Evolutionary Divergence of Arabidopsis thaliana Classical Peroxidases
E. V. Kupriyanova*, P. O. Mamoshina, and T. A. Ezhova
Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: ekupriyanova@gmail.com* To whom correspondence should be addressed.
Received July 6, 2015
Polymorphisms of 62 peroxidase genes derived from Arabidopsis thaliana were investigated to evaluate evolutionary dynamics and divergence of peroxidase proteins. By comparing divergence of duplicated genes AtPrx53-AtPrx54 and AtPrx36-AtPrx72 and their products, nucleotide and amino acid substitutions were identified that were apparently targets of positive selection. These substitutions were detected among paralogs of 461 ecotypes from Arabidopsis thaliana. Some of these substitutions are conservative and matched paralogous peroxidases in other Brassicaceae species. These results suggest that after duplication, peroxidase genes evolved under the pressure of positive selection, and amino acid substitutions identified during our study provided divergence of properties and physiological functions in peroxidases. Our predictions regarding functional significance for amino acid residues identified in variable sites of peroxidases may allow further experimental assessment of evolution of peroxidases after gene duplication.
KEY WORDS: peroxidase, Arabidopsis thaliana, gene and protein polymorphism, gene duplication, evolution, divergence of protein properties, bioinformatic analysisDOI: 10.1134/S0006297915100181
Abbreviations: Ka, level of nonsynonymous substitutions; Ks, level of synonymous substitutions; SD, segment-duplicated genes; TD, tandem-duplicated genes; ω, level of interspecific divergence (Ka/Ks ratio); ωs, level of intraspecific divergence.
Class III plant peroxidases (classical peroxidases) is an important
class of heme-containing enzymes (EC 1.11.1.7) having undiminishing
interest due to their high practical significance. Peroxidases catalyze
oxidation of various electron donors using hydrogen peroxide, but they
can also display oxygenase activity. Peroxidases exhibit broad
physiological functions by defending plants from oxidative stress and
pathogens, taking part in wound healing as well as controlling
catabolism of auxin and cell growth. Such functional variety is due to
the features of the expressed genes and location of protein products,
as well as protein structures underlying their substrate specificity
[1-4]. All classical
peroxidases are characterized by four disulfide bonds formed between
eight conservative cysteine residues and two calcium cations in each
enzyme molecule, which have three highly conservative domains (I-III),
wherein a consensus amino acid motif is similar both in dicotyledonous
and monocotyledonous plants [1, 5]. Also, conservative amino acids are present outside
these conservative domains. The majority of conservative amino acids
underlie functional activity of peroxidases. Variable amino acid
positions located around conservative domains determine substrate
specificity and features of certain peroxidases [1]. Plants were found to bear various forms of class
III peroxidases, which are encoded by dozens of genes [6, 7]. Their appearance is
explained by multiple duplications of ancestral genes [8]. Subsequent functional differences between
duplicated genes resulted from selection and inheritance of mutations
that underlie difference in protein functions and features of gene
regulation [9, 10].
Identification of amino acid differences in coding regions of duplicated genes is an important task, because they are responsible for appearance of new properties in peroxidases and determine their substrate specificity. Solving this task is complicated by the fact that these differences affect variable regions, wherein each of them is found to bear multiple amino acid substitutions, among which only some may be functionally important.
A comparison of various genes and corresponding protein products between close species (comparison of orthologous genes) as well as comparison of duplicated genes from the same species (comparison of paralogous genes) using bioinformatics methods is one of the approaches for identifying amino acid substitutions underlying differences in protein functions. Such comparative analysis allows examining divergence of duplicated genes and evaluating evolutionary processes that resulted in appearance of structural differences and might serve as grounds for functional specialization of the genes after their duplication [11]. Moreover, this analysis reveals traces of positive selections in gene sequence and changes in amino acid sequence selected during evolution. The current study investigated divergence of peroxidases by comparing peroxidase genes and corresponding proteins in Arabidopsis thaliana with genes and proteins derived from the closely-related species Arabidopsis lyrata (gene and protein sequences are available in a public database), as well as analyzing polymorphism and divergence of paralogous genes from A. thaliana to reveal potential traces of positive selection.
MATERIALS AND METHODS
Data on polymorphism of peroxidase genes from 456 ecotypes of A. thaliana were obtained from available databases [12, 13]. The genomes of five ecotypes of A. thaliana from Karelia [14] were sequenced in the Laboratory of Evolutionary Genomics, Faculty of Bioengineering and Bioinformatics, M. V. Lomonosov Moscow State University. Sequences of genes and their coding regions for each ecotype were generated using reference genome sequence and preprocessed data about genomic polymorphism. Information about polymorphic sites was filtered with a three reads supporting alternative polymorphic variant per site threshold and 10 reads coverage per polymorphic site threshold. Homologous proteins were searched in two databases – the GenBank (using BLAST programs) and PeroxiBase [12]. Multiple alignments were analyzed using ClustalW software [15]. Visualization and comparison of nucleotide and amino acid sequences were done using MultAlin software [16]. Sequences having at least 80% identity with proteins AtPrx53 and AtPrx54 from A. thaliana were used in the study. For proteins AtPrx36 and AtPrx72, identity was at least 70%.
DnaSP 5.2 [17] was used to analyze divergence level (Ka, Ks, Ka/Ks). Sliding window analysis of intraspecific divergence (ωs) and Ka across the sequence of duplicated genes from Col ecotype was done using DnaSP 5.2. Statistical processing and visualization of divergence level for 62 peroxidase genes was done using the Gnumeric Spreadsheet 1.12.20 software.
RESULTS
Examination of divergence for orthologous peroxidase genes from A. thaliana and A. lyrata. We analyzed 62 peroxidase genes that had orthologs in both species. The levels of nonsynonymous (Ka) and synonymous (Ks) substitutions and Ka/Ks (ω) ratio showing evolutionary trend (ω > 1 and < 1 determine impact of positive or negative selection on a gene, and ω = 1 determines neutral gene evolution) and revealing relative rate of gene evolution were calculated for comparing intraspecific divergence level between orthologous genes from A. thaliana and A. lyrata.
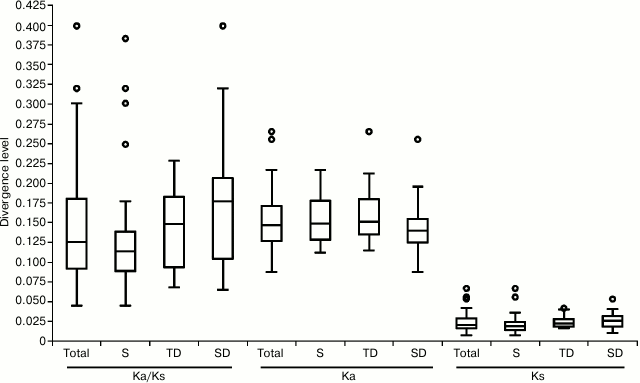
High range of variability was shown for all parameters. In particular, Ks changed from 0.087 (for Prx35) to 0.265 (Prx33), mean value – 0.153; Ka varied from 0.007 (for Prx42) to 0.067 (for Prx60), mean value – 0.024; ω ratio changed from 0.045 (for Prx42) to 0.399 (for Prx28; Fig. 1), mean value for all genes – 0.146, which, however, was lower for singleton (0.126) than for tandem-duplicated (TD, 0.146) and segment-duplicated (SD, 0.174; Fig. 1) genes. High values of ω (>0.2) were observed not only among TD Prx(23, 38, 54) and SD Prx(28, 35, 36, 56, 62, 73), but also in singleton Prx(48, 55, 60) genes.
Fig. 1. Interspecific divergence of 62 peroxidase genes. Boxplots of data on Ka, Ks, and Ka/Ks ratio for all genes (Total), only singletons (S), or tandem- or segment-duplicated genes (TD or SD, respectively) are presented. Circles mark the outliers.
The majority (10 out of 16 pairs) of duplicated genes (irrespective of duplication type) were characterized by similar value of ω, which varied between pairs. The minimum value of ω was found between orthologs of TD genes Prx(4-5), and the maximum value – for SD Prx(35-73) (Table 1). For three pairs of TD and three pairs of SD genes Prx(22-23), Prx(53-54), Prx(58-59), Prx(57-28), Prx(15-49), and Prx(36-72), values of ω for paralogs differed by 2-4-fold (Table 1), which might suggest relaxation of functional constraint for divergence of one of the paralogs.
Table 1. Divergence of peroxidase genes from
A. thaliana and A. lyrata for orthologs and
paralogs
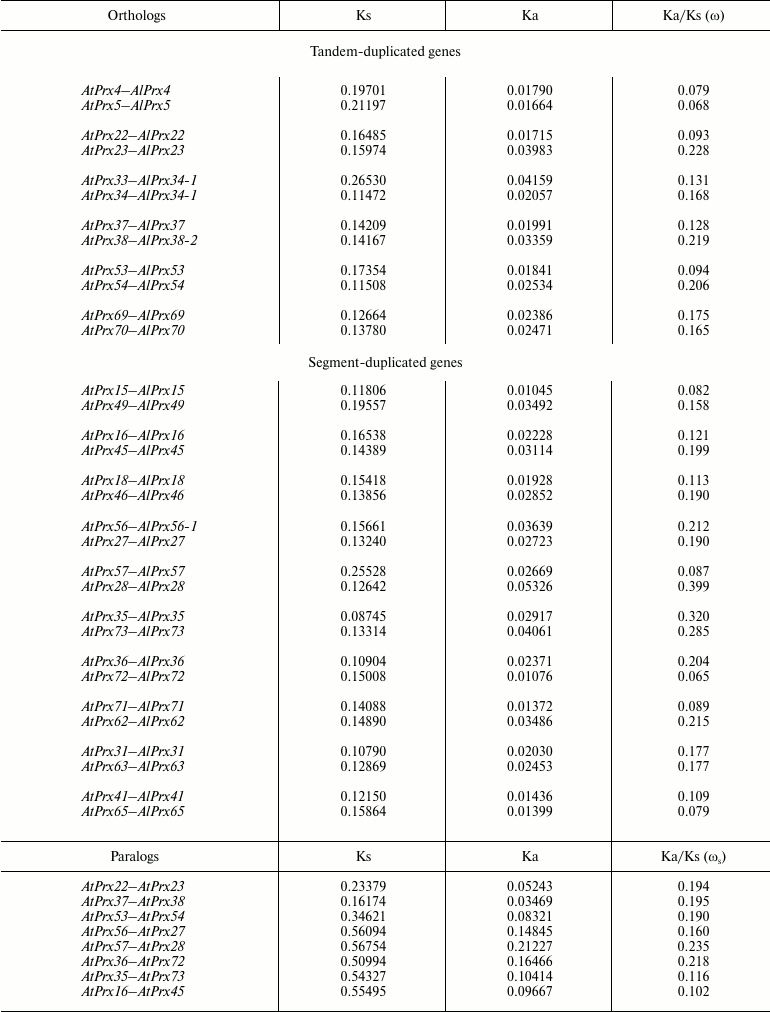
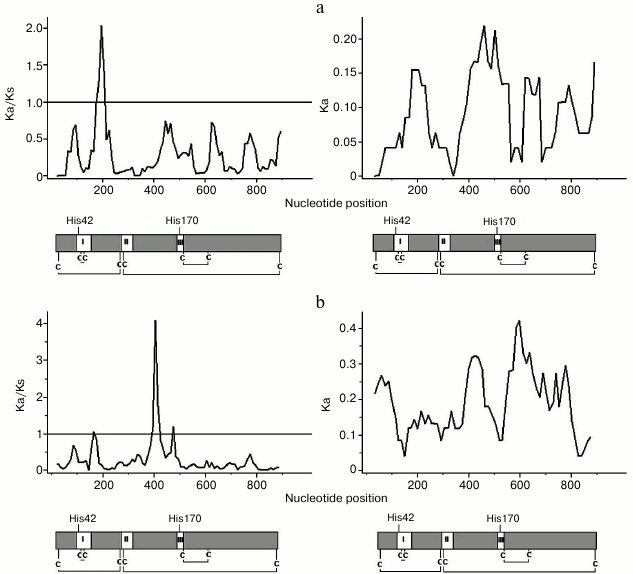
Divergence of paralogous peroxidase genes in A. thaliana. We analyzed the rate of paralog divergence ωs for eight A. thaliana gene pairs, wherein at least one gene had ω > 0.2 in comparison with A. lyrata ortholog. The ωs ranged from 0.102 to 0.235, although divergence level for synonymous and nonsynonymous sites between pairs varied by 3-6-fold (Table 1). All ωs values were much less than 1, thus suggesting that these paralogs undergo negative selection. However, average divergence values do not reflect possible changes affecting only certain domains. By comparing paralogs with each other by distribution of ωs along a gene coding sequence, some regions of sequence diversification can be found that were maintained by positive selection and that determined the appearance of new properties in encoded proteins. Thus, a distribution of ωs along the sequence of duplicated genes, wherein at least one paralog had ωs > 0.2 (Table 1), was conducted using sliding-window analysis. Regions interpreted as signature of positive selection (ωs > 1) were found only for pairs AtPrx(22-23, 53-54, 27-56, 28-57, 36-72) using a 50-bp window size and 10-bp step size. For pairs AtPrx(53-54) and AtPrx(36-72), the local maximum values of ωs were >2; therefore, the detailed distribution of ωs values along the coding part of these genes was analyzed.
For these pairs, the minimum values of ωs were found in the regions approximately corresponding to conservative domains in the protein (I-III) and adjacent gene regions. Regions with ωs > 1 had various positions (Fig. 2). It is known that high magnitude of ωs observed during this analysis might be detected upon local decrease in synonymous substitutions, when even single nonsynonymous substitutions might result in appearance of local peaks in ωs. By analyzing distribution of nonsynonymous substitutions, Ka value in paralogous genes revealed that the maximum values of Ka were observed in the same regions of the genes that had peaks of ωs (Fig. 2). Ks in the same local regions did not have the minimum value (data not shown), thus suggesting increase in ωs because of positive selection.
Fig. 2. Sliding window analysis of sequence divergence Ka/Ks (left) and Ka (right) across the coding regions of paralogs A. thaliana AtPrx53-AtPrx54 (a) and AtPrx36-AtPrx72 (b) using a window length of 50 bp and a step size of 10 bp; under X-axis – encoded proteins with delineated positions of three conservative domains in peroxidases (I-III), four disulfide bonds between eight conservative cysteine residues (C), His42, His170.
By comparing paralogs AtPrx(53-54), one peak of ωs was found in the second exon (between conservative domain I and II; Fig. 2a), which corresponded to seven amino acid substitutions in the protein: A51↔G51, I53↔L53, S59↔G59, dual substitution P68↔G68 and A69↔P69 as well as A71↔V71 and A74↔T74. Substitutions at position 51, 68, and 74 might affect protein properties, as features of polymorphic amino acids differ (hereinafter, they are called “meaningful substitutions”). In addition, the substitution A74↔T74 might also be functionally significant, which results in appearance of a potential glycosylation site in peroxidase AtPrx54.
For duplicated genes AtPrx(36-72), high value of ωs > 4 was found in exon 3 (between conservative domains II and III; Fig. 2b). In contrast to the other paralogs, significant differences in position of the maximum values of ωs were found for this pair upon changing size of the window and step. In case one, a high peak (position 400-430 bp) and a small peak with ωs < 2 to the right (~480 bp) were found by applying a 50-bp-window and 10-bp-step size (Fig. 2b), then comparison of this pair with 100-bp-window and 10-bp-step size revealed a single wide peak (360-480 bp) that had maximum value shifted towards position ~450 bp. Therefore, for this pair, substitutions in genes within a wide region were analyzed that corresponded to two peaks at 50-bp-window and 10-bp-step size or a wide peak upon changing parameters of the analysis. Within this region, changes resulting in 15 substitutions were found: E128↔G128, I132↔S132, triple substitution M135↔N135, E136↔N136, N137↔D137, then – substitutions: A140↔S140, dual substitution N142↔E142, N143↔S143, substitutions L145↔F145, M151↔K151, dual substitution N153↔K153 and F154↔R154 as well as substitutions T161↔V161, A165↔S165, L167↔S167, 10 of which were meaningful. The dual substitution N142↔E142, N143↔S143 caused appearance of a potential glycosylation site in protein AtPrx72.
Analysis of intraspecific allelic variation of paralogous genes AtPrx53, AtPrx54, AtPrx36, and AtPrx72. The local regions with high divergence of paralogs suggested positions of nonsynonymous substitutions, which might be the targets for positive selection and determine differences in structure and function of peroxidases. To assure that these substitutions were typical not only for ecotype Col, whose alleles were used for the abovementioned analysis, occurrence of such nucleotide and amino acid substitutions (as predicted traces of positive selection) in alleles and corresponding proteins from 461 ecotypes of A. thaliana were investigated.
By analyzing alleles for two pairs of paralogs, three new nonsynonymous substitutions were found in the examined regions. In particular, alleles AtPrx54 from 192 ecotypes were found to have a triplet AGC coding for S63 substituted for AGG (R63; Table 2), in three alleles of gene AtPrx36 – triplet ATG (M135) substituted for ACG (T135; Table 3), and in 47 alleles of gene AtPrx36 – triplet ATG (M151) substituted for CTG (L151). However, all nonsynonymous substitutions found upon comparing paralogs from Col were present in all alleles from the remaining 460 ecotypes (Tables 2 and 3). In all gene pairs, amino acid substitutions were found that occurred not due to one but to two nucleotide substitutions in the codon. For paralogs AtPrx(53-54), such substitutions were detected at positions 51, 59, 68, and 69 (Table 2), for paralogs AtPrx(36-72) – at positions 135, 136, 142, 143, 154, and 161 (Table 3). Moreover, silent and synonymous nucleotide substitutions were present in these regions as well.
Table 2. Polymorphic nucleotides in
alleles of paralogs AtPrx53-AtPrx54 from 461 ecotypes of A.
thaliana and corresponding amino acid substitutions in gene and
protein regions with high divergence level
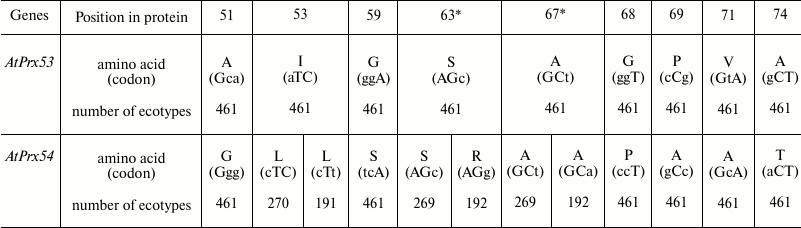
* Positions of synonymous substitutions.
Table 3. Polymorphic nucleotides in alleles
of paralogs AtPrx36-AtPrx72 from 461 ecotypes of A.
thaliana and corresponding amino acid substitutions in gene and
protein regions with high divergence level
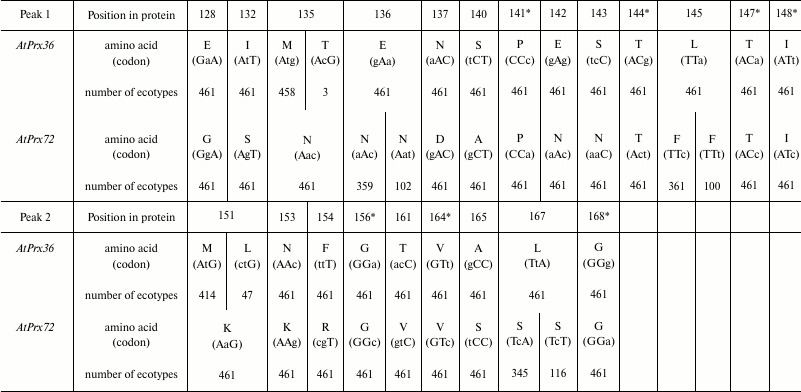
* Positions of synonymous substitutions.
Analysis of identified amino acid substitutions in homologous peroxidases from other species. In case the detected amino acid substitutions corresponding to the regions in genes with signature of positive selection were found in homologous peroxidases from other species, these data might be considered as additional evidence of selection acting on these sites that underlies a subsequent divergence of paralogs. By comparing homologous proteins Prx53-Prx54, it was found that three substitutions able to affect protein properties were conserved. In particular, there were substitution A51↔G51 and substitution of P68 (or similar V68 or A68) for G68. Substitution A74↔T74 results in appearance of a potential glycosylation site in the majority of homologs Prx54 (excepting homologs Prx54 from Armoracia rusticana and Thellungiella halophila lacking this substitution). These three residues in certain combination mark orthologous proteins Prx53 (A51…G68…A74) and Prx54 (G51…P68/V68/A68…T74) (Fig. 3a).
Fig. 3. Fragments of multiple alignment of amino acid sequences of peroxidases homologous to AtPrx53 and AtPrx54 (a) and AtPrx36 and AtPrx72 (b). I – conservative domain. Conservative amino acids are underlined below the alignment, and variable residues are highlighted in bold, Arabic numerals denote positions of amino acids in accordance with enumeration proposed by Welinder [1]. Small frames depict glycosylation sites. Arrows designate substitutions found in peroxidases of most examined species. The following proteins were used from PeroxiBase: A. lyrata AlyPrx(53, 54, 36, 72), A. rusticana AruPrx(53-1, 54), B. rapa BrPrx(53-1Aa, 54-1Aa, 36-1Aa, 72-1Aa), B. napus BnPrx(54-1, 36-1), T. halophila ThPrx(53, 54, 36, 72), A. alpina AalpPrx36; proteins from GenBank: B. napus CDX98949.1, E. halophilum ABB45838.1, and proteins from NCBI: C. sativa XP_010423310.1 and XP_010452613.1, C. rubella XP_006288143.1, E. salsugineum XP_006399146.1, C. sativa XP_010484652.1, E. salsugineum XP_006403967.1, C. sativa XP_010426640.1, C. rubella XP_006292998.1.
Likewise, a number of meaningful substitutions in proteins AtPrx36 and AtPrx72 typical for A. thaliana were conservative in homologous proteins of other species. A combination of three amino acid residues M135-G136-N137 is typical for orthologs Prx36 from both species of Arabidopsis, Capsella rubella, and Camelina sativa. Residue N137 is also present in orthologs Prx36 from Arabis alpina, Eutrema salsugineum, Brassica rapa, and B. napus, but at positions 135-136 there are N135-N136 typical for Prx72 or N135-D136 found in E. salsugineum and Thellungiella halophila. All orthologs of Prx72 possess N135-N136-D137. Orthologs of Prx36 from both species of Arabidopsis, C. sativa, and A. alpina are characterized by S140 or T140 with similar properties (in C. rubella), whereas proteins from E. salsugineum, T. halophila, B. rapa, and B. napus possess L or V at this position bearing properties similar to R, which marks position 140 in orthologs Prx72 (Fig. 3b). Orthologs Prx36 and Prx72 also clearly differed at position S143↔N143. Such substitution of similar amino acid residues is accompanied by appearance of a potential glycosylation site NNT in all orthologs Prx72, whereas among orthologs Prx36 the glycosylation site NST was found only in proteins from A. alpina, E. salsugineum, and T. halophila.
Also, it is worth mentioning other amino acids located closer to the proximal H170. In particular, M151↔K151, which distinguishes proteins Prx36 of both species of Arabidopsis, C. rubella, C. sativa, E. salsugineum, and T. halophila from proteins Prx72. Homologs of Prx36 from A. alpina and Brassica contain in this position K, which is typical to Prx72. A substitution N153 (or Y153 in A. alpina with similar properties) in proteins Prx36 for K153 is present in Prx72. A substitution T161↔V161 also differs proteins Prx36 and Prx72, although homologs Prx36 from both species of Brassica contain in this position I having properties similar to V. Proteins Prx36 and Prx72 from all examined species are distinguished by substitutions A165↔S165, L167↔S167. Thus, among 10 potentially significant substitutions revealed while comparing orthologs AtPrx36 and AtPrx72, eight of them distinguish the majority of homologs Prx36 and Prx72 from Brassicaceae, and only five of them distinguish all examined homologs Prx36 from all homologs Prx72.
DISCUSSION
Comparison of 62 orthologous genes from peroxidases A. thaliana and A. lyrata demonstrated that the average level of divergence for solitary genes is lower than for duplicated genes, thereby confirming the idea that duplicated genes evolve faster compared to singletons [18]. The average level of intraspecific divergence ω (Fig. 1) was found to be substantially higher than for genes controlling response to biotic stresses and wounding (the genes involved in pathways related to salicylic acid, ethylene, and jasmonic acid [19]), wherein peroxidase genes are also referred to. In terms of the level of intraspecific divergence, peroxidase genes are inferior only to R-genes, whose variability is mainly related to the presence of leucine-rich domains (LRR) and is determined by positive and balancing selection acting on these regions [20, 21].
The level of intraspecific divergence for all orthologous peroxidase genes including duplicated ones was substantially lower than 1 (Fig. 1), thus suggesting an impact of negative selection on all genes. However, significant differences in divergence level (by ~9-fold; Fig. 1) between the genes suggest that some of them evolved under significantly relaxed selective constraint. By comparing intraspecific divergence of paralogs, it was found that differences in evolution rate between pairs of duplicated genes was lower (by 2-3-fold; Table 1). This might be explained if rapid evolution of duplicated genes (which could result in appearance of functional differences between paralogs) would be time-limited, and then the rate of evolution would return to the level preceding gene duplication [9, 22].
Nonetheless, a comparison of paralogs using a sliding window analysis revealed traces of positive selection in terms of locally elevated divergence level observed in certain regions of the genes for two pairs of paralogs (Fig. 2). The significance of identified nucleotide substitutions for functional divergence of examined paralogs was confirmed by assessing their intraspecific conservatism. It was found that nonsynonymous substitutions corresponding to the regions with elevated divergence were present in all 461 ecotypes of A. thaliana and distinguished paralogous genes from each other. In both pairs of duplicated genes, it was found that they contained dual nucleotide substitutions in one codon and dual (and even triple) substitutions of adjacent amino acids (Tables 2 and 3), which additionally suggest the action of positive selection on these local regions in the gene and protein [23]. All these substitutions occurred in variable regions of the protein, which might be ameliorated or disappear owing to mutations and recombination unless they were functionally important. It is considered that species A. thaliana was rapidly spread over the world from a center of origin, which is reflected by the pattern of genome polymorphism. However, even by taking into consideration demographic factors, it should be admitted that 100% fixation of substitutions within a variable region of gene and protein in 461 ecotypes of A. thaliana derived from different habitats is unusual and is a weighty argument in favor of their functional significance. It seems that such substitutions occurred after gene duplication and were under the action of positive selection, because they cause the appearance of new properties in peroxidases encoded by paralogous genes that are important for plants.
Many substitutions revealed between proteins AtPrx53-AtPrx54 and AtPrx36-AtPrx72 turned out to be conservative and were present in homologous peroxidases from other Brassicaceae (Fig. 3). This suggests that they appeared after gene duplication in ancestor Brassicaceae and became subjects to positive selection. Peroxidases Prx53-Prx54 from Brassicaceae were found to bear three sites of conservative substitutions (51, 68, 74), which might alter protein properties, such as substitution A51↔G51 apparently influencing efficacy of Ca2+ binding by flanking residues D50 and S52; substitution P68 (in some proteins – V68 or A68) for G68 located close to N70 from the active site. Horseradish peroxidase contains at this position F68, which together with other F residues (41, 42, 143, 179) forms a channel for hydrogen peroxide entering to its active site. Size and hydrophobicity of the channel control access to heme iron [2]. It is possible that substitution of nonpolar hydrophobic P68 (V68 or A68) for the polar G68 residue affects the conformation of the substrate-binding site and accessibility of the active site in other peroxidases. In addition, substitution A74↔T74 may also affect protein function, which results in appearance of a potential glycosylation site in the majority of Prx54 homologs.
Gene AtPrx53 and the corresponding peroxidase (ATPA2) have been examined by many investigators. Although the gene has a moderate expression level, the peroxidase it encodes is a major protein among anionic peroxidases [24, 25] that controls cell elongation and response to stress [26]. So far, functions of AtPrx54 have not been sufficiently studied. Our investigation revealed amino acid substitutions that perhaps played a major role in divergence of peroxidases such as Prx53 and Prx54 after duplication of the ancestor gene from Brassicaceae. However, even within species A. thaliana, natural selection continues to increase the variety of peroxidase isoforms, as gene AtPrx53 is represented by two major haplogroups in A. thaliana, which are maintained via balancing selection and code for proteins differing in electrophoretic mobility [27, 28].
In peroxidases Prx36-Prx72, substitutions that might underlie divergence of their functions occurred within a region that corresponds to the α-helix D′ of many peroxidases (131-138 a.a.). Homologs of peroxidase Prx72 bear amino acids N135-N136-D137 in this region, whereas peroxidases P36 from both Arabidopsis species, Capsella rubella, and Camelina sativa — M135-E136-N137. Previous studies done with peroxidase AtPrx72 (pI 8.6) suggest that like other major S-type peroxidases, it also lacks α-helix D′, which presumably affects substrate specificity [29, 30]. Moreover, it also lacks motif 138-IPS (I138, P139, S140), which determines conformation and hydrophobicity of substrate-binding site in anionic peroxidase AtPrx53 (ATPA2) [31], although it is present in anionic peroxidase AtPrx36 (pI 4.7) and its immediate homologs (Fig. 3b). In AtPrx53, this motif is fixed by helix D′, whose structure and position depend on the presence of the heme prosthetic group [31]. Due to substitutions A140↔S140, homologs of Prx72 have motif 138-IPS changed to 138-IPA. In peroxidase Prx72, the abovementioned three residues 135-NND together with preceding residue S134 form motif 134-SNND considered as one of the four possible phosphorylation sites for casein kinase II [30]. Peroxidase Prx36 lacks this motif. Proteins Prx36 and Prx72 are also clearly distinguished at position S143↔N143, which determines appearance of potential glycosylation site NNT in all orthologs of Prx72 (another potential glycosylation site NST is present in orthologs of Prx36 in A. alpina, E. salsugineum, and T. halophila). Finally, properties of peroxidases can be affected by substitutions A165↔ S165, L167↔S167, which similarly to the above noted ones clearly distinguish orthologous proteins Prx36 and Prx72. These residues belong to a motif 162-DLVSLSGSHTI (S165 and S167 are underlined) also present in AtPrx72 and serving as a proximal ligand for the heme group [30]. This motif contains H170 residue, which creates a hydrogen bond with D247. Perhaps, significant changes in this motif within homologs of Prx36 (162-DLVALLGSHTI: substitutions are underlined) influence the heme binding at the active site of peroxidases.
It is emphasized that the above-noted substitutions occurred within a small region of the protein and might result from positive selection of genetic changes underlying divergence of properties and functions of paralogous proteins. Undoubtedly, such changes are not alone, and proteins with common origin due to a single gene duplication acquired many more differences than were discussed in this article. Among them, there might be changes supported by natural selection, but their traces already faded away. Footprints of such selection found by us are related to those important structural elements of proteins commonly originating due to a single gene duplication, which are noted among the major differences between S- and G-peroxidases possessing various substrate specificity [32]. Moreover, it was demonstrated that structural elements of S-peroxidases are of ancient origin, being found even in Physcomitrella and Marchantia [29]. These elements are typical to Prx72, but altered in Prx36, suggesting that in the ancestral form of Brassicaceae the Prx72 gene was just a parental copy that was maintained unaltered by negative selection, whereas the Prx36 gene was a daughter copy that was transferred into a novel genetic surrounding and rapidly evolved. The level of interspecific gene divergence ω (Table 1) also favors this idea. It was found that the magnitude of ω for AtPrx36 gene was 3-fold higher than for the AtPrx72 gene, pointing at relaxed negative selection on the AtPrx36 gene.
Protein AtPrx36 lost many characteristics typical of S-peroxidases, including those found in AtPrx72 that ensure its participation in lignification and strengthening of cell walls [30]. However, the AtPrx36 gene did not follow along a pseudogenization pathway. Like the AtPrx72 gene, it is under pressure of negative selection (level of interspecific divergence ω < 1; Table 1). Stable maintenance of changes revealed within exon 3 in the population (461 ecotypes), which distinguish paralogs AtPrx36 and AtPrx72, also point to impact of negative selection as well as importance of changes fixed by selection in acquiring new properties by AtPrx36. Protein AtPrx36 is mainly found in internodes [4] and developing seeds [33], whereas AtPrx72 – in root endodermis [4]. In contrast to AtPrx72, which stiffens cell walls via controlling lignin polymerization [30], AtPrx36 loosens cell walls, apparently contributing to disruption of polysaccharide bonds, which is required for cell elongation during growth of plants and seed germination [4]. So far, a substrate for AtPrx36 in plants has not been identified. Perhaps the protein changes found in our study just underlie such shift in substrate specificity.
In conclusion, it should be noted that examination of polymorphism of genes and their products can only identify traces of positive selection in proteins and attract to them attention of biochemists. To confirm functional significance of the revealed changes and their impact on protein properties, it is necessary to conduct experimental investigations.
This study was performed with financial support from the Russian Science Foundation (grant No. 14-50-00029) and Research Project “Genetic Organization of Plant Genome” (No. 13-2-01).
REFERENCES
1.Welinder, K. G., Justesen, A. F., Kjaersgard, I.
V., Jensen, R. B., Rasmussen, S. K., Jespersen, H. M., and Duroux, L.
(2002) Structural diversity and transcription of class III peroxidases
from Arabidopsis thaliana, Eur. J. Biochem., 269,
6063-6081.
2.Gazaryan, I. G., Khushpul’yan, D. M., and
Tishkov, V. I. (2006) Features and mechanism of action for plant
peroxidases, Uspekhi Biol. Khim., 46, 303-323.
3.Almagro, L., Gomez Ros, L. V., Belchi-Navarro, S.,
Bru, R., Ros Barcelo, A., and Pedreno, M. A. (2009) Class III
peroxidases in plant defense reactions, J. Exp. Bot., 60,
377-390.
4.Francoz, E., Ranocha, P., Nguyen-Kim, H., Jamet,
E., Burlat, V., and Dunand, C. (2015) Roles of cell wall peroxidases in
plant development, Phytochemistry, 112, 15-21.
5.Mathe, C., Barre, A., Jourda, C., and Dunand, C.
(2010) Evolution and expression of class III peroxidases, Arch.
Biochem. Biophys., 500, 58-65.
6.Passardi, F., Longet, D., Penel, C., and Dunand, C.
(2004) The class III peroxidase multigenic family in rice and its
evolution in land plants, Phytochemistry, 65,
1879-1893.
7.Rawal, H. C., Singh, N. K., and Sharma, T. R.
(2013) Conservation, divergence, and genome-wide distribution of PAL
and POX a gene families in plants, Int. J. Genom.; DOI:
10.1155/2013/678969.
8.Duroux, L., and Welinder, K. G. (2003) The
peroxidase gene family in plants: a phylogenetic overview, J. Mol.
Evol., 57, 397-407.
9.Lynch, M., and Conery, J. S. (2000) The
evolutionary fate and consequences of duplicate genes, Science,
290, 1151-1155.
10.Moore, R. C., and Purugganan, M. D. (2003) The
early stages of duplicate gene evolution, Proc. Natl. Acad. Sci.
USA, 100, 15682-15687.
11.Kurbidaeva, A., Novokreshchenova, M., and Ezhova,
T. (2015) ICE genes in Arabidopsis thaliana: clinal
variation in DNA polymorphism and sequence diversification, Biol.
Plant., 59, 245-252.
12.Fawal, N., Li, Q., Savelli, B., Brette, M.,
Passaia, G., Fabre, M., Mathe, C., and Dunand, C. (2013) PeroxiBase: a
database for large-scale evolutionary analysis of peroxidases,
Nucleic Acids Res., 41, 441-444.
13.Cao, J., Schneeberger, K., Ossowski, S., Gunther,
T., Bender, S., Fitz, J., Koenig, D., Lanz, C., Stegle, O., Lippert,
C., Wang, X., Ott, F., Muller, J., Alonso-Blanco, C., Borgwardt, K.,
Schmid, K. J., and Weigel, D. (2011) Whole-genome sequencing of
multiple Arabidopsis thaliana populations, Nat. Genet.,
43, 956-963.
14.Kurbidaeva, A. S., Zaretskaya, M. V., Soltabaeva,
A. D., Novokreshchenova, M. G., Kupriyanova, E. V., Fedorenko, O. M.,
and Ezhova, T. A. (2013) Genetic mechanisms of plant adaptation in
Arabidopsis thaliana to extreme environment at northern boundary
of the habitat, Genetika, 49, 943-952.
15.Thompson, J. D., Higgins, D. G., and Gibson, T.
J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific gap
penalties and weight matrix choice, Nucleic Acids Res.,
22, 4673-4680.
16.Corpet, F. (1988) Multiple sequence alignment
with hierarchical clustering, Nucleic Acids Res., 16,
10881-10890.
17.Rozas, J., Sanchez-DelBarrio, J. C., Messeguer,
X., and Rozas, R. (2003) DnaSP, DNA polymorphism analyses by the
coalescent and other methods, Bioinformatics, 19,
2496-2497.
18.Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I.,
and Koonin, E. V. (2002) Selection in the evolution of gene
duplications, Genome Biol., 3.
19.Bakker, E. G., Traw, M. B., Toomajian, C.,
Kreitman, M., and Bergelson, J. (2008) Low levels of polymorphism in
genes that control the activation of defense response in Arabidopsis
thaliana, Genetics, 178, 2031-2043.
20.Bakker, E. G., Toomajian, C., Kreitman, M., and
Bergelson, J. (2006) A genome-wide survey of R gene polymorphisms in
Arabidopsis thaliana, Plant Cell, 18,
1803-1818.
21.Guo, Y. L., Fitz, J., Schneeberger, K., Ossowski,
S., Cao, J., and Weigel, D. (2011) Genome-wide comparison of
nucleotide-binding site-leucine-rich repeat-encoding genes in
Arabidopsis, Plant Physiol., 157, 757-769.
22.Rosello, O. P., and Kondrashov, F. A. (2014)
Long-term asymmetrical acceleration of protein evolution after gene
duplication, Genome Biol. Evol., 6, 1949-1955.
23.Bazykin, G. A., Kondrashov, F. A., Ogurtsov, A.
Y., Sunyaev, S., and Kondrashov, A. S. (2004) Positive selection at
sites of multiple amino acid replacements since rat-mouse divergence,
Nature, 429, 558-562.
24.Nielsen, K. L., Indiani, C., Henriksen, A., Feis,
A., Becucci, M., Gajhede, M., Smulevich, G., and Welinder, K. G. (2001)
Differential activity and structure of highly similar peroxidases.
Spectroscopic, crystallographic, and enzymatic analyses of lignifying
Arabidopsis thaliana peroxidase A2 and horseradish peroxidase
A2, Biochemistry, 40, 11013-11021.
25.Lebedeva, O. V., Ezhova, T. A., Musin, S. M.,
Radyukina, N. L., and Shestakov, S. V. (2003) PXD gene controls
development of three isoforms of anionic peroxidases in Arabidopsis
thaliana, Izv. Akad. Nauk. Ser. Biol., 2,
159-168.
26.Jin, J., Hewezi, T., and Baum, T. J. (2011)
Arabidopsis peroxidase AtPRX53 influences cell elongation and
susceptibility to Heterodera schachtii, Plant Signal.
Behav., 6, 1778-1786.
27.Kupriyanova, E. V., Ezhova, T. A., and Shestakov,
S. V. (2007) Dimorphic DNA variation in the anionic peroxidase gene
AtPrx53 of Arabidopsis thaliana, Genes Genet.
Syst., 82, 377-385.
28.Kupriyanova, E. V., Ezhova, T. A., Lebedeva, O.
V., and Shestakov, S. V. (2006) Interspecific polymorphism of
peroxidase genes localized in chromosome 5 of Arabidopsis
thaliana, Izv. Akad. Nauk. Ser. Biol., 33,
353-362.
29.Gomez Ros, L. V., Gabaldon, C., Pomar, F.,
Merino, F., Pedreno, M. A., and Barcelo, A. R. (2007) Structural motifs
of syringyl peroxidases predate not only the
gymnosperm–angiosperm divergence but also the radiation of
tracheophytes, New Phytol., 173, 63-78.
30.Herrero, J., Fernandez-Perez, F., Yebra, T.,
Novo-Uzal, E., Pomar, F., Pedreno, M. A., Cuello, J., Guera, A.,
Esteban-Carrasco, A., and Zapata, J. M. (2013) Bioinformatic and
functional characterization of the basic peroxidase 72 from
Arabidopsis thaliana involved in lignin biosynthesis,
Planta, 237, 1599-1612.
31.Ostergaard, L., Telium, K., Mirza, O., Mattson,
O., Petersen, M., Welinder, K. G., Mundy, J., Gajhede, M., and
Henriksen, A. (2000) Arabidopsis ATPA2 peroxidase. Expression and
high-resolution structure of a plant peroxidase with implications for
lignification, Plant Mol. Biol., 44, 231-243.
32.Ros Barcelo, A., Gomez-Ros, L. V., and Carrasco,
A. E. (2007) Looking for syringyl peroxidases, Trends Plant
Sci., 12, 486-491.
33.Kunieda, T., Shimada, T., Kondo, M., Nishimura,
M., Nishitani, K., and Hara-Nishimura, I. (2013) Spatiotemporal
secretion of PEROXIDASE36 is required for seed coat mucilage extrusion
in Arabidopsis, Plant Cell, 25, 1355-1367.