REVIEW: Role of Small Noncoding RNAs in Bacterial Metabolism
T. L. Azhikina1*, D. V. Ignatov1, E. G. Salina2, M. V. Fursov2, and A. S. Kaprelyants2
1Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia; E-mail: tatazhik@ibch.ru2Bach Institute of Biochemistry, Federal Research Centre “Fundamentals of Biotechnology”, Russian Academy of Sciences, 119071 Moscow, Russia
* To whom correspondence should be addressed.
Received June 5, 2015; Revision received August 17, 2015
The study of prokaryotic small RNAs is one of the most important directions in modern molecular biology. In the last decade, multiple short regulatory transcripts have been found in prokaryotes, and for some of them functional roles have been elucidated. Bacterial small RNAs are implicated in the regulation of transcription and translation, and they affect mRNA stability and gene expression via different mechanisms, including changes in mRNA conformation and interaction with proteins. Most small RNAs are expressed in response to external factors, and they help bacteria to adapt to changing environmental conditions. Bacterial infections of various origins remain a serious medical problem, despite significant progress in fighting them. Discovery of mechanisms that bacteria employ to survive in infected organisms and ways to block these mechanisms is promising for finding new treatments for bacterial infections. Regulation of pathogenesis with small RNAs is an attractive example of such mechanisms. This review considers the role of bacterial small RNAs in adaptation to stress conditions. We pay special attention to the role of small RNAs in Mycobacterium tuberculosis infection, in particular during establishment and maintenance of latent infection.
KEY WORDS: bacteria, small noncoding RNAs, Hfq, regulation of gene expression, stress, virulence, Mycobacterium tuberculosisDOI: 10.1134/S0006297915130015
Regulatory noncoding RNAs are one of the functional elements in any prokaryotic cell. A number of cell metabolic pathways are regulated by these molecules. The first prokaryotic regulatory RNAs were found long before the discovery of eukaryotic microRNAs (miRNAs) and small interfering RNAs (siRNAs). By 2001, 11 small RNAs of E. coli were identified that were transcribed from intergenic regions of the genome, and most of them were discovered by accident [1]. A new era in the study of regulatory RNAs began in 2001-2002 with the development of revolutionary bioinformatics methods for searching small RNA candidates based on comparative analysis of genomes of closely related bacterial species. Several hundreds of bacterial small regulatory RNAs are now known.
Regulatory RNAs of bacteria can be divided into riboswitches, small noncoding RNAs, and CRISPR-RNAs. The term “riboswitches” is used for sequences located at 5′-end, or more rarely at 3′-end, region of an mRNA that can change conformation in response to environmental signals or presence of a specific ligand and thus regulate transcriptional activity. CRISPR-RNAs (clustered regularly interspaced short palindromic repeats) are sequences partially complementary to fragments of bacteriophage genomes and regions of plasmid DNAs. They render bacteria resistant to viruses and disable plasmid conjugation. To be familiarized with these types of noncoding RNAs in detail, we recommend the reviews [2-4].
Small noncoding RNAs form the most numerous group of regulatory RNAs. Their functions include modulation of RNA-polymerase activity, regulation of stability of mRNA, its translation, etc. After transcription, most small RNAs are subject to processing with the removal of extra residues at the 5′- and/or 3′-end [5]. Small noncoding RNAs can be divided into three large classes: (i) antisense RNAs interacting with target mRNAs, adjusting their translational activity and/or stability; (ii) modifying protein activity; (iii) structural RNAs that participate in so-called “housekeeping” processes. For example, 4.5S RNA and tmRNA can be included into the last group. In this review, we will focus on small antisense RNAs.
SMALL ANTISENSE RNAs
The mechanism of antisense RNA action is based on their complementary binding to mRNA-targets. Depending on mutual location of genes of small RNAs and their targets, cis- and trans-encoded antisense RNAs are distinguished.
Cis-encoded antisense RNAs. Cis-encoded antisense RNAs are encoded at the same locus as their mRNA-targets, but on the opposite genome strand. Thus, fully complementary binding is achieved. Cis-encoded transcripts participate in regulation of such processes as translation and transcription, initiation of replication, plasmid conjugation, transposition, and mRNA degradation. They also control some cellular metabolism pathways. The simplest mechanism of cis-encoded small RNA action is blocking translation via complementary binding to the ribosomal binding site on the target mRNA.
The role of cis-encoded small RNAs is insufficiently studied. It is known that some of them participate in blocking of the expression of toxic proteins. An example is the small RNA RatA found in the Bacillus subtilis transcriptome. This cis-encoded RNA controls expression of TxpA toxin. It was shown that in mutant B. subtilis strain cells deleted for promoter and 5′ leader regions of the ratA gene, TxpA level in cytoplasm is significantly increased. Despite only partial complementarity between RatA RNA and txpA gene transcript (the overlapping region reaches up to 75 nucleotides), it was established that formation of duplex between the two RNA molecules occurs without the participation of mediator proteins [6]. Later, both RNAs are degraded by ribonucleases RNase Y and RNase III [7]. Another example is the SymR-SymE system in E. coli, which consists of two genes: symR (a small RNA) and symE (a SOS-induced toxin). Upon elevation of SymE cellular concentration, synthetic activity of ribosomes drops. SymR RNA plays a role in negative regulation of this toxin gene: it complementarily binds to the mRNA transcribed from the symE gene, thus hindering translation of the latter [8].
Other cis-antisense RNAs can modulate expression within operons. For example, the small RNA GadY of E. coli triggers cleavage of gadXW mRNA transcript into gadX and gadW. GadX is a transcription factor that activates expression of glutamate decarboxylases GadA and GadB, and this scheme is a part of an E. coli defense system against acidic stress and the first described example of positive influence of small RNA on the accumulation of regulated mRNA [9]. In some cases, cis-antisense RNAs can bind mRNAs and terminate transcription after the binding site, thus hindering expression of associated genes [10].
Trans-encoded antisense RNAs. Genes of trans-encoded antisense RNAs are located in genome regions distant from a regulated gene. Lengths of these RNAs vary from 50 to 300 nucleotides. Trans-encoded RNAs are synthesized in bacteria in response to various stress factors (see more details in the third part of this review). Most are transcribed from independent promoters that do not differ significantly from promoters of other bacterial genes.
Such small RNAs are only partially complementary to targets. In view of this, every such regulatory transcript is potentially able to interact with mRNA of many genes. Since a complementary region usually does not exceed 25 nucleotides, this regulation mechanism is very sensitive to single nucleotide replacements. For example, only four nucleotide changes can affect activity of the small RNA SgrS that controls repression of the ptsG gene encoding a glucose-6-phosphate transporter [11].
Most trans-encoded antisense RNAs require special chaperones to stabilize the mRNA binding. One of the best-studied chaperones of this kind is the Hfq protein [12]. At least 40% of small RNAs bind to Hfq in E. coli [13]. Hfq was first identified as a protein essential for replication of Qβ phage in E. coli (Hfq, host factor Qβ) [14]. The amino acid sequence and structure of Hfq (a hexameric ring) indicate its resemblance to eukaryotic Sm-proteins that are components of spliceosomes [15]. Deletion of the gene of this protein leads to negative consequences for growth and viability of bacteria under various stress conditions, e.g. osmotic shock and oxidative stress. It was also established that Hfq is an essential virulence factor of pathogens belonging to genera Brucella, Vibrio, Listeria, Salmonella, etc. [16-18].
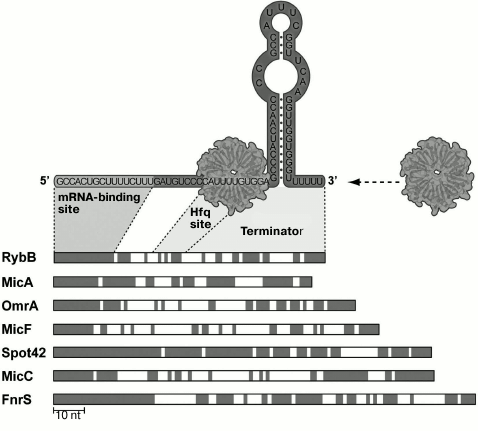
Hfq can regulate decay of some mRNAs by competing with the ribosome and exposing an RNase E cleavage site on this mRNA [19]. However, the main function of Hfq protein is its participation in trans-encoded RNA binding to target mRNAs, which also influences stability or translation of mRNA. In the structure of RNAs binding to Hfq, three domains can be distinguished (Fig. 1): a hairpin at the 3′-end provides Rho-independent transcription termination and protects a small RNA from the action of 3′ endonucleases; another domain, Hfq-binding site, provides functioning and stability of a small RNA; a third region (the so-called “seed region”) is required for binding to a target mRNA. In a complex with small RNA, Hfq binds to A/U-rich single-stranded mRNA regions, improving a complementary interaction between mRNA and small RNAs.
Fig. 1. Structural elements of Hfq-binding trans-encoded RNAs exemplified by some small RNAs of enterobacteria (modified from [133]). The most conserved areas (regions shown in gray) correspond to a small RNA region complementarily interacting with mRNA (“seed region”). Regions of Hfq binding and Rho-independent transcription terminator are indicated.
The reasons for Hfq requirement for binding of small trans-encoded RNAs to target RNAs remain unknown. There are two hypotheses. First, Hfq may be a “platform” for interaction of trans-encoded RNAs with target RNAs. In other words, binding to Hfq increases a local concentration of these transcripts, which increases probability of duplex formation between them. The second hypothesis is based on the assumption that interaction of RNAs with Hfq affects their secondary structure: in a complex with the protein, the transcripts adopt conformations that are more appropriate for the complementary interactions as compared to the free state [20].
It was established that degradation time for most trans-encoded E. coli RNAs decreases significantly in the absence of Hfq chaperone. Formation of an RNA–protein complex protects short regulatory transcripts from degradation by ribonucleases, RNase E in particular. This enzyme has an endonuclease activity and cleaves single-stranded regions of an RNA, thus it performs not only degradation, but also processing of certain transcripts [21]. The participation of the RNase E in cleavage of trans-encoded RNAs was first demonstrated in studies on the physiological activity of the small RNA RyhB [22]. The C-terminal domain of RNase E can bind to the RhlB helicase, polynucleotide phosphorylase PNPase, and enolase, thus forming a protein complex, the so-called degradosome. Degradosome components assist in the complete cleavage and destruction of small RNA/mRNA duplexes [23].
There are various known mechanisms of action of Hfq-dependent trans-encoded small RNAs on target mRNAs.
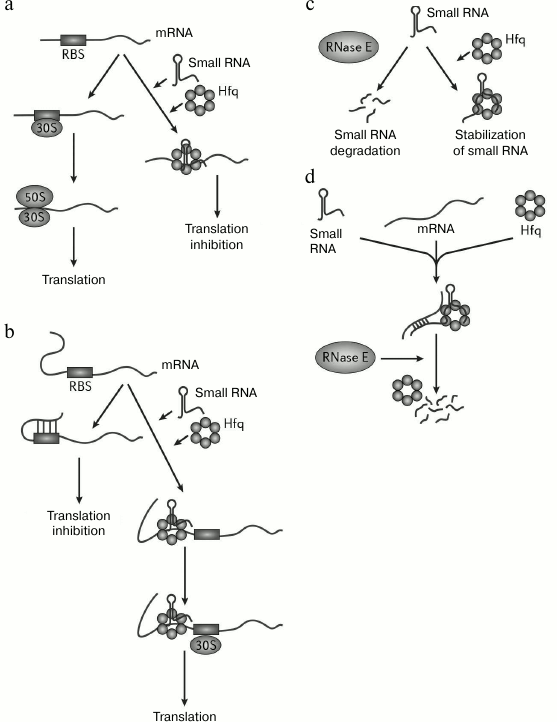
1) Inhibition of mRNA translation by blocking ribosomal binding sites with small RNA (Fig. 2a). Negative regulation of expression of the E. coli ptsG gene is an example of this kind of regulation. Small RNA SgrS with the help of Hfq blocks a ribosomal binding site on ptsG mRNA, encoding one of the glucose transporters of the phosphoenolpyruvate–phosphotransferase system. Such interaction impedes translation of this mRNA. Then, the SgrS–ptsG complex is degraded by RNase E by a mechanism that will be reviewed below [5].
2) Activation of an mRNA translation because of destruction of a secondary structure hiding the ribosomal binding site (Fig. 2b). This mechanism unusual for small RNAs is exemplified by the functioning of the σ-factor RpoS. Translation of this factor is regulated by small RNAs DsrA and RprA that interact with a 5′ leader sequence of rpoS mRNA. As a result, formation of a secondary structure that hides a ribosomal binding site becomes impossible, and translation levels of rpoS mRNA increases [24].
3) Stabilization of an mRNA caused by complementary interaction of an mRNA with a small RNA (Fig. 2c). It has been repeatedly shown that small RNAs are very labile while not stabilized by binding to Hfq. So, the half-life for the small RNA LhrA (Listeria monocytogenes) in wild type strains is more than 30 min, while it drops to 3 min in an hfq mutant strain [25]. Small RNAs MicA, GlmY, RyhB, and SgrS are shortened from the 3′-end in the absence of Hfq [26].
4) Small RNA-induced decay of an mRNA (Fig. 2d). Such mechanism was first demonstrated for the trans-encoded small RNA RyhB, triggering degradation of several mRNAs. Binding of RyhB to target mRNAs causes their degradation by RNase E, which forms various ribonucleoprotein complexes with Hfq and small RNAs using its C-terminal domain. Such complexes function as initiators for degradation of these mRNAs [27]. Despite the fact that complete degradation of an mRNA is the most frequent result, RyhB binding can lead to selective degradation of a polycistronic mRNA. It is supposed that in this case mRNAs carry additional information that determines their fate upon RyhB binding. In this system, Hfq protein increases efficiency of binding of a small RNA/target mRNA (changes the mRNA secondary structure that hinders binding), stabilizes small RNAs, protects mRNAs from degradation in the absence of RyhB, and attracts RNase E upon complex formation between RyhB and mRNA.
Fig. 2. Mechanisms of action of Hfq (modified from [12]). a) Complex of Hfq with trans-encoded small RNA blocks the ribosomal binding site (RBS); b) complex of Hfq with trans-encoded small RNA inhibits formation of a secondary structure in the 5′-UTR (untranslated mRNA region) that blocks the ribosomal binding site; c) complex of Hfq with trans-encoded small RNA protects small RNA from degradation by ribonucleases; d) complex of Hfq with trans-encoded small RNA can induce cleavage of RNA–RNA duplexes formed by a small RNA and an mRNA.
It is worth mentioning that an Hfq protein was not found in all bacteria. For example, its presence is not shown for such ε-proteobacteria as Helicobacter pylori and Campylobacter jejuni [28], though these species encode many small RNAs. Hfq or its counterparts were also not found in mycobacteria [29]. It is suggested that the presence of long regions of complementarity in the target mRNA molecule as well as an increased concentration of short regulatory RNAs under certain conditions can increase likelihood of binding of these RNAs with their target RNAs in the absence of Hfq. There are examples of such regulation, for instance, small RNAs of Staphylococcus aureus have several trans-encoded targets and interact with them without the participation of Hfq or its counterparts [30]. In hfq mutant strains of Vibrio cholerae, expression of an ompA gene is blocked by the trans-encoded small RNA VrrA [31]. In addition to Hfq, some other proteins can also act as an RNA-chaperone, e.g. E. coli protein ProQ [32] and YbeY of Sinorhizobium meliloti [33].
SMALL NONCODING RNA TARGETS AND THEIR ROLE IN REGULATION OF
CELLULAR PROCESSES
Despite the fact that the number of discovered small RNAs in bacterial cells reaches several hundred, only for some are molecular targets and consequences of interaction of these small RNAs with corresponding targets known. Recently published reviews contain comprehensive information regarding this subject [34, 35]. Summarizing the available data, it becomes clear that small RNAs in bacterial cells are involved both in regulation of various metabolic links in bacterial cells, and in certain processes of adaptation to environmental changes. The most represented group of characterized bacterial small noncoding RNAs comprises small RNAs regulating cell response to various stress factors. Examples of such small RNAs are listed below.
Nutrient deficiency. In E. coli cells, the RNA-binding regulatory protein CsrA plays an important role under carbon source starvation. CsrA binds to mRNAs of several genes including a repressor of glycogen synthesis, cstA protein, and inhibits their translation [36]. The CsrA protein as such is inhibited by binding of small RNAs CsrB and CsrC, whose secondary structures contain hairpins mimicking CsrA binding sites on its target mRNAs [37]. The competition for CsrA between these small RNAs and target mRNAs leads to repression of glycolysis and activation of gluconeogenesis [38]. Transcription of the csrB and csrC genes is triggered by two-component regulators BarA–UvrB upon a shift to a nutrient-depleted environment [39]. Homologs of CsrB and CsrC (RsmY, RsmZ) are found in many bacterial genera (Salmonella, Erwinia, Yersinia, Vibrio, etc.), in which they affect secondary metabolism by interacting with CsrA counterparts [40-42].
Under nutrient deficiency, in bacteria belonging to genera Staphylococcus, Macrococcus, and Bacillus, transcription of the small noncoding RNA RsaE is induced at the end of exponential growth. The secondary structure of this small RNA contains two hairpins divided by a 17-nucleotide sequence. Because of this, RsaE RNA can prevent formation of ribosomal complex on two target mRNAs, opp3B and opp3A, within a single locus opp3 that encodes proteins of the system of peptide and amino acid transport. RsaE also decreases activity of key proteins of the tricarboxylic acid cycle (e.g. [https://www.google.ru/search?newwindow=1&q=succinyl-CoA+synthetase+suB&spell=1&sa=X&ved=0CBkQBSgAahUKEwjxjo290pfIAhWhSXIKHYqVCpg succinyl-CoA synthetase ]SucB) and the purine biosynthesis cycle, which promotes adaptation of S. aureus cells to low concentration of nutrients [30, 43].
During amino acid starvation or in response to bactericidal action of polymyxins disrupting the structure of a cell wall in Salmonella enterica cells, the activation of stress sigma factor σS occurs, which controls synthesis of the small noncoding RNA SdsR. SdsR, in a complex with Hfq protein, decreases expression levels of the outer membrane protein OmpD [44], which is the prevailing porin in S. enterica [45]. Presumably, decrease in the outer membrane permeability prevents “leakage” of low molecular weight compounds (including amino acids) from cells.
Hfq-associated small noncoding RNA GcvB plays an important role in the physiological response of E. coli and S. typhimurium to amino acid starvation. Transcription of the gcvB gene is activated by GcvA protein at high intracellular levels of glycine and is repressed under glycine deficiency. Small RNA GcvB represses synthesis of OppA and DppA proteins (components of a transport system for small peptides, polar and branched-chain amino acids, toxins, and antibiotics). Thus, it can prevent influx of toxic compounds into the cell [46-52]. Interestingly, small noncoding RNA GcvB contains two sequences that bind to a corresponding target mRNA [51-53].
Under glucose limitation, cellular cAMP levels in E. coli and S. typhimurium cells is increased, which activates expression of the small noncoding RNA CyaR (earlier known as RyeE) by the activator protein CRP (cAMP receptor protein). At the same time, Hfq-associated CyaR represses expression of the ompX gene encoding a protein that stimulates bacterial adhesion [54-56]. Repression of OmpX protein expression apparently improves “metabolic economy” by reducing excessive biosynthetic pathways [55].
Under iron deficiency and in case of inactivation of an iron uptake regulator Fur, which is a global iron-dependent transcription repressor, in Shigella dysenteriae cells an increase in expression of a small noncoding RNA RyhB is observed. This RNA in a complex with Hfq represses expression of a transcription activator VirB that, in turn, reduces levels of synthesis of a sodB gene mRNA encoding a superoxide dismutase. In E. coli cells, Hfq-associated RyhB also represses expression of sdhCDAB operon that encodes a succinate dehydrogenase and acnA and fumA genes that encode the tricarboxylic acid cycle enzymes aconitase and fumarase. This “balances” central metabolic pathways, including iron-containing and non-containing enzymes and ftnA and bfr genes that encode ferritin [57-59].
In iron-reach media, Fur protein in Pseudomonas aeruginosa cells represses expression of two small regulatory RNAs encoded by prrF1 and prrF2 genes. These small RNAs repress genes of an antABC operon carrying the genes for enzymes that cleave anthranilate, which is a signal quinolone precursor in Pseudomonas (PQS). Small PrrF RNAs are functional homologs of small noncoding RNA RyhB, as they reduce levels of sodB, sdhCDAB, ftnA, and bfr mRNAs [60].
In Neisseria meningitidis cells an Hfq-associated, small RNA NrrF was discovered that reduces expression levels of sdhCDAB genes during cultivation in iron-limited media similarly to RyhB in S. dysenteriae and PrrF in P. aeruginosa [61, 62]. Under the iron deficiency conditions in B. subtilis cells expression rate of a small noncoding RNA FsrA is increased that also blocks synthesis of a succinate dehydrogenase SdhCDAB [63].
Acidic stress. In the stationary phase, acidification of E. coli culture medium occurs. Under this circumstance, noncoding RNA GadY expression levels are increased, leading to an increase in synthesis rate of the mRNA of transcription activator GadX. GadX, in turn, activates transcription of genes gadA and gadB encoding glutamate dehydrogenases – proteins that reduce intracellular hydrogen ion concentration [9]. Survival of E. coli cells at low pH of the medium is also promoted by noncoding RNA GcvB, as it positively regulates transcription of the rpoS gene encoding the stress sigma factor σS [64].
Excessive accumulation of glucose 6-phosphate. Under certain conditions, in bacterial cell excessive accumulation of glucose 6-phosphate (G6P) or the non-metabolized glucose analog methyl-glucoside 6-phosphate (MG6P) occurs, which causes growth arrest [65] and cell death [66]. Hfq-associated small noncoding RNA SgrS (earlier known as RyaA) under glucose-phosphate stress in E. coli inhibits synthesis of PtsG protein, which is one of the main glucose transporters of the bacterial phosphoenolpyruvate–phosphotransferase system (PTS), preventing further G6P or MG6P accumulation in the cell [11, 67]. Regulation is achieved through a complementary interaction between SgrS and ptsG mRNA leading to translation inhibition and consequent degradation of the complex by RNase E [27, 68]. In addition, small RNA SgrS encodes SgrT peptide, which also inhibits activity of PtsG [5]. Another target regulated by SgrS RNA at posttranscriptional level is the manXYZ operon of a PTS system that encodes glucose and mannose protein transporters [69].
Aerobic-anaerobic shift stress. Hfq-associated small noncoding RNA FnrS, whose expression rate is increased under shift of E. coli from aerobic to anaerobic conditions, inhibits expression of genes encoding enzymes involved in respiration: malate dehydrogenase MaeA, ethanol dehydrogenase/reductase AdhP, D-lactate dehydrogenase Dld, which are required for aerobic growth of cells in lactate-containing media. The expression of some other genes is inhibited – those encoding isoenzyme of phosphoglycerate mutase Gpm that converts 3-phosphoglycerate into 2-phosphoglycerate, mRNA of sodB encoding superoxide dismutase that protects cells from superoxide radicals, and, finally, two mRNAs encoding enzymes participating in folic acid metabolism – dihydroneopterin triphosphate epimerase FolX and a GTP-dependent cyclohydrolase I FolE [70].
Under anaerobic conditions in Neisseria meningitidis cells, synthesis of the small noncoding RNA AniS occurs, which is triggered by transcription activator FNR. Hfq-associated small noncoding RNA AniS inhibits expression of the NMB0214 gene that encodes oligopeptidase PrlC. The exact cellular function of the PrlC protein remains unknown. However, it was shown that this protein is involved in processes of protein export and degradation and cell cycle regulation in E. coli [71-73].
Under oxygen deficiency in stationary phase, global regulator ANR-mediated activation of synthesis of the small noncoding RNA PhrS occurs in P. aeruginosa cells. The Hfq-bound small RNA PhrS is an activator of PqsR protein synthesis, which is a signal quinolone receptor [74].
Oxidative stress. In response to oxidative stress in E. coli cells, the small noncoding RNA OxyS is produced, which inhibits translation of the gene of the transcription activator of formate metabolism fhlA. Besides, OxyS represses expression of the rpoS gene [75-78].
Stationary phase stress. In the stationary phase, the number of transcripts of the ompA gene encoding an outer membrane protein is reduced in E. coli or S. typhimurium cells. This process is linked to expression of the Hfq-associated small noncoding RNA MicA, which causes degradation of ompA mRNA by duplex formation with the latter [79, 80]. Under these conditions, expression of the small noncoding RNA RybB is observed, which inhibits translation of outer membrane proteins OmpC and OmpW [81-83].
In V. cholerae cells in the stationary phase, synthesis of the σE-dependent small noncoding RNA VrrA is activated, which inhibits translation of ompA mRNA. It was also demonstrated that VrrA reduces V. cholerae virulence by inhibiting expression of the tcpA gene encoding a subunit of toxin-associated pili [31].
Quorum sensing. Besides the above-mentioned functions, small RNAs participate in quorum sensing – the ability of bacteria to exchange information between cells by means of extracellular signal molecules, autoinducers, in response to a change in environmental conditions. Vibrio cholerae cells respond to autoinducers using a two-component system associated with a membrane kinase that acts as a signal receptor [84]. Every such receptor transmits information to LuxU protein that, in turn, transmits signal to regulatory protein LuxO [85-87]. At low cell density, LuxO is phosphorylated and activates transcription of five small noncoding RNAs: Qrr1, Qrr2, Qrr3, Qrr4, and Qrr5 [88]. These small RNAs inhibit activity of their own regulator LuxO and translation of three targets that belong to the global pathogenicity regulation network in V. cholerae: the hapR gene encoding a transcription factor reducing activity of virulence genes [89], the aphA gene encoding a transcription factor inducing the expression of virulence genes [90], and the vca0939 gene encoding a protein that stimulates biofilm formation [91]. At high cell density, dephosphorylation of LuxO protein occurs. As a result, activation of small RNAs Qrr becomes impossible.
Synthesis of virulence factors in S. aureus cells is also regulated by means of a quorum sensing system. The small protein RAP (RNAIII-activating protein) acts an autoinducer. In the middle of the exponential growth phase, the concentration of RAP secreted by bacteria is increased, and this induces phosphorylation of its protein target TRAP (target of RNAIII-activating protein). TRAP phosphorylation leads to activation of the agr operon-comprising gene of the small RNA RNAIII. RNAIII induces expression of numerous virulence factors including α-, β-, γ-, and δ-hemolysins [92].
Role of small RNAs in development of infection caused by bacterial pathogens
Interaction of a pathogenic microorganism with a host can be imagined as a special case of a combination of different stress factors towards the pathogen. Therefore, it is no coincidence that small RNAs, as it was recently found, play an important role in development of the pathological process. It was found that in S. aureus culture in the early stationary phase, expression of the noncoding RNA SpdR is increased, which inhibits expression of Sbi protein that evades the action of the host’s immune system [93]. Small noncoding RNA RivX of Streptococcus pyogenes is co-expressed with a gene encoding regulatory protein RivR. This small RNA stimulates expression of genes of the Mga-regulon, which in turn activates expression of 10% of the genes of S. pyogenes genome, including virulence genes of peptidase ScpA, a secreted inhibitor the complement Sic, and the fibronectin-binding protein Fba, and collagen-like protein SclA [94, 95]. In S. pyogenes, small noncoding RNA FasX inhibits expression of two adhesin fibronectin-binding proteins (FBP54 and MRP) and positively affects activity of two secreted virulence factors (streptokinase and streptolysin S) [96]. FasX also controls the interaction of S. pyogenes cells with epithelial cells of the larynx [97, 98].
During mutational analysis of the pol locus (pleiotropic effect locus) of S. pyogenes carrying the streptolysin S gene (segA), data were obtained indicating that pol mRNA as such (regardless of translation) can be a regulator of expression a number of genes encoding virulence factors. Interestingly, regulation of expression of a number of genes occurs at a level of transcription (for instance, genes emm, sic, and nga), while expression of some other virulence factors (e.g. multifunctional protein SpeB) – at the posttranscriptional level [99].
In the pathogenic bacterium Shigella flexneri, the small noncoding RNA RnaG is a negative translation regulator for icsA mRNA encoding an outer membrane protein that facilitates colonization of the host by this bacterium [100].
Small RNAs in MYCOBACTERIA and their role in regulation of stress response and persistence
Characteristics of small RNAs. Small RNAs of mycobacteria, whose most important representative is Mycobacterium tuberculosis, attract special attention. The main feature of tuberculosis is prevalence of its latent form. Approximately 30% of the Earth’s population are carriers of latent M. tuberculosis infection and live under the constant risk of rapid development of acute infection [99]. The transition of the bacterial pathogen into metabolic shutdown (latency) occurs probably due to action of various stress factors caused by the host’s immune system during the active immune response. Reactivation of the latent form happens under the influence of incompletely understood environmental factors or low immune status, e.g. in patients with HIV [101, 102]. The molecular mechanisms of the reactivation of latent tuberculosis also remain obscure. Since small noncoding RNAs participate in an adaptive response to environmental stress conditions, one can assume that they play a role in the transition into a dormant state and development of latent infection.
Using high-throughput sequencing and computer algorithms, several dozen small RNAs were found in some species of mycobacteria [103-111]. However, determination of the role of these small RNAs in physiology of mycobacteria is a more difficult task. Only a few works have been published that elucidate the function of small RNAs in mycobacteria. The history of discovery and detailed list of known small RNAs in various species of mycobacteria can be found in a recent review by Haning and coworkers [107]. The description of different types of noncoding RNAs in M. tuberculosis, discussion of the absence of Hfq protein in M. tuberculosis, and the role of small RNAs in stress response and pathogenesis in M. tuberculosis can be found in a review by Arnvig and coauthors [29]. In our review, we focus on data related to functioning of intergenic small RNAs of M. tuberculosis, their role in infection development, and generation of the dormant state.
Recently, unified nomenclature was proposed for designation of the small noncoding RNAs in M. tuberculosis [112]. However, its use is not currently widespread. The nomenclature is based on relative position of loci encoding small RNAs among adjacent genes in the bacterial chromosome. In this connection, if the small RNA gene is located in the “minus” strand of the genome, a “–c” suffix as added to its name (for “complement”). Cis-encoded small RNAs are designated according to the protein gene with which they overlap. For instance, antisense RNA encoded on a “minus” strand of genome and overlapping gene Rv0539 is referred to as ncRv0539c. Trans-encoded small RNAs are designated according to name of the protein gene situated upstream of the small RNA gene. Number “1” added prior to the gene number indicates that this small RNA is a trans-encoded one. For example, a small RNA encoded on the “plus” strand of the genome downstream of the Rv0243 gene should be named ncRv10243. If several small RNAs are found in one locus relative to adjacent genes, denomination of each is supplemented with a Latin alphabet letter.
It is worth mentioning that in the literature there are designations that do not correspond to this nomenclature as they were proposed before its invention for M. tuberculosis small RNAs. So, for example, the small noncoding RNA Mcr11 reported in 2010 [106] is referred to as MTS0997 [104] and ncrMT1302 [113] in literature. All the variant designations of M. tuberculosis small RNAs found in the literature will be used below in this review.
The MTS194 RNA gene (F6, ncRv10243) was localized between genes Rv0243 and Rv0244, whose products are involved in lipid degradation [29]. Transcription of MTS194 is controlled by SigF, an auxiliary sigma factor activated during starvation [114]. Under oxidative stress caused by adding hydrogen peroxide to the medium, and during acidification of the medium, expression of MTS194 is increased [105]. Overexpression of MTS194 RNA in cells lowers the growth rate of M. tuberculosis cells, but it does not influence growth of Mycobacterium smegmatis cells [105]. Though MTS194 targets are still unknown, the most recent data indicate a role of MTS194 in stress response.
The Mcr7 RNA gene is located between genes Rv2395 and PE_PGRS41. Expression of this small RNA is controlled by the two-component signal system PhoPR [115]. By now, Mcr7 is the only small RNA in M. tuberculosis for which mRNA target was determined. Mcr7 RNA binds to tatC gene mRNA and hinders its translation. Binding occurs due to partial complementarity between the small RNA and a portion of the mRNA comprising predicted ribosomal binding site and first six codons. Gene tatC encodes a transmembrane protein, a component of the secretory complex Tat (twin arginine translocation). In M. tuberculosis, this complex secretes a number of proteins having a specific signal sequence with two arginines, e.g. the immunodominant complex Ag85 [116] and β-lactamase BlaC [117]. The putative regulation mechanism seems to be as follows: the two-component PhoPR system modulates expression of the small RNA Mcr7, which in turn inhibits translation of tatC mRNA. In the absence of TatC protein, secretory complex Tat becomes inactive, which results in reduced secretion of a number of proteins that are substrates for this complex [115].
The MTS0997 RNA gene (Mcr11, ncrMT1302, ncRv11264c) was localized in the region between genes Rv1264 and Rv1265. The expression of MTS0997 is increased during transition from exponential growth phase to stationary phase [104, 106, 113]. Furthermore, MTS0997 expression is significantly decreased during acidification of the medium, which probably shows the role of this small RNA in the stress response to low pH [113]. Interestingly, products of genes that flank MTS0997 participate in cAMP metabolism: Rv1264 encodes an adenylyl cyclase that is activated at low pH [118], and expression of Rv1265 is regulated by the cAMP-binding protein Cmr [119]. These data show the role of MTS0997 RNA in regulation with participation of cAMP [106, 113]. Expression of MTS0997 is apparently regulated by cAMP, though details of this regulation have not been elucidated. It is established that addition of cAMP to the medium reduces expression of MTS0997 in bacteria in the exponential growth phase and induces its expression in the stationary phase [113]. Furthermore, deletion of a functional copy of the neighboring gene Rv1264 encoding a pH-dependent adenylyl cyclase causes a significant decrease in MTS0997 expression in the exponential and late stationary growth phases [113]. Possible involvement of the small RNA MTS0997 in regulation with participation of cAMP is of great interest, as cAMP plays an important role in pathogenesis of M. tuberculosis [120].
The MTS1338 RNA gene (ncRv11733) is localized in an intergenic region between genes Rv1733c and Rv1734c on the complementary DNA strand. MTS1338 is a part of the DosR regulon: there are three sites for DosR regulator binding between the transcription start points of genes MTS1338 and Rv1733c. Besides, dosR gene knockout significantly reduces expression of MTS1338 RNA [104]. DosR is a regulatory component in a two-component system that is activated under hypoxia and the action of nitric oxide [121]. DosR and the genes it activates play a key role in transition of M. tuberculosis to the dormant state under hypoxia [122]. MTS1338 RNA is virtually not expressed in the exponential growth phase, but it is one of the most common transcripts during the transition to the stationary phase [104]. Significant induction of expression in the stationary growth phase and DosR-mediated regulation show that MTS1338 may play a role in generation of dormant M. tuberculosis cells and the latent form of tuberculosis [29].
The MTS2822 RNA gene (B11, ncRv13660c) was localized between genes Rv3660c and Rv3661. MTS2822 contains a so-called 6C motif consisting of two hairpins whose loops contain six and seven cytosine nucleotide residues arranged in sequence [123]. Small RNAs containing the 6C motif are widespread in Actinobacteria. However, their function is still unknown. A sequence common for SigA-promoters is situated in front of the MTS2822 transcription start point. Expression of the MTS2822 gene is elevated under oxidative stress and low pH. Hyperexpression of MTS2822 is lethal for M. tuberculosis, while in M. smegmatis it causes changes in cell morphology and reduces growth rate [105]. This could be evidence of a role of MTS2822 in regulation of cell wall synthesis or cell division [29].
The MTS2823 RNA gene (Mpr4, Ms1, and ncRv13660c) is located between genes Rv3661 and Rv3662c. The chromosomal locus containing closely located genes of small RNAs MTS2822 and MTS2823 is conserved in most mycobacteria species. MTS2823 is efficiently expressed in the exponential growth phase, while its concentration in cells is even increased in the stationary growth phase. Hyperexpression of the small RNA MTS2823 in M. tuberculosis results in somewhat decreased growth rate and a positive regulation of genes Rv2035 (putative activator of HspG protein) and Rv3229 (acyl desaturase), and strong repression of transcription of a number of genes, including energy metabolism genes, among which genes prpC and prpD are the most repressed [107]. These genes encode, correspondingly, methylcitrate synthase and methylcitrate dehydratase. These proteins participate in detoxification of metabolites – products of cholesterol and fatty acid decay with uneven number of carbon atoms, which are in turn one of the most important carbon sources during survival of bacteria inside macrophages [124]. MTS2823 RNA was first found during a bioinformatics search for homologs of the small RNA 6S [125]. The 6S RNA is widespread in various bacterial species, and its structure is reminiscent of the structure of an “open” promoter. Due to this fact, 6S RNA binds to RNA-polymerase containing σ-factor A (σA). This interaction hinders RNA-polymerase binding to promoter sequences and reduces its transcriptional activity [126]. Hnilicova and coworkers showed that in M. smegmatis the small RNA Ms1, a homolog of MTS2823, also binds to the RNA-polymerase. However, in distinction from the 6S RNA, Ms1 interacts with RNA-polymerase that does not contain factor σA. The interaction of Ms1 with RNA-polymerase does not impede binding with σA. However, σA is able to either displace Ms1 or hinder Ms1 binding to RNA-polymerase [127]. These data reveal the principally different mechanism of Ms1 action as compared to 6S RNA. A hypothesis was proposed that Ms1 could stabilize RNA-polymerase not bound to σA in the stationary phase and in the dormant state. One can also assume that upon binding to RNA-polymerase, Ms1 changes its affinity to alternative sigma factors [127].
Small RNAs in dormant M. tuberculosis cells. Recently, it was shown that under potassium limitation in a culture, M. tuberculosis cells pass to the dormant state featuring very low level of metabolic activity and temporal inability to form colonies (“nonculturability”) [128]. A sharp decrease in general transcription is also common for dormant cells. However, it is not true for a number of small RNAs, which may indicate their relative stability and involvement in maintenance of M. tuberculosis dormancy and latent infection. The most common small RNAs in dormant nonculturable cells are MTS0997, MTS1338, and MTS2823. Maximum accumulation for MTS2823 was demonstrated in the initial steps of M. tuberculosis transition to dormancy, while the concentration of MTS0997 and MTS1338 was stably high in various stages of the dormant state, including its late stage (Ignatov et al., unpublished). It was shown that overexpression of MTS0997 and MTS1338 in M. tuberculosis cells leads to substantial decrease in cell growth rate. This is especially apparent in the case of MTS1338 (Ignatov et al., unpublished). Furthermore, during analysis of transcription profile of dormant cells, the accumulation of cis-encoded small RNAs ncRv0539c (an antisense RNA for mRNA Rv0539), ncRv1162c (an antisense RNA for narH mRNA), and ncRv12659 (an antisense RNA for mRNA Rv2660c) (Ignatov et al., unpublished) was found. It was shown that ncRv12659 might be synthesized in large quantities in cells in response to nutrition shortage [129, 130]. However, the role of this transcript in regulation of physiological processes remains unknown.
Small RNAs in M. tuberculosis during development of infection. Studying the expression of small RNAs in M. tuberculosis during development of infection may provide important information regarding their role in pathogenesis. Lung infection in mice is perhaps the most widespread infection model. Several works have been published where expression levels for M. tuberculosis small RNAs were determined using such methods as real-time PCR and northern blot hybridization. Arnvig and coauthors demonstrated that expression of small RNAs MTS0997, MTS1338, and MTS2823 is substantially increased upon infection in mice. It was calculated that the number of MTS2823 transcripts during infection is approximately 10% of the amount of ribosomal RNA, and transcripts of MTS2823 are the most represented in the cell [104].
Ignatov and coauthors studied expression of MTS0997, MTS1338, and MTS2822 upon infection in two mouse lines: mouse line B6 is resistant to M. tuberculosis infection, while infecting inbred line I/St leads to death of the animals in 3-4 months. It was found that genetic features of these mouse lines and different development of the disease have only marginal influence on expression of these three small RNAs. Expression of MTS0997, MTS1338, and MTS2822 is increased upon infecting the animals as compared to growth in a culture, and it remains equally high at different stages of the disease. It is worth mentioning that in lungs of mice of line B6, expression of all the three small RNAs is reduced in the late stages of the infection, which may be explained by transition to a chronic infection [131].
Houghton and coauthors studied expression of ncRv12659 upon infecting mice. It was shown that in mouse lungs a shortened form of the transcript is synthesized to a higher degree [129]. This may be related to a premature termination of the ncRv12659 transcription.
Increased expression of small RNAs MTS0997, MTS1338, MTS2822, and ncRv12659 during development of infection indicates their possible role in pathogenesis of tuberculosis.
Thus, we conclude that small RNAs undoubtedly play a role in adaptation of pathogens (in particular, tuberculosis) upon host infection, and, thus, in pathogenesis. It should be noted that only the first steps have been made in this direction, and determination of the role of small RNAs in host/bacterial cell relationship requires extensive studies.
Regulatory mechanisms of microbial pathogens facilitate their survival under environmental stress conditions, in particular, in an infected host, which allows them to avoid the action of the host immune system on the pathogen. These facts indicate that newly discovered small noncoding RNA “world” has novel global cellular regulators [13] participating in adaptive response of bacteria to changing environmental conditions [107]. Finding the adaptive role of small noncoding RNAs in a cell may serve as a key to understanding of regulation of bacterial stress response, including the transition to the dormant state and reactivation of dormant cells, which is important for comprehension of pathogenesis of a number of latent infections.
Though the overall number of small noncoding RNAs with documented function is still not very large, diversity of processes in which their participation is experimentally established indicates that the considered regulation level covers vast areas of cellular metabolism. It is very likely that further studies will reveal participation of small RNAs in other cellular processes, which would allow referring this type of regulation to global regulations. Thus, the small noncoding RNA pool has to be considered as entering the hierarchy of levels of cellular regulation along with regulation at the transcription level, translation level, and posttranslational modification [35]. An apparent advantage and peculiarity of this level of regulation is its transient nature, which is achieved due to absence of the translation process [132]. Special flexibility of this regulation by means of small noncoding RNAs is achieved owing to fast decay of small RNAs in a complex with target, which prevents accumulation of an effector RNA after the action of a stimulus. In general, this allows a cell to respond instantly and efficiently to changing environmental factors by adjusting cellular metabolism.
This work was supported by the Russian Foundation for Basic Research (projects Nos. 13-04-40071-comfi, 13-04-40072-comfi, 15-04-05286-a, 15-04-04563-a) and the Program of Presidium of the Russian Academy of Sciences “Molecular and Cell Biology”.
REFERENCES
1.Livny, J. (2007) Efficient annotation of bacterial
genomes for small, noncoding RNAs using the integrative computational
tool sRNAPredict2, Methods Mol. Biol., 395, 475-488.
2.Montange, R. K., and Batey, R. T. (2008)
Riboswitches: emerging themes in RNA structure and function, Annu.
Rev. Biophys., 37, 117-133.
3.Garst, A. D., Edwards, A. L., and Batey, R. T.
(2011) Riboswitches: structures and mechanisms, Cold Spring Harb.
Perspect. Biol., 3.
4.Barrangou, R., and Horvath, P. (2012) CRISPR: new
horizons in phage resistance and strain identification, Annu. Rev.
Food Sci. Technol., 3, 143-162.
5.Wadler, C. S., and Vanderpool, C. K. (2007) A dual
function for a bacterial small RNA: SgrS performs base pairing
dependent regulation and encodes a functional polypeptide, Proc.
Natl. Acad. Sci. USA, 104, 20454-20459.
6.Silvaggi, J. M., Perkins, J. B., and Losick, R.
(2005) Small untranslated RNA antitoxin in Bacillus subtilis,
J. Bacteriol., 187, 6641-6650.
7.Saramago, M., Barria, C., Dos Santos, R. F., Silva,
I. J., Pobre, V., Domingues, S., Andrade, J. M., Viegas, S. C., and
Arraiano, C. M. (2014) The role of RNases in the regulation of small
RNAs, Curr. Opin. Microbiol., 18, 105-115.
8.Kawano, M., Aravind, L., and Storz, G. (2007) An
antisense RNA controls synthesis of an SOS-induced toxin evolved from
an antitoxin, Mol. Microbiol., 64, 738-754.
9.Opdyke, J. A., Kang, J. G., and Storz, G. (2004)
GadY, a small-RNA regulator of acid response genes in Escherichia
coli, J. Bacteriol., 186, 6698-6705.
10.Stork, M., Di Lorenzo, M., Welch, T. J., and
Crosa, J. H. (2007) Transcription termination within the iron
transport-biosynthesis operon of Vibrio anguillarum
requires an antisense RNA, J. Bacteriol., 189,
3479-3488.
11.Kawamoto, H., Morita, T., Shimizu, A., Inada, T.,
and Aiba, H. (2005) Implication of membrane localization of target mRNA
in the action of a small RNA: mechanism of post-transcriptional
regulation of glucose transporter in Escherichia coli, Genes
Dev., 19, 328-338.
12.Vogel, J., and Luisi, B. F. (2011) Hfq and its
constellation of RNA, Nat. Rev. Microbiol., 9,
578-589.
13.Gottesman, S., and Storz, G. (2011) Bacterial
small RNA regulators: versatile roles and rapidly evolving variations,
Cold Spring Harb. Perspect. Biol., 3.
14.Su, Q., Schuppli, D., Tsui, H. C., Winkler, M.
E., and Weber, H. (1997) Strongly reduced phage Qβ replication,
but normal phage MS2 replication in an Escherichia coli K12
mutant with inactivated Qβ host factor (hfq) gene,
Virology, 227, 211-214.
15.Wagner, E. G. (2013) Cycling of RNAs on Hfq,
RNA Biol., 10, 619-626.
16.Bojer, M. S., Jakobsen, H., Struve, C., Krogfelt,
K. A., and Lobner-Olesen, A. (2012) Lack of the RNA chaperone Hfq
attenuates pathogenicity of several Escherichia coli pathotypes
towards Caenorhabditis elegans, Microbes Infect.,
14, 1034-1039.
17.Chao, Y., and Vogel, J. (2010) The role of Hfq in
bacterial pathogens, Curr. Opin. Microbiol., 13,
24-33.
18.Oliva, G., Sahr, T., and Buchrieser, C. (2015)
Small RNAs, 5′ UTR elements and RNA-binding proteins in
intracellular bacteria: impact on metabolism and virulence, FEMS
Microbiol. Rev., 39, 331-349.
19.Folichon, M., Arluison, V., Pellegrini, O.,
Huntzinger, E., Regnier, P., and Hajnsdorf, E. (2003) The poly(A)
binding protein Hfq protects RNA from RNase E and exoribonucleolytic
degradation, Nucleic Acids Res., 31, 7302-7310.
20.De Lay, N., Schu, D. J., and Gottesman, S. (2013)
Bacterial small RNA-based negative regulation: Hfq and its accomplices,
J. Biol. Chem., 288, 7996-8003.
21.Waters, L. S., and Storz, G. (2009) Regulatory
RNAs in bacteria, Cell, 136, 615-628.
22.Masse, E., Escorcia, F. E., and Gottesman, S.
(2003) Coupled degradation of a small regulatory RNA and its mRNA
targets in Escherichia coli, Genes Dev., 17,
2374-2383.
23.Aiba, H. (2007) Mechanism of RNA silencing by
Hfq-binding small RNAs, Curr. Opin. Microbiol., 10,
134-139.
24.Majdalani, N., Vanderpool, C. K., and Gottesman,
S. (2005) Bacterial small RNA regulators, Crit. Rev. Biochem. Mol.
Biol., 40, 93-113.
25.Christiansen, J. K., Larsen, M. H., Ingmer, H.,
Sogaard-Andersen, L., and Kallipolitis, B. H. (2004) The RNA-binding
protein Hfq of Listeria monocytogenes: role in stress tolerance
and virulence, J. Bacteriol., 186, 3355-3362.
26.Andrade, J. M., Pobre, V., Matos, A. M., and
Arraiano, C. M. (2012) The crucial role of PNPase in the degradation of
small RNAs that are not associated with Hfq, RNA, 18,
844-855.
27.Morita, T., Maki, K., and Aiba, H. (2005) RNase
E-based ribonucleoprotein complexes: mechanical basis of mRNA
destabilization mediated by bacterial noncoding RNAs, Genes
Dev., 19, 2176-2186.
28.Valentin-Hansen, P., Eriksen, M., and Udesen, C.
(2004) The bacterial Sm-like protein Hfq: a key player in RNA
transactions, Mol. Microbiol., 51, 1525-1533.
29.Arnvig, K., and Young, D. (2012) Non-coding RNA
and its potential role in Mycobacterium tuberculosis
pathogenesis, RNA Biol., 9, 427-436.
30.Bohn, C., Rigoulay, C., Chabelskaya, S., Sharma,
C. M., Marchais, A., Skorski, P., Borezee-Durant, E., Barbet, R.,
Jacquet, E., Jacq, A., Gautheret, D., Felden, B., Vogel, J., and
Bouloc, P. (2010) Experimental discovery of small RNAs in
Staphylococcus aureus reveals a riboregulator of central
metabolism, Nucleic Acids Res., 38, 6620-6636.
31.Song, T., Mika, F., Lindmark, B., Liu, Z.,
Schild, S., Bishop, A., Zhu, J., Camilli, A., Johansson, J., Vogel, J.,
and Wai, S. N. (2008) A new Vibrio cholerae sRNA modulates
colonization and affects release of outer membrane vesicles, Mol.
Microbiol., 70, 100-111.
32.Chaulk, S. G., Smith Frieday, M. N., Arthur, D.
C., Culham, D. E., Edwards, R. A., Soo, P., Frost, L. S., Keates, R.
A., Glover, J. N., and Wood, J. M. (2011) ProQ is an RNA chaperone that
controls ProP levels in Escherichia coli, Biochemistry,
50, 3095-3106.
33.Pandey, S. P., Minesinger, B. K., Kumar, J., and
Walker, G. C. (2011) A highly conserved protein of unknown function in
Sinorhizobium meliloti affects sRNA regulation similar to Hfq,
Nucleic Acids Res., 39, 4691-4708.
34.Romby, P., and Charpentier, E. (2010) An overview
of RNAs with regulatory functions in Gram-positive bacteria, Cell.
Mol. Life Sci., 67, 217-237.
35.Michaux, C., Verneuil, N., Hartke, A., and Giard,
J. C. (2014) Physiological roles of small RNA molecules,
Microbiology, 160, 1007-1019.
36.Dubey, A. K., Baker, C. S., Suzuki, K., Jones, A.
D., Pandit, P., Romeo, T., and Babitzke, P. (2003) CsrA regulates
translation of the Escherichia coli carbon starvation
gene, cstA, by blocking ribosome access to the cstA transcript,
J. Bacteriol., 185, 4450-4460.
37.Babitzke, P., and Romeo, T. (2007) CsrB sRNA
family: sequestration of RNA-binding regulatory proteins, Curr.
Opin. Microbiol., 10, 156-163.
38.Pernestig, A. K., Georgellis, D., Romeo, T.,
Suzuki, K., Tomenius, H., Normark, S., and Melefors, O. (2003) The
Escherichia coli BarA-UvrY two-component system is needed for
efficient switching between glycolytic and gluconeogenic carbon
sources, J. Bacteriol., 185, 843-853.
39.Jonas, K., and Melefors, O. (2009) The
Escherichia coli CsrB and CsrC small RNAs are strongly induced
during growth in nutrient-poor medium, FEMS Microbiol. Lett.,
297, 80-86.
40.Altier, C., Suyemoto, M., and Lawhon, S. D.
(2000) Regulation of Salmonella enterica serovar
typhimurium invasion genes by csrA, Infect.
Immun., 68, 6790-6797.
41.Julio, S. M., Heithoff, D. M., and Mahan, M. J.
(2000) ssrA (tmRNA) plays a role in Salmonella enterica serovar
typhimurium pathogenesis, J. Bacteriol., 182,
1558-1563.
42.Heroven, A. K., Bohme, K., and Dersch, P. (2012)
The Csr/Rsm system of Yersinia and related pathogens: a
post-transcriptional strategy for managing virulence, RNA Biol.,
9, 379-391.
43.Geissmann, T., Chevalier, C., Cros, M. J.,
Boisset, S., Fechter, P., Noirot, C., Schrenzel, J., Francois, P.,
Vandenesch, F., Gaspin, C., and Romby, P. (2009) A search for small
noncoding RNAs in Staphylococcus aureus reveals a conserved
sequence motif for regulation, Nucleic Acids Res., 37,
7239-7257.
44.Frohlich, K. S., Papenfort, K., Berger, A. A.,
and Vogel, J. (2012) A conserved RpoS-dependent small RNA controls the
synthesis of major porin OmpD, Nucleic Acids Res., 40,
3623-3640.
45.Santiviago, C. A., Toro, C. S., Hidalgo, A. A.,
Youderian, P., and Mora, G. C. (2003) Global regulation of the
Salmonella enterica serovar typhimurium major porin,
OmpD, J. Bacteriol., 185, 5901-5905.
46.Pulvermacher, S. C., Stauffer, L. T., and
Stauffer, G. V. (2008) The role of the small regulatory RNA GcvB in
GcvB/mRNA posttranscriptional regulation of oppA and dppA in
Escherichia coli, FEMS Microbiol. Lett., 281,
42-50.
47.Pulvermacher, S. C., Stauffer, L. T., and
Stauffer, G. V. (2009) Role of the Escherichia coli Hfq protein
in GcvB regulation of oppA and dppA mRNAs, Microbiology,
155, 115-123.
48.Pulvermacher, S. C., Stauffer, L. T., and
Stauffer, G. V. (2009) Role of the sRNA GcvB in regulation of cycA in
Escherichia coli, Microbiology, 155, 106-114.
49.Pulvermacher, S. C., Stauffer, L. T., and
Stauffer, G. V. (2009) The small RNA GcvB regulates sstT mRNA
expression in Escherichia coli, J. Bacteriol.,
191, 238-248.
50.Sharma, C. M., Darfeuille, F., Plantinga, T. H.,
and Vogel, J. (2007) A small RNA regulates multiple ABC transporter
mRNAs by targeting C/A-rich elements inside and upstream of
ribosome-binding sites, Genes Dev., 21, 2804-2817.
51.Sharma, C. M., Papenfort, K., Pernitzsch, S. R.,
Mollenkopf, H. J., Hinton, J. C., and Vogel, J. (2011) Pervasive
posttranscriptional control of genes involved in amino acid metabolism
by the Hfq-dependent GcvB small RNA, Mol. Microbiol., 81,
1144-1165.
52.Urbanowski, M. L., Stauffer, L. T., and Stauffer,
G. V. (2000) The gcvB gene encodes a small untranslated RNA involved in
expression of the dipeptide and oligopeptide transport systems in
Escherichia coli, Mol. Microbiol., 37,
856-868.
53.Stauffer, L. T., and Stauffer, G. V. (2012) The
Escherichia coli GcvB sRNA uses genetic redundancy to control
cycA expression, ISRN Microbiol., doi: 10.5402/2012/636273.
54.De Lay, N., and Gottesman, S. (2009) The
Crp-activated small noncoding regulatory RNA CyaR (RyeE) links
nutritional status to group behavior, J. Bacteriol., 191,
461-476.
55.Johansen, J., Eriksen, M., Kallipolitis, B., and
Valentin-Hansen, P. (2008) Down-regulation of outer membrane proteins
by noncoding RNAs: unraveling the cAMP-CRP- and σE-dependent
CyaR-ompX regulatory case, J. Mol. Biol., 383, 1-9.
56.Papenfort, K., Pfeiffer, V., Lucchini, S.,
Sonawane, A., Hinton, J. C., and Vogel, J. (2008) Systematic deletion
of Salmonella small RNA genes identifies CyaR, a conserved
CRP-dependent riboregulator of OmpX synthesis, Mol. Microbiol.,
68, 890-906.
57.Masse, E., and Gottesman, S. (2002) A small RNA
regulates the expression of genes involved in iron metabolism in
Escherichia coli, Proc. Natl. Acad. Sci. USA, 99,
4620-4625.
58.Masse, E., Vanderpool, C. K., and Gottesman, S.
(2005) Effect of RyhB small RNA on global iron use in Escherichia
coli, J. Bacteriol., 187, 6962-6971.
59.Vecerek, B., Moll, I., and Blasi, U. (2007)
Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive
decoding, EMBO J., 26, 965-975.
60.Wilderman, P. J., Sowa, N. A., Fitzgerald, D. J.,
Fitzgerald, P. C., Gottesman, S., Ochsner, U. A., and Vasil, M. L.
(2004) Identification of tandem duplicate regulatory small RNAs in
Pseudomonas aeruginosa involved in iron homeostasis,
Proc. Natl. Acad. Sci. USA, 101, 9792-9797.
61.Mellin, J. R., Goswami, S., Grogan, S., Tjaden,
B., and Genco, C. A. (2007) A novel fur- and iron-regulated small RNA,
NrrF, is required for indirect fur-mediated regulation of the
sdhA and sdhC genes in Neisseria
meningitidis, J. Bacteriol., 189, 3686-3694.
62.Metruccio, M. M., Fantappie, L., Serruto, D.,
Muzzi, A., Roncarati, D., Donati, C., Scarlato, V., and Delany, I.
(2009) The Hfq-dependent small noncoding RNA NrrF directly mediates
Fur-dependent positive regulation of succinate dehydrogenase in
Neisseria meningitidis, J. Bacteriol., 191,
1330-1342.
63.Gaballa, A., Antelmann, H., Aguilar, C., Khakh,
S. K., Song, K. B., Smaldone, G. T., and Helmann, J. D. (2008) The
Bacillus subtilis iron-sparing response is mediated by a
Fur-regulated small RNA and three small, basic proteins, Proc. Natl.
Acad. Sci. USA, 105, 11927-11932.
64.Jin, Y., Watt, R. M., Danchin, A., and Huang, J.
D. (2009) Small noncoding RNA GcvB is a novel regulator of acid
resistance in Escherichia coli, BMC Genomics, 10,
165.
65.Englesberg, E., Anderson, R. L., Weinberg, R.,
Lee, N., Hoffee, P., Huttenhauer, G., and Boyer, H. (1962)
L-Arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient
mutants of Escherichia coli, J. Bacteriol., 84,
137-146.
66.Irani, M. H., and Maitra, P. K. (1977) Properties
of Escherichia coli mutants deficient in enzymes of glycolysis,
J. Bacteriol., 132, 398-410.
67.Vanderpool, C. K., and Gottesman, S. (2004)
Involvement of a novel transcriptional activator and small RNA in
post-transcriptional regulation of the glucose phosphoenolpyruvate
phosphotransferase system, Mol. Microbiol., 54,
1076-1089.
68.Maki, K., Morita, T., Otaka, H., and Aiba, H.
(2010) A minimal base-pairing region of a bacterial small RNA SgrS
required for translational repression of ptsG mRNA, Mol.
Microbiol., 76, 782-792.
69.Rice, J. B., and Vanderpool, C. K. (2011) The
small RNA SgrS controls sugar-phosphate accumulation by regulating
multiple PTS genes, Nucleic Acids Res., 39,
3806-3819.
70.Durand, S., and Storz, G. (2010) Reprogramming of
anaerobic metabolism by the FnrS small RNA, Mol. Microbiol.,
75, 1215-1231.
71.Trun, N. J., and Silhavy, T. J. (1989) PrlC, a
suppressor of signal sequence mutations in Escherichia coli, can
direct the insertion of the signal sequence into the membrane, J.
Mol. Biol., 205, 665-676.
72.Jiang, X., Zhang, M., Ding, Y., Yao, J., Chen,
H., Zhu, D., and Muramatu, M. (1998) Escherichia coli
prlC gene encodes a trypsin-like proteinase regulating the cell
cycle, J. Biochem., 124, 980-985.
73.Jain, R., and Chan, M. K. (2007) Support for a
potential role of E. coli oligopeptidase A in protein
degradation, Biochem. Biophys. Res. Commun., 359,
486-490.
74.Sonnleitner, E., Gonzalez, N., Sorger-Domenigg,
T., Heeb, S., Richter, A. S., Backofen, R., Williams, P., Huttenhofer,
A., Haas, D., and Blasi, U. (2011) The small RNA PhrS stimulates
synthesis of the Pseudomonas aeruginosa quinolone signal,
Mol. Microbiol., 80, 868-885.
75.Altuvia, S., Weinstein-Fischer, D., Zhang, A.,
Postow, L., and Storz, G. (1997) A small, stable RNA induced by
oxidative stress: role as a pleiotropic regulator and antimutator,
Cell, 90, 43-53.
76.Altuvia, S., Zhang, A., Argaman, L., Tiwari, A.,
and Storz, G. (1998) The Escherichia coli OxyS regulatory RNA
represses fhlA translation by blocking ribosome binding, EMBO
J., 17, 6069-6075.
77.Argaman, L., and Altuvia, S. (2000) fhlA
repression by OxyS RNA: kissing complex formation at two sites results
in a stable antisense-target RNA complex, J. Mol. Biol.,
300, 1101-1112.
78.Zhang, A., Altuvia, S., Tiwari, A., Argaman, L.,
Hengge-Aronis, R., and Storz, G. (1998) The OxyS regulatory RNA
represses rpoS translation and binds the Hfq (HF-I) protein, EMBO
J., 17, 6061-6068.
79.Rasmussen, A. A., Eriksen, M., Gilany, K.,
Udesen, C., Franch, T., Petersen, C., and Valentin-Hansen, P. (2005)
Regulation of ompA mRNA stability: the role of a small regulatory RNA
in growth phase-dependent control, Mol. Microbiol., 58,
1421-1429.
80.Udekwu, K. I., Darfeuille, F., Vogel, J.,
Reimegard, J., Holmqvist, E., and Wagner, E. G. (2005) Hfq-dependent
regulation of OmpA synthesis is mediated by an antisense RNA, Genes
Dev., 19, 2355-2366.
81.Papenfort, K., Bouvier, M., Mika, F., Sharma, C.
M., and Vogel, J. (2010) Evidence for an autonomous 5′ target
recognition domain in an Hfq-associated small RNA, Proc. Natl. Acad.
Sci. USA, 107, 20435-20440.
82.Papenfort, K., Pfeiffer, V., Mika, F., Lucchini,
S., Hinton, J. C., and Vogel, J. (2006) σE-dependent small RNAs
of Salmonella respond to membrane stress by accelerating global
Omp mRNA decay, Mol. Microbiol., 62, 1674-1688.
83.Johansen, J., Rasmussen, A. A., Overgaard, M.,
and Valentin-Hansen, P. (2006) Conserved small non-coding RNAs that
belong to the σE regulon: role in down-regulation of outer
membrane proteins, J. Mol. Biol., 364, 1-8.
84.Miller, M. B., Skorupski, K., Lenz, D. H.,
Taylor, R. K., and Bassler, B. L. (2002) Parallel quorum sensing
systems converge to regulate virulence in Vibrio
cholerae, Cell, 110, 303-314.
85.Bassler, B. L., Wright, M., and Silverman, M. R.
(1994) Sequence and function of LuxO, a negative regulator of
luminescence in Vibrio harveyi, Mol. Microbiol.,
12, 403-412.
86.Freeman, J. A., and Bassler, B. L. (1999)
Sequence and function of LuxU: a two-component phosphorelay protein
that regulates quorum sensing in Vibrio harveyi, J.
Bacteriol., 181, 899-906.
87.Lilley, B. N., and Bassler, B. L. (2000)
Regulation of quorum sensing in Vibrio harveyi by LuxO and
σ-54, Mol. Microbiol., 36, 940-954.
88.Bardill, J. P., Zhao, X., and Hammer, B. K.
(2011) The Vibrio cholerae quorum sensing response is mediated
by Hfq-dependent sRNA/mRNA base pairing interactions, Mol.
Microbiol., 80, 1381-1394.
89.Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni,
R. V., Wingreen, N. S., and Bassler, B. L. (2004) The small RNA
chaperone Hfq and multiple small RNAs control quorum sensing in
Vibrio harveyi and Vibrio cholerae, Cell,
118, 69-82.
90.Rutherford, S. T., Van Kessel, J. C., Shao, Y.,
and Bassler, B. L. (2011) AphA and LuxR/HapR reciprocally control
quorum sensing in vibrios, Genes Dev., 25, 397-408.
91.Hammer, B. K., and Bassler, B. L. (2007)
Regulatory small RNAs circumvent the conventional quorum sensing
pathway in pandemic Vibrio cholerae, Proc. Natl. Acad. Sci.
USA, 104, 11145-11149.
92.Korem, M., Gov, Y., Kiran, M. D., and Balaban, N.
(2005) Transcriptional profiling of target of RNAIII-activating
protein, a master regulator of staphylococcal virulence, Infect.
Immun., 73, 6220-6228.
93.Chabelskaya, S., Gaillot, O., and Felden, B.
(2010) A Staphylococcus aureus small RNA is required for
bacterial virulence and regulates the expression of an immune-evasion
molecule, PLoS Pathog., 6, e1000927.
94.Leday, T. V., Gold, K. M., Kinkel, T. L.,
Roberts, S. A., Scott, J. R., and McIver, K. S. (2008) TrxR, a new
CovR-repressed response regulator that activates the Mga virulence
regulon in group A Streptococcus, Infect. Immun.,
76, 4659-4668.
95.Roberts, S. A., and Scott, J. R. (2007) RivR and
the small RNA RivX: the missing links between the CovR regulatory
cascade and the Mga regulon, Mol. Microbiol., 66,
1506-1522.
96.Kreikemeyer, B., Boyle, M. D., Buttaro, B. A.,
Heinemann, M., and Podbielski, A. (2001) Group A streptococcal growth
phase-associated virulence factor regulation by a novel operon (Fas)
with homologies to two-component-type regulators requires a small RNA
molecule, Mol. Microbiol., 39, 392-406.
97.Klenk, M., Koczan, D., Guthke, R., Nakata, M.,
Thiesen, H. J., Podbielski, A., and Kreikemeyer, B. (2005) Global
epithelial cell transcriptional responses reveal Streptococcus
pyogenes Fas regulator activity association with bacterial
aggressiveness, Cell Microbiol., 7, 1237-1250.
98.Mangold, M., Siller, M., Roppenser, B.,
Vlaminckx, B. J., Penfound, T. A., Klein, R., Novak, R., Novick, R. P.,
and Charpentier, E. (2004) Synthesis of group A streptococcal virulence
factors is controlled by a regulatory RNA molecule, Mol.
Microbiol., 53, 1515-1527.
99.Dye, C. (2006) Global epidemiology of
tuberculosis, Lancet, 367, 938-940.
100.Giangrossi, M., Prosseda, G., Tran, C. N.,
Brandi, A., Colonna, B., and Falconi, M. (2010) A novel antisense RNA
regulates at transcriptional level the virulence gene icsA of
Shigella flexneri, Nucleic Acids Res., 38,
3362-3375.
101.Corbett, E. L. (2003) HIV and
tuberculosis: surveillance revisited, Int. J. Tuberc. Lung
Dis., 7, 709.
102.Corbett, E. L., Watt, C. J., Walker, N., Maher,
D., Williams, B. G., Raviglione, M. C., and Dye, C. (2003) The growing
burden of tuberculosis: global trends and interactions with the HIV
epidemic, Arch. Intern. Med., 163, 1009-1021.
103.Ignatov, D., Malakho, S., Majorov, K.,
Skvortsov, T., Apt, A., and Azhikina, T. (2013) RNA-Seq analysis of
Mycobacterium avium non-coding transcriptome, PLoS One,
8, e74209.
104.Arnvig, K. B., Comas, I., Thomson, N. R.,
Houghton, J., Boshoff, H. I., Croucher, N. J., Rose, G., Perkins, T.
T., Parkhill, J., Dougan, G., and Young, D. B. (2011) Sequence-based
analysis uncovers an abundance of noncoding RNA in the total
transcriptome of Mycobacterium tuberculosis, PLoS
Pathog, 7, e1002342.
105.Arnvig, K. B., and Young, D. B. (2009)
Identification of small RNAs in Mycobacterium tuberculosis,
Mol. Microbiol., 73, 397-408.
106.DiChiara, J. M., Contreras-Martinez, L. M.,
Livny, J., Smith, D., McDonough, K. A., and Belfort, M. (2010) Multiple
small RNAs identified in Mycobacterium bovis BCG are also
expressed in Mycobacterium tuberculosis and Mycobacterium
smegmatis, Nucleic Acids Res., 38, 4067-4078.
107.Haning, K., Cho, S. H., and Contreras, L. M.
(2014) Small RNAs in mycobacteria: an unfolding story, Front. Cell.
Infect. Microbiol., 4, 96.
108.Li, S. K., Ng, P. K., Qin, H., Lau, J. K., Lau,
J. P., Tsui, S. K., Chan, T. F., and Lau, T. C. (2013) Identification
of small RNAs in Mycobacterium smegmatis using heterologous Hfq,
RNA, 19, 74-84.
109.Miotto, P., Forti, F., Ambrosi, A., Pellin, D.,
Veiga, D. F., Balazsi, G., Gennaro, M. L., Di Serio, C., Ghisotti, D.,
and Cirillo, D. M. (2012) Genome-wide discovery of small RNAs in
Mycobacterium tuberculosis, PLoS One, 7,
e51950.
110.Pellin, D., Miotto, P., Ambrosi, A., Cirillo,
D. M., and Di Serio, C. (2012) A genome-wide identification analysis of
small regulatory RNAs in Mycobacterium tuberculosis by RNA-Seq
and conservation analysis, PLoS One, 7, e32723.
111.Tsai, C. H., Baranowski, C., Livny, J.,
McDonough, K. A., Wade, J. T., and Contreras, L. M. (2013)
Identification of novel sRNAs in mycobacterial species, PLoS
One, 8, e79411.
112.Lamichhane, G., Arnvig, K. B., and McDonough,
K. A. (2013) Definition and annotation of (myco)bacterial non-coding
RNA, Tuberculosis (Edinb.), 93, 26-29.
113.Pelly, S., Bishai, W. R., and Lamichhane, G.
(2012) A screen for non-coding RNA in Mycobacterium tuberculosis
reveals a cAMP-responsive RNA that is expressed during infection,
Gene, 500, 85-92.
114.Hartkoorn, R. C., Sala, C., Uplekar, S., Busso,
P., Rougemont, J., and Cole, S. T. (2012) Genome-wide definition of the
SigF regulon in Mycobacterium tuberculosis, J.
Bacteriol., 194, 2001-2009.
115.Solans, L., Gonzalo-Asensio, J., Sala, C.,
Benjak, A., Uplekar, S., Rougemont, J., Guilhot, C., Malaga, W.,
Martin, C., and Cole, S. T. (2014) The PhoP-dependent ncRNA Mcr7
modulates the TAT secretion system in Mycobacterium
tuberculosis, PLoS Pathog., 10, e1004183.
116.Wiker, H. G., and Harboe, M. (1992) The antigen
85 complex: a major secretion product of Mycobacterium
tuberculosis, Microbiol. Rev., 56, 648-661.
117.Flores, A. R., Parsons, L. M., and Pavelka, M.
S., Jr. (2005) Genetic analysis of the β-lactamases of
Mycobacterium tuberculosis and Mycobacterium smegmatis
and susceptibility to β-lactam antibiotics, Microbiology,
151, 521-532.
118.Dittrich, D., Keller, C., Ehlers, S., Schultz,
J. E., and Sander, P. (2006) Characterization of a Mycobacterium
tuberculosis mutant deficient in pH-sensing adenylate cyclase
Rv1264, Int. J. Med. Microbiol., 296, 563-566.
119.Gazdik, M. A., Bai, G., Wu, Y., and McDonough,
K. A. (2009) Rv1675c (cmr) regulates intramacrophage and cyclic
AMP-induced gene expression in Mycobacterium tuberculosis
complex mycobacteria, Mol. Microbiol., 71, 434-448.
120.Agarwal, N., Lamichhane, G., Gupta, R., Nolan,
S., and Bishai, W. R. (2009) Cyclic AMP intoxication of macrophages by
a Mycobacterium tuberculosis adenylate cyclase,
Nature, 460, 98-102.
121.Kumar, A., Toledo, J. C., Patel, R. P.,
Lancaster, J. R., Jr., and Steyn, A. J. (2007) Mycobacterium
tuberculosis DosS is a redox sensor and DosT is a hypoxia
sensor, Proc. Natl. Acad. Sci. USA, 104, 11568-11573.
122.Honaker, R. W., Leistikow, R. L., Bartek, I.
L., and Voskuil, M. I. (2009) Unique roles of DosT and DosS in DosR
regulon induction and Mycobacterium tuberculosis dormancy,
Infect. Immun., 77, 3258-3263.
123.Weinberg, Z., Barrick, J. E., Yao, Z., Roth,
A., Kim, J. N., Gore, J., Wang, J. X., Lee, E. R., Block, K. F.,
Sudarsan, N., Neph, S., Tompa, M., Ruzzo, W. L., and Breaker, R. R.
(2007) Identification of 22 candidate structured RNAs in bacteria using
the CMfinder comparative genomics pipeline, Nucleic Acids Res.,
35, 4809-4819.
124.Chang, J. C., Miner, M. D., Pandey, A. K.,
Gill, W. P., Harik, N. S., Sassetti, C. M., and Sherman, D. R. (2009)
igr genes and Mycobacterium tuberculosis cholesterol
metabolism, J. Bacteriol., 191, 5232-5239.
125.Panek, J., Krasny, L., Bobek, J., Jezkova, E.,
Korelusova, J., and Vohradsky, J. (2011) The suboptimal structures find
the optimal RNAs: homology search for bacterial non-coding RNAs using
suboptimal RNA structures, Nucleic Acids Res., 39,
3418-3426.
126.Barrick, J. E., Sudarsan, N., Weinberg, Z.,
Ruzzo, W. L., and Breaker, R. R. (2005) 6S RNA is a widespread
regulator of eubacterial RNA polymerase that resembles an open
promoter, RNA, 11, 774-784.
127.Hnilicova, J., Jirat Matejckova, J., Sikova,
M., Pospisil, J., Halada, P., Panek, J., and Krasny, L. (2014) Ms1, a
novel sRNA interacting with the RNA polymerase core in mycobacteria,
Nucleic Acids Res., 42, 11763-11776.
128.Salina, E. G., Waddell, S. J., Hoffmann, N.,
Rosenkrands, I., Butcher, P. D., and Kaprelyants, A. S. (2014)
Potassium availability triggers Mycobacterium tuberculosis
transition to, and resuscitation from, non-culturable (dormant) states,
Open Biol., 4, doi: 10.1098/rsob.140106.
129.Houghton, J., Cortes, T., Schubert, O., Rose,
G., Rodgers, A., De Ste Croix, M., Aebersold, R., Young, D. B., and
Arnvig, K. B. (2013) A small RNA encoded in the Rv2660c locus of
Mycobacterium tuberculosis is induced during starvation and
infection, PLoS One, 8, e80047.
130.Uplekar, S., Rougemont, J., Cole, S. T., and
Sala, C. (2013) High resolution transcriptome and genome wide dynamics
of RNA polymerase and NusA in Mycobacterium tuberculosis,
Nucleic Acids Res., 41, 961-977.
131.Ignatov, D. B., Timoshina O. Yu., Logunova, N.
N., Skvortsov, T. A., and Azhikina, T. L. (2014) Expression of
Mycobacterium tuberculosis small RNA in mice models of
tuberculosis, Bioorg. Khim., 40, 253-256.
132.Beisel, C. L., and Storz, G. (2011) The base
pairing RNA spot 42 participates in a multioutput feedforward loop to
help enact catabolite repression in Escherichia coli,
Mol. Cell, 41, 286-297.
133.Storz, G., Vogel, J., and Wassarman, K. M.
(2011) Regulation by small RNAs in bacteria: expanding frontiers,
Mol. Cell, 43, 880-891.