Influence of SkQ1 on Expression of Nrf2 Gene, ARE-Controlled Genes of Antioxidant Enzymes and Their Activity in Rat Blood Leukocytes under Oxidative Stress
V. V. Vnukov, O. I. Gutsenko, N. P. Milutina*, I. V. Kornienko, A. A. Ananyan, A. O. Danilenko, S. B. Panina, A. A. Plotnikov, and M. S. Makarenko
Southern Federal University, Academy of Biology and Biotechnology, Department of Biochemistry and Microbiology, 344090 Rostov-on-Don, Russia; E-mail: natmilut@rambler.ru* To whom correspondence should be addressed.
Received July 27, 2015; Revision received August 13, 2015
The study demonstrated that oxidative stress induced by hyperoxia (0.5 MPa for 90 min) resulted in reduction of mRNA levels of transcription factor Nrf2 and Nrf2-induced genes encoding antioxidant enzymes (SOD1, CAT, GPx4) in peripheral blood leukocytes of rats. The changes in gene expression profiles under hyperoxia were accompanied by disbalance of activity of antioxidant enzymes in the leukocytes, namely activation of superoxide dismutase and inhibition of catalase, glutathione peroxidase, and glutathione-S-transferase. Pretreatment of rats with SkQ1 (50 nmol/kg for five days) significantly increased mRNA levels of transcription factor Nrf2 and Nrf2-induced genes encoding antioxidant enzymes SOD2 and GPx4 and normalized the transcriptional activity of the SOD1 and CAT genes in the leukocytes in hyperoxia-induced oxidative stress. At the same time, the activity of catalase and glutathione peroxidase was increased, and the activity of superoxide dismutase and glutathione-S-transferase returned to the control level. It is hypothesized that protective effect of SkQ1 in hyperoxia-induced oxidative stress can be realized via a direct antioxidant property and the stimulation of the Keap1/Nrf2 redox-sensitive signaling system.
KEY WORDS: mitochondria-targeted antioxidant, leukocytes, gene expression, antioxidant enzymes, hyperoxiaDOI: 10.1134/S0006297915120081
Abbreviations: ARE, antioxidant response element; HBO, hyperbaric oxygen (therapy); ROS, reactive oxygen species.
Oxidative stress and oxidative damage to cell structures are involved in
pathological processes including disease and extreme conditions,
adaptation, and aging [1-3].
Modeling of disease conditions has been applied for the investigation
of oxidative stress mechanisms. One widely used experimental model is
hyperoxia [4, 5]. According to
various data, mitochondrial dysfunction plays an important role in the
development of oxidative stress induced by hyperoxia [6, 7]. At the same time,
hyperbaric oxygen (HBO) therapy is widely used in medical practice for
the treatment of various disorders [8].
Pharmacological agents with antioxidant properties have been applied
for the correction of redox homeostasis disturbance. Among these
substances, mitochondria-targeted antioxidants of the SkQ family have
been confirmed to be highly efficient [9, 10].
Many studies have confirmed that the Keap1/Nrf2/ARE redox-sensitive signaling system plays the central role in cellular mechanisms of protection against oxidative/electrophilic stress [11]. Inducers of this system have been revealed in different studies [12]. SkQ1 (10-(6′-plastoquinonyl)decyltriphenylphosphonium) containing in its structure a plastoquinone with quinone groups in para-position has been proven to be one of such inducers [13].
The goal of the present study was to investigate the influence of mitochondrial-targeted antioxidant SkQ1 on gene expression of transcription factor Nrf2 and Nrf2-controlled genes encoding antioxidant enzymes in the blood leukocytes of rats under oxidative stress induced by hyperoxia.
MATERIALS AND METHODS
Mature male rats (Rattus norvegicus) weighing 180-200 g were used for this study. The experimental animals were divided into three groups. Group I (n = 10-23) included control animals, which were kept under the standard vivarium conditions. Animals of group II (n = 10-18) were exposed to HBO treatment (0.5 MPa for 90 min) and decapitated 12 h after the HBO session. The experimental animals were placed in the HBO chamber (volume 60 liters) with carbon dioxide absorber. After 3 min of pure oxygen ventilation, compression of 0.5 MPa was set up in the HBO chamber (compression and decompression was performed with a rate of 0.2 MPa/min); isopression continued for 90 min. This HBO procedure (0.5 MPa for 90 min) has been shown to cause acute oxidative stress [14]. Group III (n = 10-22) included animals that were pretreated with 50 nmol SkQ1/kg body weight daily for five days, then exposed to HBO (0.5 MPa for 90 min) 1 h after the last SkQ1 supplementation, and decapitated 12 h after the HBO session. The calculated SkQ1 dose was diluted in 100 µl 0.2% ethanol in distilled water and administered into the cheek pouch of the rats.
All experiments on the laboratory animals were performed in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (Strasbourg, March 18, 1986).
The expression of mRNA was analyzed by reverse transcription and following polymerase chain reaction (RT-PCR) with gene-specific primers. Blood (50 µl) was sampled for analysis. Total RNA was isolated by guanidinium thiocyanate–phenol–chloroform extraction using a Ribo-zol-B kit (InterLabService, Russia). The quality of extracted RNA was assessed by electrophoresis in a 1.2% agarose gel; the quantity of RNA was assessed by absorbance at 260 nm. Reverse transcription was performed using a cDNA synthesis kit (Syntol, Russia) containing MMLV-RT (Moloney murine leukemia virus reverse transcriptase), primer Random-6, deoxyribonucleotide triphosphates (dNTP), and RNase inhibitor.
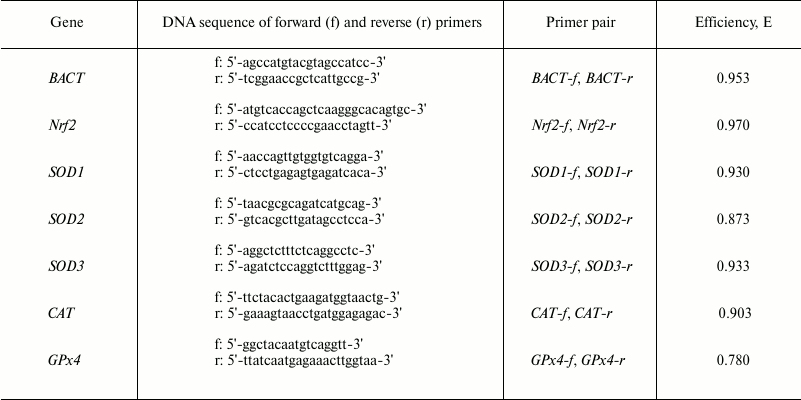
Gene expression patterns of transcription factor Nrf2 (Nrf2), superoxide dismutase isoforms Cu/ZnSOD (SOD1), MnSOD (SOD2), extracellular SOD – ECSOD (SOD3), catalase (CAT), and glutathione peroxidase 4 (GPx4) were analyzed by real-time PCR using intercalating dye EvaGreen (Molecular Probes, USA) and a real-time PCR kit (Syntol). Real-time PCR was performed using an iQ5 real-time PCR detection system (BioRad Laboratories, USA). BACT (β-actin) served as an internal control gene. Primer BLAST and Primer 3 software were used to design target-specific primers. The sequences of the specific primers are presented in the table. To evaluate the efficiency (E) of the primer pairs, PCR was performed with serial dilutions of the cDNA (1 : 1, 1 : 2, 1 : 4, 1 : 8), then average ΔCt values were calculated. The efficiencies of the primer pairs are presented in the table.
Primers for real-time PCR and the efficiency of used primer pairs
Real-time PCR was conducted under the following conditions: the 1st step at 95°C for 300 s, the 2nd step at 58(60)°C for 50 s (fluorescence detection), the 3rd step at 95°C for 15 s, then the 2nd step was repeated. Steps 2 and 3 were repeated 40 times; after that, the fluorescence intensity was plotted as a function of time. The melting curve was analyzed to determine the specificity of the amplification.
The software package Bio-Rad IQ5 Optical System Software Version 2.0 was used to analyze the PCR amplification plots. The real-time PCR data were analyzed using iCycler IQ5 software (BioRad). Relative quantification of gene expression level was performed by the ΔCt method using the amount of cDNA of the comparison gene.
The mononuclear cell suspension was used for the following biochemical analyses and was separated by centrifugation of whole blood in a Ficoll–Verografin density gradient (ρ = 1.077) according to the method of Boyum [15]. The cells were washed three times and suspended in Tris-buffered saline (pH 7.4).
Superoxide dismutase (SOD) activity was evaluated based on the ability of SOD to inhibit nitroblue tetrazolium reduction by superoxide generated during autoxidation of adrenaline [16]. Catalase activity was measured using the reaction of hydroperoxide with ammonium molybdate [17]. Glutathione peroxidase (GPx) activity assay was based on the intensity of oxidation of reduced glutathione triggered by tert-butyl hydroperoxide [18], and glutathione-S-transferase (GST) activity was evaluated using the reaction between reduced glutathione and 1-chloro-2,4-dinitrobenzene [19]. Myeloperoxidase (MPO) activity was measured using spectrophotometry [20], and the NADPH-oxidase activity assay was based on the reduction of 2,6-dichlorophenolindophenol in the presence of NADPH [21].
All statistical calculations were carried out with Biostat software (Version 2009 Professional – 5.8.4.3 Analyst Soft). The Kolmogorov–Smirnov and Lilliefors test was used to assess the normality of distribution. If the data were not normally distributed, the nonparametric Mann–Whitney U-test was applied for comparing two samples. If the distribution was normal, Student’s t-test for small sample sizes was applied. All p-values of less than 0.05 were regarded as significant. A tendency towards statistical significance was postulated in case of 0.05 < p < 0.1.
RESULTS
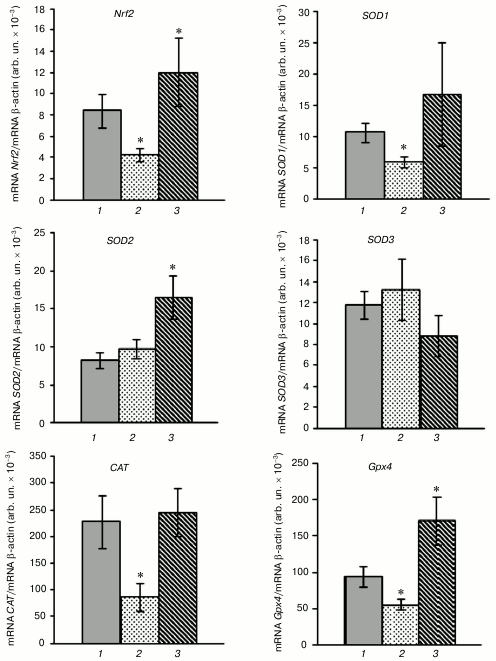
HBO-induced (0.5 MPa for 90 min) oxidative stress resulted in decreased expression level of transcription factor Nrf2 by 33%. Nrf2 is known to play a critical role in cell protection against oxidative stress in hyperoxia [22, 23]. This fact is confirmed by the decrease in mRNA level of transcription factor Nrf2 compared with the control (Fig. 1). The reduction in the expression level of ARE-controlled genes encoding antioxidant enzymes – superoxide dismutase 1 (SOD1), catalase (CAT), and mitochondrial glutathione peroxidase 4 (GPx4) – was demonstrated in the leukocytes of rats in hyperoxia concurrently with the inhibition of transcriptional activity of the Nrf2 gene. The SOD1 mRNA level decreased by 44%, CAT mRNA – by 62%, and GPx4 mRNA – by 40% in the leukocytes compared with normal values (Fig. 1). The differences in expression level of the SOD2 and SOD3 genes in the leukocytes were not statistically significant between the group with these conditions and the control group.
Fig. 1. Influence of SkQ1 on mRNA level of Nrf2 gene and genes encoding antioxidant enzymes in rat blood leukocytes after HBO-induced oxidative stress (M ± m; number of animals per group is 13-26; statistically significant differences between experimental and control group (p < 0.05) are marked by asterisks (*)). 1) Control; 2) HBO; 3) SkQ1 + HBO.
Pretreatment of rats with SkQ1 for five days before the HBO session significantly increased the transcriptional activity of the Nrf2, SOD2, and GPx4 genes after hyperoxia. This is confirmed by the increase in Nrf2 mRNA level by 47%, and SOD2 and GPx4 mRNA – by 99 and 80%, respectively, in the rat leukocytes compared with the control. The expression of the SOD1 and CAT genes was within the normal range and was decreased in HBO-induced stress in leukocytes of animals that were not pretreated with SkQ1 (Fig. 1). It was shown previously that SkQ1 stimulates the expression of the transcription factor Nrf2 gene and Nrf2-controlled genes (SOD1, SOD2, CAT, GPx4) in rat blood leukocytes under physiological conditions [13].
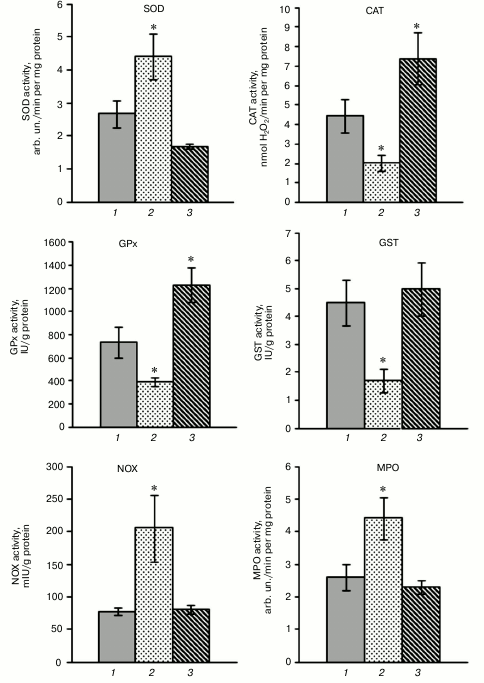
The investigation of the activity of antioxidant enzymes demonstrated disturbance of their functioning in rat blood leukocytes after a HBO session (0.5 MPa for 90 min; Fig. 2). The superoxide dismutase (SOD) activity increased by 66% in the leukocytes after hyperoxia, and at the same time the activities of catalase, glutathione peroxidase (GPx), and glutathione-S-transferase (GST) decreased by 54, 47, and 62%, respectively, which promoted the activation of free radical oxidation.
Fig. 2. Influence of SkQ1 on activity of pro- and antioxidant enzymes in blood mononuclear fraction of rats after HBO-induced oxidative stress (M ± m; number of animals per group is 13-26; statistically significant differences between experimental and control group (p < 0.05) are marked by asterisks (*)). 1) Control; 2) HBO; 3) SkQ1 + HBO.
The decreased activity of catalase and GPx in the leukocytes after the HBO session is in accordance with the decrease in expression level of the CAT and GPx4 genes. The increased total SOD activity in the leukocytes was accompanied by inhibition of SOD1 gene expression and normal transcriptional activity of the SOD2 and SOD3 genes.
Pretreatment with SkQ1 for five days before the HBO session resulted in normalization of SOD and GST activity after hyperoxia, activation of catalase activity by 67% and GPx activity by 79% in rat blood leukocytes (Fig. 2). These results demonstrate that the pretreatment with SkQ1 contributes to normalization of activity of antioxidant enzymes (SOD, GST) or activation of enzymes (catalase, GPx) after hyperoxia. In addition, total SOD activity stayed within the normal range after the HBO session in leukocytes of rats that were pretreated with SkQ1, and the expression level of the SOD2 gene significantly increased, but expression level of the SOD1 and SOD3 genes did not differ from control. The activation of catalase was accompanied by constant expression level of the CAT gene in the leukocytes of rats pretreated with SkQ1 before the HBO session, and increase in total GPx activity coincided with the increase in transcriptional activity of the GPx4 gene. It was shown previously [13] that the administration of SkQ1 promotes significant activation of antioxidant enzymes catalase, GPx, and GST under physiological conditions, whereas total SOD activity is maintained at a constant level.
The pretreatment with SkQ1 (50 nmol/kg for five days) did not cause statistically significant changes in activity of prooxidant enzymes NADPH-oxidase and myeloperoxidase (MPO) in the blood leukocytes of rats under physiological conditions. Hyperoxia-induced oxidative stress increased the activity of NADPH-oxidase and myeloperoxidase by 164 and 68%, respectively, when compared with control (Fig. 2).
Most mammalian cells contain NADPH-oxidase (NOX) enzymatic complex that produces superoxide anion radical O2‾• by transfer of electrons from intracellular NADPH to molecular O2. Among seven known enzymatic isoforms, leukocytes express NOX1, NOX2 (mostly in the neutrophils and monocytes), NOX4, and NOX5 (T- and B-lymphocytes) [24]. A study [25] demonstrated that hyperoxia occurs together with increase in ROS/O2‾• level, and ROS generation via NADPH-oxidase is a major pathway for HBO-induced lung damage. It can be supposed that the activation of prooxidant enzymes NADPH-oxidase and myeloperoxidase in the blood leukocytes of rats is observed in hyperoxia, and that it contributes to ROS production under these conditions. The pretreatment with SkQ1 for five days before the HBO session promotes normalization of the activity of prooxidant enzymes in the blood leukocytes of rats (Fig. 2).
DISCUSSION
The results showed that acute hyperoxia-induced oxidative stress leads to suppression of expression of the Nrf2 gene and ARE-controlled genes SOD1, CAT, and GPx4. The expression of transcription factor Nrf2 (NF-E2-related factor 2) gene is known to occur in the cells of different types [12, 26, 27]. Nrf2 is a cap’n’collar (CNC) basic-region leucine zipper (bZIP) DNA-binding protein [28]. Under physiological conditions, Nrf2 level is rather low, because it binds with Keap1, is ubiquitinated by ubiquitin E3 ligase, and is decomposed by the proteasome system [27]. Activation of this system is known to be accompanied by disturbance of DLG-mediated binding of Nrf2 to Keap1, but high affinity ETGE-binding keeps the proteins together. As a result, de novo synthesized Nrf2 molecules are translocated to the nucleus, interact with Maf proteins and coactivator proteins, and bind to cis-acting antioxidant response elements (ARE) located in the promoters of redox-sensitive genes; these processes significantly increase expression of these genes [26, 27, 29].
Nrf2 has cytoplasmic and mitochondrial pools [30]. It has been demonstrated that the ternary complex containing phosphoglycerate mutase (PGAM5), Nrf2, and Keap1 is targeted to the outer membrane of mitochondria and may play an important role in signal transmission to the nucleus under the conditions of disturbed redox-homeostasis and mitochondrial dysfunction, facilitating coordination between mitochondria and the expression of Nrf2-controlled genes encoding antioxidant enzymes. Previous studies revealed that Nrf2 supports the structural and functional integrity of mitochondria, especially under conditions of oxidative stress [31].
The present study demonstrates changes in gene expression profiles in the blood leukocytes of rats under HBO-induced oxidative stress; this is in accordance with other results. Oxidative stress is known to activate gene expression programs consisting of variety of genes [27, 32]. Research [23] has shown that hyperoxia (95-99% O2) leads to the stimulation of expression of 175 genes and inhibition of transcriptional activity of approximately 100 genes. Chromosome 2 has been identified to include locus 1, susceptible to hyperoxia, and containing candidate gene Nrf2 that encodes transcription factor Nrf2 [22]. Nrf2-knockout homozygous mice (Nrf2–/–) were shown to be more susceptible to lung oxygen toxicity when compared to wild-type animals (Nrf2+/+). A study [33] demonstrated that Nrf2 promotes survival and inhibits the suppression of alveolus growth in Nrf2+/+ neonatal mice exposed to hyperoxia (at 80-90% O2 for 24 h) when compared to Nrf2–/– animals. The stimulation of Nrf2-controlled genes glutathione peroxidase 2 and NAD(P)H:quinone oxidoreductase was observed in the lungs of Nrf2+/+ mice, in contrast to transcription factor knockout mice.
Considering possible causes of the inhibition of transcriptional activity of the Nrf2 gene under acute hyperoxia-induced oxidative stress, it should be noted that the promoter of the Nrf2 gene has two ARE-like sequences, which points to the capacity to activate its own expression [34] and significantly increase susceptibility and activity of signal pathway Keap1/Nrf2/ARE. To date, at least two alternative pathways of Nrf2 regulation have been reported – Keap1-dependent and Keap1-independent, including the phosphorylation of a transcription factor by various protein kinases (PKC, PI3K/Akt, GSK-3β, JNK), interaction with other protein partners (p21, caveolin-1), and epigenetic modification (miRNA, promoter methylation) [35]. All these processes are potentially important for the activation of Nrf2 and may be disturbed in HBO-induced oxidative stress. Evaluating the contribution of different changes in the inhibition of transcriptional activity of Nrf2, it should be noted that acute hyperoxia-induced oxidative stress could change cellular redox-homeostasis via thiol oxidation of critical redox-sensitive proteins, including Keap1 and Nrf2 that play the key role. Some studies [36, 37] have shown that mouse Keap1 contains ~25 cysteine residues, and Nrf2 has only seven cysteines; they are highly conserved and play a crucial role in the functioning of the signal system Keap1/Nrf2 signaling system. Research [36] has revealed that mutations of Nrf2 cysteines enhanced Nrf2–Keap1 association, ubiquitination of Nrf2, and proteasomal degradation of Nrf2. Besides, mutations at Cys119, Cys235, and Cys506 have been demonstrated to reduce the binding of Nrf2 to endogenous ARE and to coactivator CBP/p300 [36].
Evidently, suppression of various pathways of Nrf2 expression can occur under conditions of HBO-induced oxidative stress. The main pathways are expected to be the disturbances of the “cysteine code” [36] of transcription factor Nrf2 and Keap1 via oxidative modification of cysteine residues in the proteins by increased level of ROS under hyperoxia [38]. This process may cause conformational changes in Nrf2 and Keap1 that enhances ubiquitination and proteasomal degradation of Nrf2. This prevents nuclear translocation of Nrf2 and subsequent activation of expression of Nrf2 and Nrf2-controlled genes encoding antioxidant enzymes. Oxidative modifications of cysteine residues in the proteins Keap1 and Nrf2 under hyperoxia may be linked with enhanced production of hypochlorite and superoxide anion radical due to activation of MPO and NADPH-oxidase demonstrated in the present study. Reports [7, 8] have revealed that mitochondrial dysfunction significantly triggers ROS overproduction in hyperoxia. Experiments involving transgenic animals have shown that Nrf2 plays a crucial role in the regulation of mitochondrial redox homeostasis [31]. Deficiency of transcription factor Nrf2 has been shown to disturb the functioning of complex I and to lead to ROS overproduction.
Disbalance of functioning of antioxidant enzymes may intensify free radical oxidation in hyperoxia. Inhibition of catalase, GST, and GPx is observed at the same time with SOD activation, which results in hydroperoxide accumulation, onset of the Fenton and Osipov reactions, and overproduction of toxic hydroxyl radicals.
Pretreatment with SkQ1 for five days before an HBO session enhanced transcriptional activity of the Nrf2 gene and SOD2 and GPx4 genes, their protein products having mitochondrial localization in the blood leukocytes of rats. In addition, SkQ1 administration resulted in normalization of expression levels of SOD1 and CAT that were decreased under oxidative stress conditions in animals that which were not pretreated with SkQ1.
A study [13] showed previously that under physiological conditions SkQ1 is a positive mediator of transcriptional activity of Nrf2 and Nrf2-controlled genes encoding antioxidant enzymes, which enhances the antioxidant potential of leukocytes in normoxia. The protective effect of SkQ1 in HBO-induced oxidative stress might be realized via direct antioxidant properties or indirectly by stimulation of the Keap1/Nrf2/ARE signaling system. The administration of mitochondria-targeted antioxidant and changes in expression profiles of Nrf2 and Nrf2-controlled genes encoding antioxidant enzymes occur together with changes in their activity in the blood leukocytes of rats in hyperoxia. Under these conditions, the activity of SOD and GST was found to be normal and the activity of catalase and GPx was found to increase. Pretreatment with SkQ1 normalized the activity of prooxidant enzymes NADPH-oxidase and myeloperoxidase, which was significantly higher in hyperoxia.
The present study shows that pretreatment with SkQ1 (50 nmol/kg for five days) before HBO enhances the expression of the Nrf2 gene and Nrf2-controlled genes encoding antioxidant enzymes (SOD2, GPx4) and normalizes the transcriptional activity of SOD1 and CAT genes in hyperoxia-induced oxidative stress. It also increases the activity of catalase and GPx and normalizes SOD and GST activity in hyperoxia-induced oxidative stress.
This work was financially supported by the Ministry of Education and Science of the Russian Federation (project No. 213.01-11/2014-32).
REFERENCES
1.Valko, M., Leibfritz, D., Moncol, J., Cronin, M.
T., Mazur, M., and Telser, J. (2007) Free radicals and antioxidants in
normal physiological functions and human disease, Int. J. Biochem.
Cell Biol., 39, 44-84.
2.Sies, H. (2015) Oxidative stress: a concept in
redox biology and medicine, Redox Biol., 4, 180-183.
3.Zhivotovsky, B., and Orrenius, S. (2010) Cell death
mechanisms: cross-talk and role in disease, Exper. Cell Res.,
316, 1374-1383.
4.Ay, H., Topal, T., Ozler, M., Uysal, B., Korkmaz,
A., Oter, S., Ogur, R., and Dundar, K. (2007) Persistence of hyperbaric
oxygen-induced oxidative effects after exposure in rat brain cortex
tissue, Life Sci., 80, 2025-2029.
5.Berkelhamer, S. K., Kim, G. A., Radder, J. E.,
Wedgwood, S., Czech, L., Steinhorn, R. H., and Schumacker, P. T. (2013)
Developmental differences in hyperoxia-induced oxidative stress and
cellular responses in the murine lung, Free Radic. Biol. Med.,
61, 51-60.
6.Mathieu, D. (2009) Handbook of Hyperbaric
Medicine [Russian translation], BINOM, Laboratoriya Znanii,
Moscow.
7.Das, K. C. (2013) Hyperoxia decreases glycolytic
capacity, glycolytic reserve and oxidative phosphorylation in MLE-12
cells and inhibits complex I and II function, but not complex IV in
isolated mouse lung mitochondria, PLoS One, 8,
e73358.
8.Resseguie, E. A., Staversky, R. J., Brookes, P. S.,
and O’Reilly, M. A. (2015) Hyperoxia activates ATM independent
from mitochondrial ROS and dysfunction, Redox Biol., 5,
176-185.
9.Skulachev, V. P. (2007) A biochemical approach to
the problem of aging: “megaproject” on membrane-penetrating
ions. The first results and prospects, Biochemistry (Moscow),
72, 1385-1396.
10.Plotnikov, E. Y., Silachev, D. N., Chupyrkina, A.
A., Danshina, M. I., Jankauskas, S. S., Morosanova, M. A., Stelmashook,
E. V., Vasileva, A. K., Goryacheva, E. S., Pirogov, Y. A., Isaev, N.
K., and Zorov, D. B. (2010) New-generation Skulachev ions exhibiting
nephroprotective and neuroprotective properties, Biochemistry
(Moscow), 75, 145-150.
11.Niture, S. K., Khatri, R., and Jaiswal, A. K.
(2014) Regulation of Nrf2 – an update, Free Radic. Biol.
Med., 66, 34-36.
12.Forman, H. J., Davies, K. J. A., and Ursini, F.
(2014) How do nutritional antioxidants really work: nucleophilic tone
and para-hormesis free radical scavenging in vivo, Free
Radic. Biol. Med., 66, 24-35.
13.Vnukov, V. V., Gutsenko, O. I., Milutina, N. P.,
Ananyan, A. A., Danilenko, A. O., Panina, S. B., and Kornienko, I. V.
(2015) Influence of SkQ1 on expression of Nrf2 transcription factor
gene, ARE-controlled genes of antioxidant enzymes, and their activity
in rat blood leukocytes, Biochemistry (Moscow), 80,
586-591.
14.Lukash, A. I., Vnukov, V. V., Ananyan, A. A.,
Milutina, N. P., and Kvasha, P. N. (1996) Metal-Containing
Substances of Blood Plasma in Hyperbaric Oxygenation (Experimental and
Clinical Aspects) [in Russian], RSU Publishers, Rostov-on-Don.
15.Boyum, A. (1968) Separation of leukocytes from
blood and bone marrow, Scand. J. Clin. Lab. Invest. Suppl.,
97, 77-89.
16.Sirota, N. V. (1999) New approach in studies of
adrenalin autoxidation and its use in measurements of superoxide
dismutase activity, Vopr. Med. Khim., 3, 14-15.
17.Korolyuk, M. A., Ivanova, L. I., Maiorova, I. G.,
and Tokarev, V. E. (1988) Method for measurements of catalase activity,
Lab. Delo, 1, 16-19.
18.Moin, V. M. (1986) Simple and specific approach
for determination of glutathione peroxidase activity in erythrocytes,
Lab. Delo, 12, 724-727.
19.Habig, W. H., Pabst, M. J., and Jacoby, W. B.
(1974) Glutathione-S-transferase: the first step in mercapturic acid
formation, J. Biol. Chem., 249, 7130-7139.
20.Saidov, M. Z., and Pinegin, B. V. (1991)
Spectrophotometric method of myeloperoxidase assay in phagocytic cells,
Lab. Delo, 3, 56-59.
21.Dluzhevskaya, T. S., Pogorelova, T. N., and
Afonin, A. A. (1989) NADPH-oxidase activity in determination of newborn
health status, Pediatriya, 3, 44-47.
22.Cho, H.-Y., Jedlicka, A. E., Reddy, S. P.,
Kensler, T. W., Yamamoto, M., Zhang, L. Y., and Kleeberger, S. R.
(2002) Role of NRF2 in protection against hyperoxic lung injury in
mice, Am. J. Respir. Cell Mol. Biol., 26, 175-182.
23.Cho, H.-Y., Reddy, S. P., De Biase, A., Yamamoto,
M., and Kleeberger, S. R. (2005) Gene expression profiling of
NRF2-mediated protection against oxidative injury, Free Radic. Biol.
Med., 38, 325-343.
24.Pendyala, S., and Natarajan, V. (2010) Redox
regulation of Nox proteins, Respir. Physiol. Neurobiol.,
174, 265-271.
25.Pendyala, S., Gorshkova, I. A., Usatyuk, P. V.,
He, D., Pennathur, A., Lambeth, J. D., Thannickal, V. J., and
Natarajan, V. (2009) Role of Nox4 and Nox2 in hyperoxia-induced
reactive oxygen species generation and migration of human lung
endothelial cells, Antioxid. Redox. Signal., 11,
747-764.
26.Kaspar, J. W., Niture, S. K., and Jaiswal, A. K.
(2009) Nrf2: INrf2 (Keap1) signaling in oxidative stress, Free
Radic. Biol. Med., 47, 1304-1309.
27.Ma, Q. (2013) Role of Nrf2 in oxidative stress
and toxicity, Annu. Rev. Pharmacol. Toxicol., 53,
401-426.
28.Hayes, J. D., and Dinkova-Kostova, A. T. (2014)
The Nrf2 regulatory network provides an interface between redox and
intermediary metabolism, Trends Biochem. Sci., 39,
199-216.
29.Taguchi, K., Motohashi, H., and Yamamoto, M.
(2011) Molecular mechanisms of the Keap1–Nrf2 pathway in stress
response and cancer evolution, Genes Cells, 16,
123-140.
30.Lo, S.-C., and Hannink, M. (2008) PGAM5 tethers a
ternary complex containing Keap1 and Nrf2 to mitochondria, Exp. Cell
Res., 14, 1789-1803.
31.Dinkova-Kostova, A. T., and Abramov, A. Y. (2015)
The emerging role of Nrf2 in mitochondrial function, Free Radic.
Biol. Med., doi: 10.1016/j.freeradbiomed.2015.04.036.
32.Ma, Q. (2010) Transcriptional responses to
oxidative stress: pathological and toxicological implications,
Pharmacol. Ther., 125, 376-393.
33.McGrath-Morrow, S., Lauer, T., Yee, M., Neptune,
E., Podowski, M., Thimmulappa, R. K., O’Reilly, M., and Biswal,
S. (2009) Nrf2 increases survival and attenuates alveolar growth
inhibition in neonatal mice exposed to hyperoxia, Am. J. Physiol.
Lung Cell. Mol. Physiol., 296, 565-573.
34.Kwak, M.-K., Itoh, K., Yamamoto, M., and Kensler,
T. W. (2002) Enhanced expression of the transcription factor Nrf2 by
cancer chemopreventive agents: role of antioxidant response
element-like sequences in the nrf2 promoter, Mol. Cell Biol.,
22, 2883-2892.
35.Bryan, H. K., Olayanju, A., Goldring, C. E., and
Park, B. K. (2013) The Nrf2 cell defense pathway: Keap1-dependent and
-independent mechanisms of regulation, Biochem. Pharmacol.,
85, 705-717.
36.He, X., and Ma, Q. (2009) NRF2 cysteine residues
are critical for oxidant/electrophile-sensing, Kelch-like
ECH-associated protein-1-dependent ubiquitination-proteasomal
degradation, and transcription activation, Mol. Pharmacol.,
76, 1265-1278.
37.Vnukov, V. V., Milutina, N. P., Ananyan, A. A.,
Danilenko, A. O., Gutsenko, O. I., and Verbitsky, E. V. (2013) The
influence of plastoquinone cation derivative –
10(6′-plastoquinonyl)decyltriphenylphosphonium (SkQ1) – on
the apoptosis intensity and structural state of rat lymphocyte
membranes under oxidative stress induced by the hyperbaric oxygenation,
Vestnik SSC RAN, 9, 78-86.
38.Takaya, K., Suzuki, T., Motohashi, H., Onodera,
K., Satomi, S., Kensler, T. W., and Yamamoto, M. (2012) Validation of
the multiple sensor mechanism of the Keap1–Nrf2 system, Free
Radic. Biol. Med., 53, 817-827.