seen that the d(CACTGG-CC-(GGGT)4-TA-CCAGTG) oligonucleotide is the strongest topoisomerase inhibitor, its melting temperature being 12°C lower than Tmelt of non-substituted d(GGGT)4.
Fig. 8. Inhibition of topoisomerase I activity by oligonucleotides: a) d(GGGT)4; b) d(CACTGG-CC-(GGGT)4-TA-CCAGTG); c) d(GGT)4. Curves of the relationship between enzymatic activity inhibition rate and the logarithm of concentration of the oligonucleotides are built according to the electrophoresis data (Fig. 7) using ImageJ software.
DISCUSSION
Using physicochemical methods, we demonstrated that d(GGGT)4 and d(GGT)4 oligonucleotides mimicking one strand of the microsatellite repeats generate in buffer A propeller-type parallel-stranded intramolecular G-quadruplexes with Tmelt more than 85 and 40°C, respectively. The pattern of temperature dependence of the UV absorbance at 295 nm (hypochromic effect occurrence) was an indication of a quadruplex structure. Absence of Tmelt dependence on concentration of the oligonucleotide indicated intramolecular complex formation in case of d(GGGT)4 and d(GGT)4. Parallel topology of formed G4 was proven using CD spectroscopy. Our data do not contradict the literature data, which suggest that intramolecular quadruplexes with three mononucleotide loops possess parallel topology irrespective of nucleotide composition of the loops [34, 35]. It is worth mentioning that published data regarding structure of the G4 formed by d(GGGT)4 were contradictory for a long time. It was described as an antiparallel quadruplex stabilized by two G•G•G•G tetrads and one G•T•G•T tetrad [36]. Only recently, using a molecular dynamics method [37], NMR-spectroscopy [31], thermodynamic analysis [38], and fluorescence [39], it was shown that the d(GGGT)4 sequence considered as an aptamer for binding to HIV-1 integrase folds into a parallel G4. Extremely high thermodynamic stability that preserves the quadruplex structure even in the presence of the complementary strand is a hallmark of this quadruplex [39].
In a 2014 work [40], it was pointed out that addition of two nucleotide residues GC to the 5′-end of d(GGGTGGGTGGGTGGG) does not affect the parallel topology of the formed G4, but causes its destabilization. In our work, hexanucleotide complementary regions were attached to both ends of the analogous sequence through the short linkers. For the first time, it was shown that duplex domain formation does not occur in this case, and G4-motif-flanking regions significantly (by more than 12°C) destabilize the quadruplex structure. The observed effect is caused by steric incompatibility of duplex domain with a parallel quadruplex structure: because of its formation, duplex-forming strands are separated in space and cannot hybridize (Fig. 3a). In contrast to the d(CACTGG-CC-(GGGT)4-TA-CCAGTG), during intramolecular folding of the thrombin-binding aptamer d(CACTGGTAGGTTGGTGTGGTTGGGGCCAGTG) an antiparallel G-quadruplex is formed that coexists with DNA duplex. At that, both structural domains significantly influence each other [29].
Secondary structure generated by the d(GGT)4 oligonucleotide is poorly studied. Earlier, it was described as self-associate that is only formed in the presence of potassium and magnesium ions [41]. In a recent work, d(GGT)4 sequence was a part of an oligonucleotide (antitumor aptamer), in which it was connected with four TGG-repeats through dinucleotide linker [42]. Using NMR-spectroscopy, CD, and chromatographic methods, the authors showed that this oligonucleotide folds into eight monomeric parallel quadruplex structures that are in equilibrium with each other. These observations do not contradict our results, according to which d(GGT)4 forms several different G4 with parallel topology and all of them were products of intramolecular folding. It is worth mentioning that stable quadruplexes having two G-tetrads usually possess antiparallel topology, for example, G4 formed by thrombin-binding aptamers [29, 43] or some telomere repeats [44]. Thus, characterized in our work bi-G-tetrad parallel quadruplexes with Tmelt 40°C are of interest. It should be mentioned that parallel G4 formed by extended microsatellite loci could form the pearl-necklace monomorphic G4 in the G-rich strand as it was stated for minisatellite repeats [45]. In our work, using UV melting, minimal quantity of monovalent ions was determined that is required for full folding of the d(GGT)4 oligonucleotide into a parallel quadruplex. It is known that G-quadruplexes are sensitive to the kind and concentration of cations present in the environment [46]. Monovalent metal ions cause the most stabilizing effect, especially K+, as its size allows it to move into G4 cavity and to be located between neighboring G-tetrads [35, 47].
In the present work, in three sets of experiments concentration of potassium ions varied significantly, while sodium ion concentration was constant and comprised 0, 20, and 120 mM for different experimental sets. It was shown that assembly of perfect G4 does not occur if the buffer solution only contains K+ in the 5-750 µM concentration range. Under the experimental conditions, DNA carries a charge of –1 per each internucleotide phosphate residue, i.e. the concentration of negative charge on the d(GGT)4 oligonucleotide (at 9.4·10–6 M concentration) was approximately 103 µM. It is obvious that volume concentration of ions that did not exceed 750 µM was not high enough for shielding of the phosphate group charges and for specific binding in the quadruplex cavity. However, formation of partially assembled G4 due to propensity of G-tracts to form G-tetrads was registered at low temperatures even in the absence of metal ions (Fig. 5a); this phenomenon was also observed during antiparallel bi-G-tetrad quadruplex assembly [48]. Using sets of d(GGT)4 solutions in buffers C and D with constant concentrations of sodium ions that participate in shielding of the charge of the phosphate groups, we have shown that threshold concentrations of K+ that facilitate intramolecular folding of the oligonucleotides into the G-quadruplex depend on Na+ concentration: 103 mM K+ in the presence of 20 mM NaCl and 14 mM K+ in the presence of 120 mM NaCl. Further increase in K+ concentration caused stabilization of the G4 (Fig. 5, b and c).
Secondary structures generated by the repeated GT sequence-containing oligonucleotides were studied in works by Kaluzhny and collaborators [49, 50]. According to them, oligonucleotides fold into unstable quadruplex structures, and ability to form G4 was shown to depend on the length of the repeat. However, CD and fluorescence data did not allow making unequivocal conclusions about existence of quadruplexes, in particular, for d(GT)12 oligonucleotide. In our work, it was shown that oligodeoxyribonucleotides containing GT-repeats do not form perfect G-quadruplexes. Stability of the partially assembled G4-like structures did not depend on the dinucleotide repeat length, presence of the flanking sequences, and concentration of monovalent ions. Absence of inhibitory effect of these oligonucleotides on topoisomerase I also suggests that they do not generate stable quadruplexes under enzymatic reaction conditions, and topoisomerase does not initiate assembly of such structures.
It is known that numerous cellular proteins specifically recognize G-quadruplexes [4] causing their unwinding [51-53], stabilization [54-57], or even initiation of the quadruplex assembly (in this case proteins act as molecular chaperons) [48, 58]. We focused on topoisomerase I, which is a widespread multifunctional enzyme that regulates DNA topology during transcription, replication, chromosome condensation, and recombination by means of reversible cleavage of one DNA duplex strand and relaxation of supercoiled DNA [59]. It was shown that topoisomerase I is one of the important intracellular targets for antitumor drugs [21]. Topoisomerase I can recognize and bind to certain non-canonical DNA forms – Holiday junctions, DNA triplexes, G-quadruplexes [19, 20, 60, 61]. In particular, affinity of the enzyme to single strand oligonucleotides that form G4 is much higher than affinity to random oligonucleotides [19, 20, 62]. It was shown that inter- and intramolecular G4 can inhibit topoisomerase I-mediated DNA cleavage [19]. This effect was also observed for the oligonucleotides having repeated G-tracts, but unable to form stable quadruplex structures [19, 20]. Moreover, as it was shown, topoisomerase I is able to induce assembly of intermolecular parallel G4 by oligonucleotides containing five successive guanosines [62]. However, a set of published DNA and RNA models was random, and antiparallel quadruplexes prevailed among intramolecular G4. For the first time, we correlated stability of the parallel G4 generated by microsatellite repeats and their ability to inhibit supercoiled plasmid DNA relaxation by topoisomerase I.
We have shown strong inhibitory effect of parallel G-quadruplexes formed by the oligonucleotides d(GGT)4 and d(GGGT)4 on topoisomerase I activity. At that, the inhibitory effect appeared at much lower concentrations of the quadruplex “traps” (nano(micro)molar) than for previously studied G4 [20] (Figs. 7 and 8). The ability of the d(GGT)4 and d(GGGT)4 oligonucleotides to inhibit topoisomerase I activity correlated well with stability of the corresponding G-quadruplexes and with number of the guanine residues in the repeat. In work [62], where influence of the d(GGGGGT)4 and d(GGGT)4 oligonucleotides on the topoisomerase activity was dissected, it was also shown that the longer G-tract in the repeated sequence, the stronger the inhibitory effect. However, comparison of our data with literature data was complicated, because in [62] type and stability of G4 was not addressed and G4 structure formed by d(GGGT)4 was mistakenly described as antiparallel. Furthermore, approaches used to estimate topoisomerase I activity in our work and in [62] were different.
Comparative study of d(GGGT)4 and its derivative having hexanucleotide fragments at the 5′- and 3′-ends of the G4-motif revealed that the d(CACTGG-CC-(GGGT)4-TA-CCAGTG) inhibits topoisomerase more efficiently, though Tmelt of its G4 is significantly lower than Tmelt of the d(GGGT)4-generated quadruplex. This apparent discrepancy might be explained by increased dynamic flexibility of the system, which contains quadruplex-destabilizing elements, allowing the enzyme to adjust its structure to the oligonucleotide ligand-binding site features more easily than the structure of extremely stable d(GGGT)4. In work [20], it was shown that G4-motifs that are flanked with duplex-generating regions possess higher inhibition ability, and the strongest inhibitory effect was observed for the duplex domains containing 6 bp. However, G4 in this work possessed antiparallel topology allowing formation of the duplex that was able to support quadruplex structure (Fig. 3b). Therefore, reasons for the elevated inhibitory effect of such composite oligonucleotides might differ from the ones stated by us.
As it was shown, oligonucleotides containing GT-repeats did not inhibit topoisomerase I irrespective of their length and presence of flanking fragments. They behaved as a control random oligomer (Fig. 7, d and e). Apparently, the enzyme did not recognize partially formed G4 possessing low thermodynamic stability and did not facilitate conformational shift to the quadruplex structures as it was seen for oligonucleotides containing several successive guanine residues [62].
Thus, it was shown that d(GGGT)4 and d(GGT)4 specifically bind to topoisomerase I and inhibit supercoiled DNA relaxation. At that, extent of influence of the oligonucleotides on activity of the topoisomerase correlated with stability of the corresponding G-quadruplexes. However, in case of greatly stable G4 generated by d(GGGT)4, factors that decrease thermodynamic stability of the structure, increase inhibitory activity of the ligand. Thus, of the studied set of oligonucleotides, the d(CACTGG-CC-(GGGT)4-TA-CCAGTG), which has G4-motif flanking sequences that do not hybridize in case of parallel topology of quadruplexes, appeared to be the strongest inhibitor. When the number of guanosine residues in the repeats was decreased to one (d(GT)n), imperfect (because of alternate thymidine residues interference) G-quadruplexes with low thermodynamic stability were not recognized by topoisomerase I, and the presence of the enzyme did not shift equilibrium to the formation of complete quadruplex conformation by the d(GT)n oligonucleotides.
This work was supported by RFBR-DFG grant 14-04-91343.
REFERENCES
1.Brandstrom, M., Bagshaw, A. T., Gemmell, N. J., and
Ellegren, H. (2008) The relationship between microsatellite
polymorphism and recombination hot spots in the human genome, Mol.
Biol. Evol., 25, 2579-2587.
2.Boland, C. R., and Goel, A. (2010) Microsatellite
instability in colorectal cancer, Gastroenterology, 138,
2073-2087.
3.Neidle, S. (2009) The structures of quadruplex
nucleic acids and their drug complexes, Curr. Opin. Struct.
Biol., 19, 239-250.
4.Sissi, C., Gatto, B., and Palumbo, M. (2011) The
evolving world of protein-G-quadruplex recognition: a medicinal
chemist’s perspective, Biochimie, 93,
1219-1230.
5.Wu, Y., and Brosh, R. M., Jr. (2010) G-Quadruplex
nucleic acids and human disease, FEBS J., 277,
3470-3488.
6.Biffi, G., Tannahill, D., McCafferty, J., and
Balasubramanian, S. (2013) Quantitative visualization of DNA
G-quadruplex structures in human cells, Nat. Chem., 5,
182-186.
7.Henderson, A., Wu, Y., Huang, Y. C., Chavez, E. A.,
Platt, J., Johnson, F. B., Brosh, R. M., Jr., Sen, D., and Lansdorp, P.
M. (2014) Detection of G-quadruplex DNA in mammalian cells, Nucleic
Acids Res., 42, 860-869.
8.Biffi, G., Di Antonio, M., Tannahill, D., and
Balasubramanian, S. (2014) Visualization and selective chemical
targeting of RNA G-quadruplex structures in the cytoplasm of human
cells, Nat. Chem., 6, 75-80.
9.Hershman, S. G., Chen, Q., Lee, J. Y., Kozak, M.
L., Yue, P., Wang, L. S., and Johnson, F. B. (2008) Genomic
distribution and functional analyses G-quadruplex-forming sequences in
Saccharomyces cerevisiae, Nucleic Acids Res., 36,
144-156.
10.Mani, P., Yadav, V. K., Das, S. K., and
Chowdhury, S. (2009) Genome-wide analyses of recombination prone
regions predict role of DNA structural motif in recombination, PLos
One, 4, e4399.
11.Verma, A., Yadav, V. K., Basundra, R., Kumar, A.,
and Chowdhury, S. (2009) Evidence of genome-wide G4
DNA-mediated gene expression in human cancer cells, Nucleic Acids
Res., 37, 4104-4204.
12.Lansdorp, P. M. (2005) Major cutbacks at
chromosome ends, Trends Biochem. Sci., 30, 388-395.
13.Tarsounas, M., and Tijsterman, M. (2013) Genomes
and G-quadruplexes: for better or for worse, J. Mol. Biol.,
425, 4782-4789.
14.Balasubramanian, S., and Neidle, S. (2009)
G-quadruplex nucleic acids as therapeutic targets, Curr. Opin. Chem.
Biol., 13, 345-353.
15.McLuckie, K. I. E., Di Antonio, M., Zecchini, H.,
Xian, J., Caldas, C., Krippendorff, B.-F., Tannahill, D., Lowe, C., and
Balasubramanian, S. (2013) G-quadruplex DNA as a molecular target for
induced synthetic lethality in cancer cells, J. Am. Chem. Soc.,
135, 9640-9643.
16.De Soultrait, V. R., Lozach, P. Y., Altmeyer, R.,
Tarrago-Litvak, L., Litvak, S., and Andreola, M. L. (2002) DNA aptamers
derived from HIV-1 RNase H inhibitors are strong anti-integrase agents,
J. Mol. Biol., 324, 195-203.
17.Teng, Y., Girvan, A. C., Casson, L. K., Pierce,
W. M., Jr., Qian, M., Thomas, S. D., and Bates, P. J. (2007) AS1411
alters the localization of a complex containing protein arginine
methyltransferase 5 and nucleolin, Cancer Res., 67,
10491-10500.
18.Sun, D., and Hurley, L. H. (2009) The importance
of negative superhelicity in inducing the formation of G-quadruplex and
i-motif structures in the c-Myc promoter: implications for drug
targeting and control of gene expression, J. Med. Chem.,
52, 2863-2874.
19.Marchand, C., Pourquier, P., Laco, G. S., Jing,
N., and Pommier, Y. (2002) Interaction of human nuclear topoisomerase I
with guanosine quartet-forming and guanosine-rich single-stranded DNA
and RNA oligonucleotides, J. Biol. Chem., 277,
8906-8911.
20.Shuai, L., Deng, M., Zhang, D., Zhou, Y., and
Zhou, X. (2010) Quadruplex-duplex motifs as new topoisomerase I
inhibitors, Nucleosides, Nucleotides Nucleic Acids, 29,
841-853.
21.Moukharskaya, J., and Verschraegen, C. (2012)
Topoisomerase I inhibitors and cancer therapy, Hematol. Oncol. Clin.
North Am., 26, 507-525.
22.Rachwal, P. A., Brown, T., and Fox, K. R. (2007)
Effect of G-tract length on the topology and stability of
intramolecular DNA quadruplexes, Biochemistry, 46,
3036-3044.
23.Rachwal, P. A., Brown, T., and Fox, K. R. (2007)
Sequence effects of single base loops in intramolecular quadruplex DNA,
FEBS Lett., 581, 1657-1660.
24.Rachwal, P. A., Findlow, I. S., Werner, J. M.,
Brown, T., and Fox, K. R. (2007) Intramolecular DNA quadruplexes with
different arrangements of short and long loops, Nucleic Acids
Res., 35, 4214-4222.
25.Guedin, A., Gros, J., Alberti, P., and Mergny,
J.-L. (2010) How long is too long? Effects of loop size on G-quadruplex
stability, Nucleic Acids Res., 38, 7858-7868.
26.Nitiss, J. L., Soans, E., Rogojina, A., Seth, A.,
and Mishina, M. (2012) Topoisomerase assays: author’s manual,
Curr. Protoc. Pharmacol., Chap. 3, Unit 3.3; doi:
10.1002/0471141755.ph0303s57.
27.Karsisiotis, A. I., Hessari, N. M., Novellino,
E., Spada, G. P., Randazzo, A., and Webba da Silva, M. (2011)
Topological characterization of nucleic acid G-quadruplexes by UV
absorption and circular dichroism, Angew. Chem., 50,
10645-10648.
28.Vorlickova, M., Kejnovska, I., Sagi, J., Renciuk,
D., Bednarova, K., Motlova, J., and Kupr, J. (2012) Circular dichroism
and guanine quadruplexes, Methods, 57, 64-75.
29.Dolinnaya, N. G., Yuminova, A. V., Spiridonova,
V. A., Arutyunyan, A. M., and Kopylov, A. M. (2012) Coexistence of
G-quadruplex and duplex domains within the secondary structure of
31-mer DNA thrombin-binding aptamer, J. Biomol. Struct. Dyn.,
30, 524-531.
30.Benson, G. (1999) Tandem repeats finder: a
program to analyze DNA sequences, Nucleic Acids Res., 27,
573-580.
31.Do, N. Q., Lim, K. W., Teo, M. H., Heddi, B., and
Phan, A. T. (2011) Stacking of G-quadruplexes: NMR structure of a
G-rich oligonucleotide with potential anti-HIV and anticancer activity,
Nucleic Acids Res., 39, 9448-9457.
32.Krishnan-Ghosh, Y., Liu, D., and Balasubramanian,
S. (2004) Formation of an interlocked quadruplex dimer by d(GGGT),
J. Am. Chem. Soc., 126, 11009-11016.
33.Coletta, A., and Desideri, A. (2013) Role of the
protein in the DNA sequence specificity of the cleavage site stabilized
by the camptothecin topoisomerase IB inhibitors: a metadynamics study,
Nucleic Acids Res., 41, 9977-9986.
34.Guedin, A., De Cian, A., Gros, J., Lacroix, L.,
and Mergny, J.-L. (2008) Sequence effects in single-base loops for
quadruplexes, Biochimie, 90, 686-696.
35.Bugaut, A., and Balasubramanian, S. (2008) A
sequence-independent study of the influence of short loop lengths on
the stability and topology of intramolecular DNA G-quadruplexes,
Biochemistry, 47, 689-697.
36.Jing, N., Gao, X., Rando, R. F., and Hogan, M. E.
(1997) Potassium-induced loop conformational transition of a potent
anti-HIV oligonucleotide, J. Biomol. Struct. Dyn., 15,
573-585.
37.Li, M. H., Zhou, Y. H., Luo, Q., and Li, Z. S.
(2010) The 3D structures of G-quadruplexes of HIV-1 integrase
inhibitors: molecular dynamics simulations in aqueous solution and in
the gas phase, J. Mol. Model., 16, 645-657.
38.Kelley, S., Boroda, S., Musier-Forsyth, K., and
Kankia, B. I. (2011) HIV-integrase aptamer folds into a parallel
quadruplex: a thermodynamic study, Biophys. Chem., 155,
82-88.
39.Johnson, J., Okyere, R., Joseph, A.,
Musier-Forsyth, K., and Kankia, B. (2013) Quadruplex formation as a
molecular switch to turn on intrinsically fluorescent nucleotide
analogs, Nucleic Acids Res., 41, 220-228.
40.Mathias, J., Okyere, R., Lomidze, L.,
Gvarjaladze, D., Musier-Forsyth, K., and Kankia, B. (2014) Thermal
stability of quadruplex primers for highly versatile isothermal DNA
amplification, Biophys. Chem., 185, 14-18.
41.Chen, F. M. (1997) Supramolecular self-assembly
of d(TGG)4, synergistic effects of K+ and
Mg2+, Biophys. J., 73, 348-356.
42.Dailey, M. M., Miller, M. C., Bates, P. J., Lane,
A. N., and Trent, J. O. (2010) Resolution and characterization of the
structural polymorphism of a single quadruplex-forming sequence,
Nucleic Acids Res., 38, 4877-4888.
43.Kelly, J. A., Feigon, J., and Yeates, T. O.
(1996) Reconciliation of the X-ray and NMR structures of the
thrombin-binding aptamer d(GGTTGGTGTGGTTGG), J. Mol. Biol.,
256, 417-422.
44.Hansel, R., Lohr, F., Trantirek, L., and Dotsch,
V. (2013) High-resolution insight into G-overhang architecture, J.
Am. Chem. Soc., 135, 2816-2824.
45.Amrane, S., Adrian, M., Heddi, B., Serero, A.,
Nicolas, A., Mergny, J. L., and Phan, A. T. (2012) Formation of
pearl-necklace monomorphic G-quadruplexes in the human CEB25
minisatellite, J. Am. Chem. Soc., 134, 5807-5816.
46.Gaynutdinov, T. I., Neumann, R. D., and Panyutin,
I. G. (2008) Structural polymorphism of intramolecular quadruplex of
human telomeric DNA: effect of cations, quadruplex-binding drugs and
flanking sequences, Nucleic Acids Res., 36,
4079-4087.
47.Zhang, A. Y. Q., Bugaut, A., and Balasubramanian,
S. (2011) A sequence-independent analysis of the loop length dependence
of intramolecular RNA G-quadruplex stability and topology,
Biochemistry, 50, 7251-7258.
48.Nagatoishi, S., Tanaka, Y., and Tsumoto, K.
(2007) Circular dichroism spectra demonstrate formation of the
thrombin-binding DNA aptamer G-quadruplex under
stabilizing-cation-deficient conditions, Biochem. Biophys. Res.
Commun., 352, 812-817.
49.Kaliuzhnyi, D. N., Bondarev, F. S., Shchelkina,
A. K., Livshits, M. A., and Borisova, O. F. (2008) Intramolecular
G-quadruplexes from microsatellite d(GT)12 sequence in the
presence of K+, Mol. Biol. (Moscow), 42,
693-700.
50.Kaluzhny, D., Shchyolkina, A., Livshits, M.,
Lysov, Y., and Borisova, O. (2009) A novel intramolecular
G-quartet-containing fold of single-stranded d(GT)8 and
d(GT)16 oligonucleotides, Biophys. Chem., 143,
161-165.
51.Salas, T. R., Petruseva, I., Lavrik, O.,
Bourdoncle, A., Mergny, J. L., Favre, A., and Saintome, C. (2006) Human
replication protein A unfolds telomeric G-quadruplexes, Nucleic
Acids Res., 34, 4857-4865.
52.Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr.,
Bohr, V. A., and Hickson, I. D. (2001) The Bloom’s and
Werner’s syndrome proteins are DNA structure-specific helicases,
Nucleic Acids Res., 29, 2843-2849.
53.Cogoi, S., Shchekotikhin, A. E., and Xodo, L. E.
(2014) HRAS is silenced by two neighboring G-quadruplexes and activated
by MAZ, a zinc-finger transcription factor with DNA unfolding property,
Nucleic Acids Res., 42, 8379-8388.
54.Giraldo, R., Suzuki, M., Chapman, L., and Rhodes,
D. (1994) Promotion of parallel DNA quadruplexes by a yeast telomere
binding protein: a circular dichroism study, Proc. Natl. Acad. Sci.
USA, 91, 7658-7562.
55.Jing, N., Marchand, C., Liu, J., Mitra, R.,
Hogan, M. E., and Pommier, Y. (2000) Mechanism of inhibition of HIV-1
integrase by G-tetrad-forming oligonucleotides in vitro, J.
Biol. Chem., 275, 21460-21467.
56.Paeschke, K., Simonsson, T., Postberg, J.,
Rhodes, D., and Lipps, H. J. (2005) Telomere end-binding proteins
control the formation of G-quadruplex DNA structures in vivo,
Nat. Struct. Mol. Biol., 12, 847-854.
57.Kang, H. J., Le, T. V., Kim, K., Hur, J., Kim, K.
K., and Park, H. J. (2014) Novel interaction of the Z-DNA binding
domain of human ADAR1 with the oncogenic c-Myc promoter G-quadruplex,
J. Mol. Biol., 426, 2594-2604.
58.Baldrich, E., and O’Sullivan, C. K. (2005)
Ability of thrombin to act as molecular chaperone, inducing formation
of quadruplex structure of thrombin-binding aptamer, Anal.
Biochem., 341, 194-197.
59.Gupta, M., Fujimori, A., and Pommier, Y. (1995)
Eukaryotic DNA topoisomerases I, Biochim. Biophys. Acta,
1262, 1-14.
60.Sekiguchi, J., Cheng, C., and Shuman, S. (2000)
Resolution of a Holliday junction by vaccinia topoisomerase requires a
spacer DNA segment 3′ of the CCCTT↓ cleavage sites,
Nucleic Acids Res., 28, 2658-2663.
61.Arimondo, P. B., Moreau, P., Boutorine, A.,
Bailly, C., Prudhomme, M., Sun, J. S., Garestier, T., and Helene, C.
(2000) Recognition and cleavage of DNA by rebeccamycin- or
benzopyridoquinoxaline conjugated of triple helix-forming
oligonucleotides, Bioorg. Med. Chem., 8, 777-784.
62.Arimondo, P. B., Riou, J. F., Mergny, J. L.,
Tazi, J., Sun, J. S., Garestier, T., and Helene, C. (2000) Interaction
of human DNA topoisomerase I with G-quartet structures, Nucleic
Acids Res., 28, 4832-4838.