Double Subgenomic Promoter Control for a Target Gene Superexpression by a Plant Viral Vector
E. V. Putlyaev1#*, A. A. Smirnov1#, O. V. Karpova1, and J. G. Atabekov1,2
1Faculty of Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; fax: +7 (495) 938-0601; E-mail: putlegor@mail.ru; putlegor@gmail.com2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; fax: +7 (495) 938-3181; E-mail: okar@genebee.msu.ru
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received January 30, 2015; Revision received March 6, 2015
Several new deconstructed vectors based on a potexvirus genome sequence for efficient expression of heterologous proteins in plants were designed. The first obtained vector (AltMV-single), based on the Alternanthera mosaic virus (AltMV) strain MU genome, bears a typical architecture for deconstructed plant viral vectors, i.e. a triple gene block was deleted from the viral genome and the model gene of interest was placed under control of the first viral subgenomic promoter. To enhance the efficiency of expression, maintained by the AltMV-single, another vector (AltMV-double) was designed. In AltMV-double, the gene of interest was controlled by two viral subgenomic promoters located sequentially without a gap upstream of the target gene. It was found that AltMV-double provided a significantly higher level of accumulation of the target protein in plants than AltMV-single. Moreover, our data clearly show the requirement of the presence and functioning of both the subgenomic promoters for demonstrated high level of target protein expression by AltMV-double. Taken together, our results describe an additional possible way to enhance the efficiency of transient protein expression maintained in plants by a plant viral vector.
KEY WORDS: Alternanthera mosaic virus, viral vector, protein overexpression, subgenomic promoterDOI: 10.1134/S000629791508009X
Abbreviations: AltMV, Alternanthera mosaic virus; CP, capsid protein; d.p.i., days post-injection; f.l.t., fresh leaf tissue; hG-CSF, human granulocyte colony-stimulating factor; ORF, open reading frame; sgp, subgenomic promoter; sgRNA, subgenomic RNA; TGB, triple gene block.
A variety of technologies has been developed for production of foreign
proteins in plants, including enzymes, vaccine proteins, cytokines, and
other pharmaceuticals [1-5].
The technologies based on plants as factories for production of foreign
proteins provided several advantages including high yield, low cost,
and biosafety of the product. These technologies are safe, since plants
and animals share no common pathogens [5]. To
increase the level of the recombinant protein production, the transient
expression strategy of a foreign gene is frequently used. This strategy
implies delivery of a transgene into the plant cells by a viral vector.
Therefore, further increase in expression efficiency of plant viral
vectors is an important goal [4].
Members of the Potexvirus genus, a group of plant-infecting viruses with an RNA genome packed into flexuous rod-shaped virions [6, 7], are some of the most usually applied for construction of expression vectors among other groups of plant viruses. The genomic RNA of potexviruses is single-stranded, has positive polarity, and normally encodes five translated open reading frames (ORFs). ORF1 encodes viral RNA-dependent RNA-polymerase (replicase) and is translated from the genomic RNA itself. The other four ORFs are translated from three special subgenomic RNAs (sgRNAs) that are synthetized under control of three subgenomic promoters (sgp). ORFs 2, 3, and 4 are usually referred to as a “triple gene block” (TGB) [8], and the viral proteins they encode are called pTGB1 (translated from sgRNA1), pTGB2, and pTGB3 (both translated from sgRNA2), correspondingly. These proteins, due to their dispensability for viral replication and evident requirement for short and long distance transport of infection, are usually called “transport proteins”, though their functions appear to be more complex [9, 10]. ORF5 encodes a capsid protein (CP), the most abundant of all viral proteins (translated from sgRNA3).
During the last two decades, numerous strategies for construction of vectors based on genomes of the plant viruses with RNA genome have been developed [5]. One of the most efficient among them is the so-called “deconstructed virus” strategy (see for review [4, 5]). This concept of vector engineering implies an agrobacterium-mediated vector delivery process, e.g. agroinjection [5], which provides up to 100% efficiency level of vector delivery of cells in the agrobacterium-treated zone of plant tissue. This peculiarity allows the deletion of all viral genes responsible for cell-to-cell and systemic movement of the wild-type virus from the resulting vector. Such a reduction helps to overcome certain restrictions on size of the inserted target gene and also cuts the competition process for cell resources between an overexpressed target gene and other viral genes not involved into replication.
Recently, a new strain of Alternanthera mosaic virus (AltMV-MU, GenBank FJ822136.1), a member of the family Alphaflexiviridae, Potexvirus genus [11, 12], was described in our laboratory [13]. The AltMV-MU genomic RNA molecule (Fig. 1a) is 6606-nt-long and has an organization typical for the genus [13]. Previously, another strain of AltMV (AltMV-SP) was utilized for construction of a bipartite vector for efficient target protein expression [14]. In our current work, we have elaborated an additional improvement for a potexvirus-based “deconstructed virus” engineering strategy, namely a technique of a multiple subgenomic promoter control of a target gene, resulting in a higher yield of target protein. Several deconstructed vectors based on the AltMV-MU genome were designed to study this novel method, and a significant yield of a foreign protein was achieved in Nicotiana benthamiana plants by one of these newly constructed vectors.
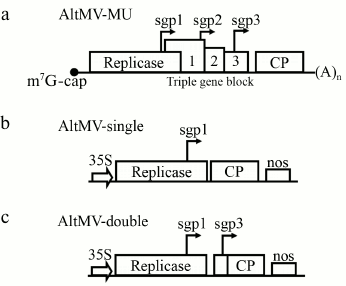
Fig. 1. Schematic presentation of the native AltMV-MU genome and AltMV-MU-derived vectors: a) AltMV-MU genome; b) AltMV-single; c) AltMV-double. sgp 1, 2, and 3 are putative subgenomic promoters; 35S – cauliflower mosaic virus 35S promoter; nos – nopaline synthase terminator; (A)n – poly(A).
MATERIALS AND METHODS
Construction of AltMV-based expression vectors. The AltMV-MU genome cDNA copy [13] was used for creation of all vectors presented in this article. The full cDNA copy of the AltMV-MU genome was previously obtained as a number of overlapping fragments, cloned into pBluescript SK+ plasmids: pAltMV19 (1-1639 bp); pAltMV12 (1292-3446 bp); pAltMV46 (3279-4488 bp); pAltMV33 (4335-5559 bp); pAltMV50 (5479-6606 bp). Sequence numbers of the first and the last bp of the corresponding fragment in the AltMV-MU full genome sequence (1-6606 bp, GenBank FJ822136.1) are parenthesized. pPVX201 [15] was used as a template for amplification of the cauliflower mosaic virus 35S promoter (35S) and nopaline-synthase terminator (nos) sequences by PCR reactions.
All preliminary DNA fragments utilized for construction of AltMV-MU vectors are listed in Table 1 with all corresponding specific components of reaction, i.e. specific DNA oligonucleotides for PCR reactions and specific restriction endonucleases. Sequences of all DNA specific oligonucleotides utilized for PCRs described in our work are listed in Table 2.
Table 1. Schematically presented steps of
obtaining of preliminary DNA fragments used in construction of AltMV-MU
based vectors

Table 2. List of names and corresponding
nucleotide sequences of DNA specific oligonucleotides used in PCR
described in the present work

To obtain the AltMV-single (Fig. 1b) construct, a fragment of the AltMV-MU genome from 4725 to 5762 bp was deleted, and the translation-initiating codon of movement protein 1 was mutated from ATG to ACG. This mutation was performed to prevent the possible initiation of translation at this starting point. The resulting sequence was cloned to pCambia1300 plasmid together with previously obtained 35S- and nos-sequences as shown on Fig. 1b. To reach this goal, four DNA fragments were cloned into XbaI-EcoRI region of pCambia1300 plasmid: Subcl1 cleaved by XbaI and SphI; Subcl2 cleaved by SphI and BamHI; PCR6 cleaved by BamHI and KpnI; Restr3. To obtain Subcl1 construct, pSL1180 plasmid was cleaved by XbaI and SphI and ligated with Restr1, Restr2, and PCR3 cleaved by XbaI and EcoRI. Another construct, Subcl2, was obtained by ligation of pSL1180 plasmid (cleaved by SphI and SpeI) with Restr4, -5, -6, -7 fragments.
The AltMV-double construct was also derived from the AltMV-MU genome sequence by deleting a fragment from 4725 to 5762 bp, mutation of transport protein 1 translation initiating codon (ATG to ACG), and cloning to pCambia1300 plasmid with 35S- and nos-sequences as described for AltMV-single. To obtain AltMV-double, the following fragments were cloned into the XbaI-EcoRI region of pCambia1300 plasmid: Subcl1 cleaved by XbaI and SphI; Subcl2 cleaved by SphI and BamHI; PCR9 cleaved by BamHI and KpnI; Restr3.
The AltMV-double* was derived from AltMV-double by a G(4683)A PCR-substitution, i.e. PCR11 was cleaved by ApaI and SpeI enzyme and then cloned into the ApaI-SpeI region of the AltMV-double construct. The purpose of the nucleotide substitution G(4683)A was to abolish the transcription activity of sgp1.
For construction of a AltMV-double-based vector for expression of human granulocyte colony-stimulating factor (AltMV-d-gcsf and hG-CSF, correspondingly), a fragment of nucleotide sequence of AltMV-double from 5961 to 6382 bp was substituted with ORF, the combination of a hg-csf gene sequence and the 3′-terminal tag, encoding six histidine residues and an enterokinase cleavage site [16], attached to the 3′-end of the target gene. The six histidine-tagged hg-csf ORF was obtained from pGEM3Z-hG-CSF bearing the corresponding gene and previously used in the work of Zvereva et al. [16]. According to Komarova et al. [17], the 3′-terminal 100 bp of the CP ORF were left downstream of the G-CSF stop codon to enhance the viral vector expression. The translation initiating ATG codon of AltMV-CP ORF was substituted with ACG. The precise scheme of construction of AltMV-d-gcsf was performed in three steps. As a first step, a pAl3016 construct was obtained: PCRA cleaved by SpeI and SalI, and PCRB cleaved by SalI and KpnI were cloned into the KpnI-SpeI region of pSL1180 plasmid. Next, pAl3017 was obtained by cloning of Restr9 and Restr11 fragments together into the AhlI and KpnI region of pAl3016 construct. Finally, to produce AltMV-d-gcsf, PCR12 was cloned into the AscII-AvrII region of pAl3017.
Agroinjection of constructed vectors into plant leaves. Agroinjection of the viral vectors into Nicotiana benthamiana leaves was performed as described in [16]. Strain GV3101 of the A. tumefaciens was transformed by the obtained constructs. Transformed bacteria were incubated overnight at 26°C in 2YT medium with the following antibiotics: rifampicin (50 mg/liter), kanamycin (50 mg/liter), and gentamycin (25 mg/liter). The overnight culture was centrifuged for 5 min at 5000g and then resuspended in agroinjection buffer (10 mM MgSO4, 10 mM MES-NaOH, pH 5.5) to resulting A600 = 0.2. To enhance the target protein yield, all studied vectors were agroinjected together with a vector, expressing the tombusvirus p19 protein – a suppressor of gene silencing [18]. The bacterial suspensions were injected into N. benthamiana leaves using a syringe without a needle.
AltMV-MU capsid protein extraction from agroinjected leaves. The leaves agroinjected with AltMV-single, AltMV-double, or AltMV-double* were collected on different days post agroinjection (d.p.i.). Agroinjected leaves were homogenized in 10 mM Tris-HCl, pH 8.0, and centrifuged at 16,000g for 15 min. The supernatant (S16 fraction) was collected for further analysis.
Subgenomic promoter transcriptional activity assay. The formation of subgenomic RNAs in the agroinjected leaves was studied by 5′-Step-Out RACE RT-PCR. The N. benthamiana leaves transformed with viral vectors were collected on d.p.i. 3, frozen by liquid nitrogen, and homogenized to powder. Total RNA samples from the homogenized leaves were extracted by TRI REAGENT (Molecular Research Center, USA) according to the protocol provided by the manufacturer. To obtain cDNA copies of viral vector subgenomic RNAs, the isolated RNA samples were studied by the 5′-Step-Out RACE RT-PCR (Mint Universal cDNA Synthesis Kit; Evrogen, Russia) according to the manufacturer’s protocol with slight modifications. In particular, the first cDNA strand was synthesized using the AltMVCPrev primer (tgtgtcgactcagtgatggtgatggtgatgctccggtggtgggaggtattga), complementary to the 3′-end sequence of the AltMV-MU CP gene. The double-stranded cDNA was amplified using the AltMV CPrev primer and the PCR primer M1 (Evrogen). The products of the 5′-Step-Out RACE RT-PCR were analyzed by electrophoresis in 1% agarose gel. The separated products of the 5′-Step-Out RACE RT-PCR were extracted from the agarose gel, and their nucleotide sequence was then determined (Evrogen).
Extraction of recombinant hG-CSF from agroinjected plant leaves. To isolate the foreign protein tagged with six histidine residues, the samples of agroinjected leaves were frozen with liquid nitrogen and homogenized to powder. The samples were then suspended in buffer A (Qiagen, Germany) for 4 h at room temperature, and then centrifuged at 16,000g for 15 min at 15°C. Aliquots of the supernatant were collected for subsequent assessment of the amount of the target protein in unpurified samples. The remaining volume of the supernatant containing the tagged protein was purified on a QIA expressionist Ni-NTA agarose column according to manufacturer’s protocol (Qiagen). The elution fractions were collected for further analyses.
Analysis of recombinant hG-CSF protein yield. The samples of unpurified and roughly purified target protein obtained from agroinjected leaves were analyzed by SDS-PAGE with Coomassie Brilliant Blue G-250 staining. Digital high-resolution photographs of the stained gels were taken with the ChemiDoc™ XRS Plus gel recording system (Bio-Rad, USA). For calculation of relative amounts of protein in corresponding bands, the acquired digital images were analyzed using TotalLab Quant (TotalLab Ltd, UK) software.
RESULTS AND DISCUSSION
Expression of AltMV CP in plants by AltMV-double. As a general design for all new AltMV-MU-based vectors, the “deconstructed” type of architecture, lacking the genes of movement proteins (Fig. 1, a-c), was chosen due to its high biosafety [5]. Initially, the deconstructed viral vector named “AltMV-single” based on the AltMV-MU genome was created. In AltMV-single, the CP gene was placed under control of sgp1 (Fig. 1b). AltMV-single has an ordinary structure for a potexvirus deconstructed vector [17]. To obtain a higher expression level, the gene of interest was placed under control of two AltMV putative subgenomic promoters 1 and 3 (sgp1 and sgp3) (Fig. 1c), which were previously described by Lim and colleagues [14]. This second vector was designated as “AltMV-double”.
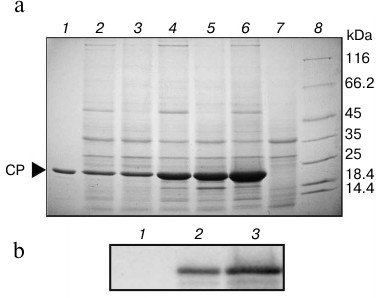
To assess the functionality of AltMV-double, both AltMV-double and AltMV-single were used for expression of the AltMV CP gene in leaves of N. benthamiana as a model gene of interest (Fig. 1, b and c). The AltMV CP accumulation was tested 8, 9, and 13 d.p.i. by SDS-PAGE (Fig. 2a) and by Western blot assay with antiserum to AltMV-MU CP (Fig. 2b). It was found that S16 fractions from leaves agroinjected with AltMV-double and AltMV-single contained a major protein band of the size expected for the AltMV-MU CP (22 kDa). The level of the CP gene expression by AltMV-double (Fig. 2a, lanes 4-6) was significantly higher than that of AltMV-single (Fig. 2a, lanes 2 and 3).
To calculate the AltMV CP yield in leaves agroinjected with AltMV-double, several units of serial dilutions of S16 fraction were examined by Coomassie-stained SDS-PAGE. According to our data, the AltMV CP yield provided by AltMV-double vector in N. benthamiana leaves ranged up to 5.5 mg per gram of fresh leaf tissue (f.l.t.). According to [4, 19], this amount represented over 50% of total soluble protein. The AltMV-MU yield from a mechanically inoculated plant was significantly lower (about 340 µg per gram, as reported by [20]). Thereby, the new approach implying the double subgenomic promoter control of a target gene was efficient for overexpression of the viral CP gene by plant viral vector in N. benthamiana.
Fig. 2. a) Time course of AltMV CP expression in Nicotiana benthamiana leaves agroinjected with AltMV-single and AltMV-double: 1) 1 µg of AltMV CP (positive control); S16 fraction from leaves agroinjected with: 2) AltMV-single, 8 d.p.i.; 3) AltMV-single, 9 d.p.i.; 4) AltMV-double, 8 d.p.i.; 5) AltMV-double, 9 d.p.i.; 6) AltMV-double, 13 d.p.i.; 7) P19 alone (negative control); 8) molecular weight markers in kDa (8-20% SDS-PAGE, Coomassie R-250 staining). b) Western blot analysis with antiserum to AltMV CP: 1) negative control – S16 fraction from leaves agroinjected with P19 alone; 2) positive control – 0.2 µg of AltMV CP; 3) S16 fraction from leaves agroinjected with AltMV-double.
Recently, Lim and colleagues [21] presented a novel vector system based on the AltMV genome with 35S and T7 promoters simultaneously functional in transcription of the Agrobacterium-delivered bipartite vector. This viral vector, though quite effective, relies on ectopic expression of T7 polymerase in addition to the AltMV-replicase, while AltMV-double expression requires the AltMV-replicase only. Wang and colleagues [22] recently presented a PVX-genome-based viral vector in which three different sgps of various origin (i.e. one sgp originating from TMV genome) were placed one after another and separated with the multi cloning site sequences cloned to the 3′-terminal region of the PVX genome. The construction (pCaPVX760-GFP) was regarded as a platform for multiple protein expression. Nevertheless, the above-mentioned feature of pCaPVX760-GFP did not look as a self-sufficient way to enhance expression of a target gene. It is also worth mentioning that no data was provided on the functionality of those three sgps, revealing whether they were cooperatively maintaining the target gene expression or not. Moreover, no quantitative estimation of target protein accumulation maintained by pCaPVX760-GFP was provided.
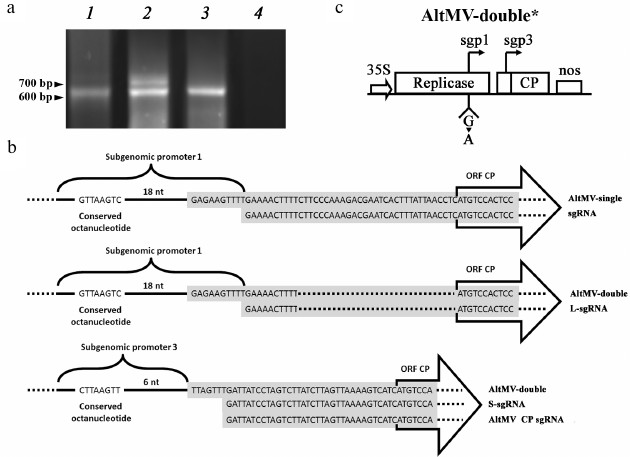
Both sgps of AltMV-double are functional. To reveal the reasons of the CP expression stimulation demonstrated by AltMV-double, a series of experiments was performed. The sgRNAs produced in plant cells during AltMV-single and AltMV-double multiplication were assayed by 5′-Step-Out RACE RT-PCR and subsequent sequencing of the obtained cDNA-copies of sgRNAs. The results shown in Fig. 3a indicated that AltMV-double simultaneously produced two sgRNAs of different length, unlike AltMV-single, that produced only a single sgRNA. Moreover, computer alignment of these RNA sequences showed (Fig. 3b) that the longer sgRNA (L-sgRNA, 740 bp) of AltMV-double had a transcription start at the putative sgp1 sequence, while transcription of the shorter one (S-sgRNA, 637 bp) started at the putative sgp3 sequence. These results imply that both putative sgp1 and sgp3 present in AltMV-double are functional.
Fig. 3. Analysis of translational activity of sgps in AltMV-double. a) Electrophoresis assay in 1% agarose of 5′-Step-Out RACE RT-PCR of RNA extracted from leaf material agroinjected with AltMV-single (1), AltMV-double (2), AltMV-double* (3), P19 as negative control (4). b) Alignment of sequenced products of 5′-Step-Out RACE RT of RNA samples from agroinjected leaves of N. benthamiana; gray box represent matching sequences of 5′-Step-Out RACE RT products; conserved octanucleotide sequence characteristic for potexvirus sgps [23] are shown. The fragments of the nucleotide sequences of AltMV-single and AltMV-double are aligned with the nucleotide sequences of cDNA-copies of the 5′-proximal regions of sgRNAs produced by these vectors during their multiplication in plant leaves. The AltMV-single nucleotide sequence is aligned with the sequence of the only sgRNA; this vector was shown to produce (upper scheme). The fragments of the AltMV-double sequence are aligned with the sequences of L- and S-sgRNA (central and lower schemes, correspondingly). Additionally, nucleotide sequence of the original AltMV sgRNA carrying the viral ORF5 alone is also added to the alignment (lower scheme) to denote its full identity with the nucleotide sequence of the S-sgRNA. c) Schematic representation of AltMV-double* construction denoting the mutated first nucleotide at the starting point of sgp1-controlled transcription.
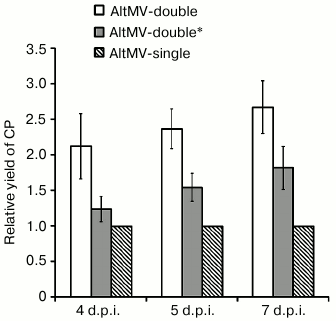
To study the requirement of two transcriptionally active subgenomic promoters for maintenance of the demonstrated high level of protein expression by AltMV-double, an additional genetic construction AltMV-double* was acquired by transcriptional inactivation of sgp1 in the sequence of AltMV-double (Fig. 3c). In AltMV-double*, the sgp1 transcriptional activity was ceased by replacement of the first guanylate residue of the L-sgRNA molecule by PCR similarly to how it was done previously for PVX in [24]. The 5′-RACE RT-PCR assay with subsequent sequencing of the PCR products showed that the resulting vector AltMV-double* produces only one, S-sgRNA product (Fig. 3a, lane 3). Comparison of the relative CP yields produced by AltMV-single, AltMV-double, and AltMV-double* constructs during 4, 5, and 7 d.p.i. (Fig. 4) showed that AltMV-double* provides a significantly higher level of expression on all three time intervals assayed than AltMV-single. At the same time, our data showed that AltMV-double* was as well significantly deficient in target protein accumulation compared to AltMV-double. These data imply that both sgps are required in their functional state to provide the high expression level of the target protein demonstrated by AltMV-double.
Fig. 4. Diagram representing the relative amounts of AltMV-MU CP accumulated by AltMV-single, AltMV-double, and AltMV-double* in agroinjected leaves of N. benthamiana and measured on 4, 5, and 7 d.p.i. The calculated yield of CP provided by AltMV-single on each analyzed day was taken as 1. Error bars represent 95% confidence intervals for the mean (n = 3).
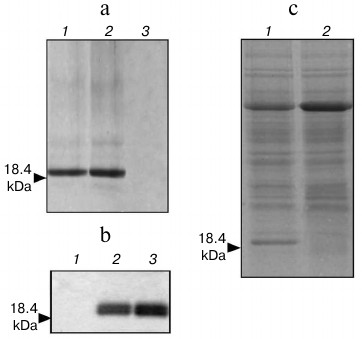
Expression of heterologous (human) protein in plants by AltMV-double. In the experiments presented above, AltMV-MU CP was used as a model protein convenient for comparison of expression efficiencies of AltMV-single and AltMV-double. Furthermore, the human granulocyte colony-stimulating factor (hG-CSF, also known as CSF3) was selected as a foreign (nonviral) protein to examine its expression in plants under control of AltMV-double vector. Recently, some vectors based on plant RNA viruses were used for expression of human cytokines in plants [16, 25]. We compared the published results with the yields of the human cytokine (hG-CSF) production in plants provided by AltMV-double. The new vector for hG-CSF in plant expression – AltMV-d-gcsf – was obtained as follows: hg-csf gene 3′-fused with the 6-His tag was cloned into the AltMV-double vector to substitute the CP gene. The hG-CSF was purified from the agroinjected leaves by immobilized metal-affinity chromatography (IMAC). Analyses of IMAC fractions by Coomassie-stained SDS-PAGE (Fig. 5a) and by Western blot with antibodies to bacterially expressed hG-CSF (Fig. 5b) revealed the presence of hG-CSF (18.4 kDa). The estimated yield of purified hG-CSF was calculated as much as 400 µg of target protein per gram f.l.t. To evaluate the efficiency of the purification process, the amount of the target protein was assayed in crude protein extracts from the agroinjected leaves (Fig. 5c). According to our results, the yield of unpurified hG-CSF was up to 590 µg per gram f.l.t. or up to 5.5% of total soluble protein. Therefore, the efficiency of purification process can be calculated as up to 73%.
Fig. 5. Analysis of hG-CSF expression in Nicotiana benthamiana leaves agroinjected with AltMV-d-gcsf. Recombinant proteins were purified from agroinjected leaves by Ni-NTA agarose IMAC and analyzed by Coomassie stained 8-20% SDS-PAGE (a): 1, 2) AltMV-d-gcsf agroinjected leaf; 3) P19 agroinjected leaf; and by Western blot assay with antiserum against E. coli-expressed hG-CSF (b): 1) P19 agroinjected leaf; 2) 0.4 µg of E. coli-expressed hG-CSF; 3) hG-CSF expressed by AltMV-d-gcsf. c) Analysis of hG-CSF amount in S16 fraction from Nicotiana benthamiana leaves agroinjected with AltMV-d-gcsf was performed by Coomassie stained 8-20% SDS-PAGE: 1) AltMV-d-gcsf agroinjected leaf; 2) P19 agroinjected leaf. Molecular mass of hG-CSF is shown by arrow.
To our knowledge, this was the highest yield of hG-CSF produced by a plant viral expression vector. The results presented in previous works were 100 µg of purified or 400 µg unrefined of hG-CSF per gram f.l.t. [16], and 19 mg of unpurified GM-CSF (granulocyte macrophage CSF) per gram of total soluble protein, that is approximately 160 µg of target protein per gram f.l.t. [25]. These data confirm that the AltMV-double is a powerful expression vector that allows reaching high levels of expression of heterologous proteins in plants. In our opinion, the technique of multiple subgenomic promoter control of the gene of interest, described in our work, is promising due to its simplicity. Yet, it is still to be studied whether this method could be applicable to subgenomic promoters of other plant viruses.
We are grateful to Dr. P. A. Ivanov for providing the cDNA copy of the AltMV-MU genome and the cDNA copy of the hG-CSF mRNA.
This work was funded by the Russian Science Foundation (grant No. 14-24-00007).
REFERENCES
1.Boehm, R. (2007) Bioproduction of therapeutic
proteins in the 21st century and the role of plants and plant cells as
production platforms, Ann. NY Acad. Sci., 1102,
121-134.
2.Larrick, J. W., and Thomas, D. W. (2001) Producing
proteins in transgenic plants and animals, Curr. Opin.
Biotechnol., 12, 411-418.
3.Mett, V., Farrance, C. E., Green, B. J., and
Yusibov, V. (2008) Plants as biofactories, Biologicals,
36, 354-358.
4.Gleba, Y. Y., Klimyuk, V., and Marillonnet, S.
(2007) Viral vectors for the expression of proteins in plants, Curr.
Opin. Biotechnol., 18, 134-141.
5.Gleba, Y. Y., Tuse, D., and Giritch, A. (2014)
Plant viral vectors for delivery by Agrobacterium, Curr. Top.
Microbiol. Immunol., 375, 155-192.
6.Verchot-Lubicz, J., Ye, C. M., and Bamunusinghe, D.
(2007) Molecular biology of potexviruses: recent advances, J. Gen.
Virol., 88, 1643-1655.
7.Park, M. R., Seo, J. K., and Kim, K. H. (2013)
Viral and nonviral elements in potexvirus replication and movement and
in antiviral responses, Adv. Virus Res., 87, 75-112.
8.Morozov, S. Y., and Solovyev, A. G. (2003) Triple
gene block: modular design of a multifunctional machine for plant virus
movement, J. Gen. Virol., 84, 1351-1366.
9.Tilsner, J., Linnik, O., Wright, K. M., Bell, K.,
Roberts, A. G., Lacomme, C., Santa Cruz, S., and Oparka, K. J. (2012)
The TGB1 movement protein of potato virus X reorganizes actin and
endomembranes into the X-body, a viral replication factory, Plant
Physiol., 158, 1359-1370.
10.Bayne, E. H., Rakitina, D. V., Morozov, S. Y.,
and Baulcombe, D. C. (2005) Cell-to-cell movement of potato potexvirus
X is dependent on suppression of RNA silencing, Plant J.,
44, 471-482.
11.Geering, A. D., and Thomas, J. E. (1999)
Characterization of a virus from Australia that is closely related to
papaya mosaic potexvirus, Arch. Virol., 144, 577-592.
12.Hammond, J., Reinsel, M. D., and Maroon-Lango, C.
J. (2006) Identification and full sequence of an isolate of
Alternanthera mosaic potexvirus infecting Phlox
stolonifera, Arch. Virol., 151, 477-493.
13.Ivanov, P. A., Mukhamedzhanova, A. A., Smirnov,
A. A., Rodionova, N. P., Karpova, O. V., and Atabekov, J. G. (2010) The
complete nucleotide sequence of Alternanthera mosaic virus
infecting Portulaca grandiflora represents a new strain distinct
from phlox isolates, Virus Genes, 42, 268-271.
14.Lim, H. S., Vaira, A. M., Reinsel, M. D., Bae,
H., Bailey, B. A., Domier, L. L., and Hammond, J. (2010) Pathogenicity
of Alternanthera mosaic virus is affected by determinants in
RNA-dependent RNA polymerase and by reduced efficacy of silencing
suppression in a movement-competent TGB1, J. Gen. Virol.,
91, 277-287.
15.Baulcombe, D. C., Chapman, S., and Santa-Cruz, S.
(1995) Jellyfish green fluorescent protein as a reporter for virus
infections, Plant J., 7, 1045-1053.
16.Zvereva, A. S., Petrovskaya, L. E., Rodina, A.
V., Frolova, O. Y., Ivanov, P. A., Shingarova, L. N., Komarova, T. V.,
Dorokhov, Y. L., Dolgikh, D. A., Kirpichnikov, M. P., and Atabekov, J.
G. (2009) Production of biologically active human myelocytokines in
plants, Biochemistry (Moscow), 74, 1187-1194.
17.Komarova, T. V., Skulachev, M. V., Zvereva, A.
S., Schwartz, A. M., Dorokhov, Yu. L., and Atabekov, J. G. (2006) New
viral vector for efficient production of target proteins in plants,
Biochemistry (Moscow), 71, 846-850.
18.Voinnet, O., Rivas, S., Mestre, P., and
Baulcombe, D. (2003) An enhanced transient expression system in plants
based on suppression of gene silencing by the p19 protein of tomato
bushy stunt virus, Plant J., 33, 949-956.
19.Marillonnet, S., Thoeringer, C., Kandzia, R.,
Klimyuk, V., and Gleba, Y. (2005) Systemic Agrobacterium
tumefaciens-mediated transfection of viral replicons for efficient
transient expression in plants, Nature Biotechnol., 23,
718-723.
20.Mukhamedzhanova, A. A., Smirnov, A. A.,
Arkhipenko, M. V., Ivanov, P. A., Chirkov, S. N., Rodionova, N. P.,
Karpova, O. V., and Atabekov, J. G. (2011) Characterization of
Alternanthera mosaic virus and its coat protein, Open Virol.
J., 5, 136-140.
21.Lim, H. S., Vaira, A. M., Domier, L. L., Lee, S.
C., Kim, H. G., and Hammond, J. (2010) Efficiency of VIGS and gene
expression in a novel bipartite potexvirus vector delivery system as a
function of strength of TGB1 silencing suppression, Virology,
402, 149-163.
22.Wang, Y., Cong, Q. Q., Lan, Y. F., Geng, C., Li,
X. D., Liang, Y. C., Yang, Z. Y., Zhu, X. P., and Li, X. D. (2014)
Development of new potato virus X-based vectors for gene
over-expression and gene silencing assay, Virus Res.,
191, 62-69.
23.Kim, K.-H., and Hemenway, C. L. (1999)
Long-distance RNA–RNA interactions and conserved sequence
elements affect potato virus X plus-strand RNA accumulation,
RNA, 5, 636-645.
24.Kim, K.-H., and Hemenway, C. (1997) Mutations
that alter a conserved element upstream of the potato virus X triple
block and coat protein genes affect subgenomic RNA accumulation,
Virology, 232, 187-197.
25.Zhou, F., Wang, M. L., Albert, H. H., Moore, P.
H., and Zhu, Y. J. (2006) Efficient transient expression of human
GM-CSF protein in Nicotiana benthamiana using potato virus X
vector, Appl. Microbiol. Biotechnol., 72, 756-762.