REVIEW: Bacterial Small Regulatory RNAs and Hfq Protein
V. N. Murina* and A. D. Nikulin
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: thyrada@rambler.ru* To whom correspondence should be addressed.
Received June 25, 2015
Small regulatory RNA (sRNA) is a unique noncoding RNA involved in regulation of gene expression in both eukaryotic and bacterial cells. This short review discusses examples of positive and negative translation regulation by sRNAs in bacteria and participation of Hfq in these processes. The importance of structure investigation of nucleotide–protein and RNA–protein complexes for designing a model of Hfq interaction with both mRNA and sRNA simultaneously is demonstrated.
KEY WORDS: Hfq, small regulatory RNA, sRNA, RNA–protein recognition, regulation of gene expressionDOI: 10.1134/S0006297915130027
After the discovery of the fact that the main genetic information is encoded by chromosomal DNA (described in [1, 2]) and development of an approach for its sequencing by Sanger and colleagues [3], DNA sequences were determined for different organisms including humans – the Human Genome Project was started in 1990 and accomplished in 2003 [4, 5]. Genome deciphering showed that not all DNA encodes proteins or ribosomal and transfer RNAs. Even before, “noncoding” regions were found in bacterial DNA. It was proposed that they do not perform any important functions in cells and are rather ballast or “junk” DNA [6]. Later, it was shown that “junk” DNA is transcribed and encodes regulatory RNAs and mobile transgenomic elements – transposons [7]. Both of these play important roles in all living organisms. In the present review, a small portion of the “junk” DNA products will be considered, e.g. regulatory RNAs and their interactions with Hfq protein, a global regulator of gene expression in Gram-negative bacteria.
DISCOVERY OF REGULATORY RNAs
In bacteria, the first regulatory RNAs were found in non-chromosomal genetic elements, namely in the plasmid ColE1 (RNAI) [8] and in the transposon Tn10 [9]. They were referred to as small regulatory RNAs (sRNAs) or noncoding RNAs (ncRNAs). Later, in one of the most studied organisms, Escherichia coli, it was shown that in many cases, regulation of mRNA translation by small regulatory RNAs is mediated by Hfq protein.
The presence of regulatory RNAs in eukaryotes was demonstrated in 1993. The eukaryotic regulatory RNA (miRNA, microRNA) lin-4 was discovered during study of the nematode Caenorhabditis elegans. This RNA was shown not to encode a protein, and it negatively regulated lin-14 mRNA translation due to partial complementary pairing with seven repeats present in the 3′-untranslated region of lin-14 mRNA [10]. Eukaryotic regulatory RNAs are now classified by length – long noncoding RNAs (lncRNAs), by action mechanism – small interfering RNAs (siRNAs), by cellular localization – small nucleolar and nuclear RNAs (snoRNAs and snRNAs) and outside the cell – exRNAs, by size – microRNAs (miRNAs), and by interaction mediators – piRNAs that act together with piwi proteins.
Regulatory RNAs in Archaea were predicted by bioinformatics methods only in 2002. In 2009, the first discovered archaeal regulatory RNA, Gö1 from Methanosarcina mazei, was found (reviewed in detail in [11] and [12]). It turned out that some sRNAs in Archaea contain short regions of open reading frames (ORFs) and are able to combine functions of messenger and regulatory RNAs [13].
ROLE OF SMALL RNAs IN BACTERIA
Within the last 30 years, more than 100 small regulatory RNAs have been discovered and studied in bacteria. From 80 to 100 regulatory RNAs were found in E. coli [14, 15]. Their studies demonstrated that regulation of bacterial vital functions is not performed solely by proteins. Most interesting was the fact that in humans and animal bacterial pathogens, small regulatory RNAs participate in their virulence and drug resistance [16]. Now the most studied small regulatory RNAs are those of Gram-negative bacteria, where the RNA translation regulation system includes such components as RNase E, degradosomes, and Hfq protein. In Gram-positive bacteria, RNase E is absent and Hfq protein does not participate as much in mRNA translation regulation. It is possible that other, not yet studied mechanisms of regulation of mRNA translation exist in Gram-positive bacteria. We will review the interaction of sRNAs with mRNAs and Hfq protein in Gram-negative bacteria.
Small regulatory RNAs are synthesized in cells in response to such cellular stresses as glucose or iron starvation, oxidative stress, radiation, unfavorable temperature, etc. The effect of small RNAs on translational activity of mRNAs is provided by mechanisms described below. To regulate translation, small regulatory RNAs utilize their partial complementarity to mRNA regions, due to which RNA duplexes are formed that cannot be read by the bacterial translational apparatus. Hence, because of the coupling between transcription and translation of bacterial mRNAs, translation stop causes termination of mRNA transcription.
Information encoded by the mRNA nucleotide sequence to amino acid sequence of protein molecules in all living organisms is translated by a macromolecular ribonucleoprotein complex – the ribosome. In prokaryotic cells, chromosomal DNA is not separated from the cytoplasm by the nuclear membrane as it is in eukaryotes. For this reason, messenger RNA becomes available to ribosomes immediately after its transcription from DNA. Thus, in bacteria, transcription and translation occur simultaneously, in other words, they are coupled. One of the forms of small regulatory RNA-mediated negative regulation of mRNA translation occurs by means of uncoupling of transcription and translation.
Accumulating in a cell, sRNA can specifically inhibit translation of a messenger RNA by forming a duplex due to partial complementarity of nucleotide sequences. In most cases, small RNA is complementary to the ribosome-binding site (RBS) on mRNA. Thus, duplex formation between a small RNA and a messenger RNA shields a region of a messenger RNA RBS to which the ribosome normally binds. As a result, the ribosome does not bind to a messenger RNA. Inhibition of translation initiation occurs, leading to transcription termination by Rho-dependent or Rho-independent mechanisms, depending on mRNA type. Irreversibility of translation–transcription processes is achieved by the enzymatic decay of a messenger RNA. RNA decay is initiated by RNase III, which cleaves mRNA–sRNA duplexes, or by RNase E that cleaves single-stranded RNA regions that are not protected by ribosomes. Cleavage of the inhibited mRNA is completed by the degradosome.
Small regulatory RNAs can regulate transcription even if they are complementary to any coding sequence of a messenger RNA (not necessarily the RBS). In this case, translation inhibition at the elongation stage occurs because of formation of a duplex between sRNA and its target mRNA, which cannot be translated by the ribosome. Then, uncoupling of transcription–translation occurs, resulting in messenger RNA degradation by the mechanism described in the previous paragraph.
Positive translation regulation by means of small regulatory RNAs can be direct or mediated by RNA mimicry. Direct positive regulation is realized by providing the ribosomes access to the start codon or by modifying the process of mRNA editing that increases its stability. Indirect positive regulation occurs when sRNA mimics mRNA, which dilutes the regulated RNA pool; thus, the amount of cleaved RNA decreases [17].
Examples of negative translation regulation. Regulation of rpoS messenger RNA translation is now most extensively studied. The rpoS gene encodes an alternative σ-factor of RNA-polymerase. It is induced under cellular stresses (starvation, acidic or basic pH, osmotic shock, stationary phase) and triggers expression of genes encoding stress-response proteins. When cellular stresses are absent, translation of rpoS mRNA is auto-inhibited due to an internal hairpin structure at the 5′-untranslated region near the start codon.
Negative translation regulation of the RpoS σ-factor is mediated by sRNA OxyS. This was the first regulatory RNA discovered, by Gisela Storz in 1985 during a study of an effect of hydrogen peroxide on processes in cells. Storz found that after hydrogen peroxide treatment, a 109-nt OxyS RNA is accumulated in cells [18]. Later, this regulatory RNA was shown to accumulate in cells under oxidative stress. It inhibits translation of messenger RNAs rpoS and fhlA, and some others, by blocking access to the ribosomal binding site [15, 19].
Since then, tens of new regulatory RNAs have been discovered, and mechanisms of their action have been investigated. One of these RNAs is RyhB RNA. The mechanism of its action is interesting because of the involvement of the additional specific iron metabolism protein Fur. Fur is a repressor of translation of six mRNAs that encode iron-binding proteins in E. coli [20]. Except for inhibition of translation of these mRNAs, they are degraded by RNase E upon binding of Hfq protein and sRNA RyhB [21].
Synthesis of ptsG mRNA encoding a membrane subunit of the glucose transporter of the phosphoenolpyruvate phosphotransferase system (EIICBGlc) is regulated similarly [22]. Under sugar-phosphate stress, when glucose 6-phosphate and its derivatives are accumulated in cells, sRNA SgrS is induced with participation of SgrR protein. SgrS RNA, in turn, forms a complex with RNase E, Hfq protein, and a partially complementary ptsG mRNA region; this leads to inhibition of initiation the mRNA translation. Then, the mRNA is degraded by RNase E [23-25].
An interesting mechanism of negative translation regulation of polycistronic templates by Hfq and sRNA chiPQ exists in E. coli. It is known that in bacterial cells transcription and translation of mRNA are coupled and proceed simultaneously. During complementary pairing of sRNA ChiX with its target mRNA chiPQ, the ribosomal binding site on the mRNA is hidden, which triggers termination of the mRNA transcription by providing access for transcription terminator Rho. Therefore, the next gene is not transcribed, and the mRNA–sRNA complex is degraded by RNase E [26].
Another mechanism exists for regulation of translation of polycistronic templates with involvement of Hfq and sRNA Spot42 in sugar metabolism. This sRNA is encoded by the spf gene, which is inhibited by the complex of receptor protein SRP with cyclic AMP (cAMP). The galETKM operon has two overlapping promoters, P1 and P2. The cAMP–CRP complex stimulates transcription from the first promoter and inhibits transcription from the second promoter P2. Promoter 1 drives expression of UDP-galactose epimerase (galE gene product) and galactokinase (galK gene product). While galactokinase is only required under glucose shortage to cleave galactose, UDP-galactose epimerase plays a role not only under glucose shortage, but it also participates in other cell growth conditions in UDP-glucose synthesis, which is a building block for the cell wall and the capsule.
Spot42 sRNA is partially complementary to the translation initiation region of the galK gene, and upon binding, it inhibits galK translation by hiding its ribosomal binding site. At the same time, it does not inhibit translation of galE, i.e. causes discoordination of expression of the gal operon genes [27, 28]. Beside the gal operon, Spot42 regulates at least 14 other operons encoding proteins required for assimilation of alternative carbon sources. Some of them are also back regulated by the cAMP–CRP complex [29].
Examples of positive translation regulation. Three sRNAs (DsrA, ArcZ, and RprA) can bind to the 5′-untranslated region of RpoS mRNA, which causes its unwinding and facilitates ribosome binding [28, 30]. The dsrA promoter is activated at lower temperatures, below 30°C [31, 32] it activates expression of σ factor RpoS, which in turn activates expression of the otsA and otsB genes. These genes regulate levels of the disaccharide trehalose (mycose), which protects cell at low (about 4°C) temperatures [33]. Furthermore, sRNA DsrA negatively regulates expression of the hns gene encoding global transcriptional regulator HNS. It is known that HNS negatively regulates genes responsible for protection of cells against osmotic stress. Hence, DsrA removes negative control by HNS and triggers expression of not only cold shock proteins, but also proteins that protect against osmotic shock. By the example of sRNA DsrA, it was shown that Hfq accelerates formation of DsrA-RpoS by a factor of 30-50 [34] and protects RpoS against degradation [35]. A second sRNA that affects translation levels of rpoS mRNA, RprA, was discovered as a suppressor in cells that lack sRNA DsrA. Nevertheless, a signal that activates sRNA RprA is not yet identified [28]. Translation inhibition by a third sRNA (ArcZ) is mediated by the ArcA/ArcB system and occurs under oxygen deficiency in anaerobic conditions, while negative control is removed in the presence of oxygen in the medium [36].
ROLE OF Hfq PROTEIN IN FUNCTIONING OF SMALL RNAs
Translation regulation of bacterial mRNAs by antisense RNAs is well documented for plasmids, phages, and transposons. In these cases, a regulatory RNA (sRNA) and a target RNA are encoded in the same chromosomal locus but in opposite directions. Hence, regulatory RNA is fully complementary to an mRNA region. Here, Hfq protein is not required for functioning sRNA [37]. Hfq is necessary for other types of regulatory RNAs that show imperfect complementarity to the target RNAs, the so-called trans-encoded RNAs. These RNAs are expressed “in trans”, i.e. genes encoding these RNAs are located far from genes encoding their target messenger RNAs. It was shown that Hfq protein accelerates duplex formation between partially complementary RNAs [38-42]. Acceleration of duplex formation can occur both due to local increase in RNA concentration in the vicinity of Hfq protein during their interaction, and due to the ability of Hfq to unwind double-stranded RNAs [28, 43, 44]. Furthermore, it is also known that Hfq protects some short-lived sRNAs from degradation by RNase E (e.g. DsrA, Spot 42, and RyhB) [41, 45, 46].
In addition to regulation of mRNA translation by means of regulatory RNAs, Hfq can directly change mRNA translation levels by affecting mRNA half-life. Destabilization of mRNA in the presence of Hfq is best studied for mRNA ompA. This mRNA encodes OmpA, a major protein of the outer membrane of E. coli. Stability of this mRNA is inversely proportional to the cell growth rate. The half-life of mRNA is determined by the 5′-untranslated region, which contains a stabilizing hairpin structure and a site for RNase E cleavage. In fast growing cells, ribosomes upon binding to the 5′-untranslated region protect it against degradation by RNase E. In slow growing cells (in the stationary phase), Hfq protein accumulates, which competes with ribosomes for the 5′-untranslated region of mRNA, impedes ribosome binding, and this results in mRNA degradation due to uncovering of the RNase E binding site [46-48].
Hfq has three RNA-binding sites that upon polymerization form two main binding regions – a U-rich RNA-binding site on the proximal side of a toroid and a region for interaction with poly(A) stretches of RNAs on the distal side. Several structures of Hfq complexes with short RNA-oligomers (PDBID 4HT8, 4HT9, 3QSU, 3RER, 3AHU, 3HSB, 3GIB, 1KQ2) have been determined, and a low resolution structure of Hfq in complex with sRNA RydC form Salmonella sp. was obtained (PDB ID 4V2S) [49].
Structures of complexes of Hfq with oligo(U) RNA fragments revealed details of interaction of U-rich RNAs with the distal surface of the protein near the central pore of a hexamer [50, 51]. These are structures of Hfq from Staphylococcus aureus in complex with AU5G RNA [52], Hfq from Salmonella typhimurium in complex with U6 RNA [53], and Hfq from E. coli in complex with AU6A RNA [54].
A uracil-binding site is formed by amino acid residues of two adjacent subunits of the hexamer. Upon binding to the protein, uracil nucleobase stacks to a benzene ring of Phe42, while the O2 and O4 atoms of uracil form hydrogen bonds with atoms of side chains of Gln8 and Gln41, correspondingly [53]. It is worth mentioning that there are certain differences between the three structures of Hfq complexes with U-rich RNAs. In the complex of Hfq from S. typhimurium, the asymmetric unit contains one monomer of the protein and one uridine with symmetrical phosphate residues. Moreover, free oxygens of the phosphates do not form bonds with the protein, being directed toward the central pore [53]. In the structure of the complex Hfq S. aureus with AU5G RNA, free oxygens of a phosphate group are faced away from the central pore and form hydrogen bonds with the NH2-group of His56 [52].
The latest structure of the complex of Hfq with U-rich RNAs [54] demonstrated that RNA binding to the proximal side of a toroid might be labile. A region of DsrA RNA containing the AU6A sequence was utilized for crystallization. It turned out that in the obtained structure not all nucleotides are localized in the uridine-binding pocket: two of six uridines (U30 and U33) do not contact the protein and face away from the protein surface. One uridine (U29) was bound close to but not exactly in the pocket where it should have formed stack with Phe42. The sugar-phosphate backbone of RNA is directed either toward the central cavity (as in Hfq complexed with the U6 RNA) and away from the central cavity (as in Hfq complexed with the AU5G RNA).
The structure of Hfq from E. coli with a 15-nt oligo(A) RNA was first determined in 2009 [55]. It showed that oligo(A) binds to the distal surface of the protein by means of a repeated tripartite motif referred to as the A-R-E RNA binding motif. Initially, it was proposed that the A-site corresponds to a specific adenine-binding pocket, the R-site corresponds to a purine-binding pocket, while the E-site is a non-discriminating nucleotide-binding site, since the nucleobase placed in this site does not interact with atoms of the protein [55]. However, later it was demonstrated that, in fact, the adenosine specificity site is the R-site [54, 56]. At the same time, another purine, guanine, does not bind to Hfq either in the R-site or in the A-site; but cytidine can bind to the “purine specificity” R-site [53, 57].
The distal A-binding site of E. coli Hfq is situated between adjacent subunits of the Hfq toroid. Upon RNA binding to the protein, hydrogen bonds are formed between Gln33 and the N7 and N6 atoms of the adenine base, the Gln53 residue and N1 atom of the adenine base, and the Lys31 residue and phosphate group of the adenosine. In addition, a hydrophobic contact arises between Leu32 and the adenine base [55].
The binding of RNA to the protein R-site is realized through stacking interactions between Tyr25 and the adenine base and through hydrophobic interactions of the latter with Leu26, Ile30, and Leu32. Change of Tyr25 to alanine causes 100-fold decrease in affinity between RNA and Hfq [58]. Hydrophobic interactions are supplemented with formation of hydrogen bonds between the side chain of Gln52 and the N6 atom of the adenine base, between Thr61 and N1 of the adenine base, and between Asn28 and the N3 and Nδ atoms of the adenine base [55].
In Gram-negative bacteria, binding of A-rich oligo RNAs occurs almost identically. Simultaneous binding of AU6A and A7 in a complex with the E. coli protein revealed that formation of the triple complex does not affect the position of RNA or the protein structure [59].
However, nothing was known about structure of RNA/Hfq complexes from Gram-positive bacteria. In Gram-positive bacteria, Hfq does not play such an important role, e.g. hfq gene knockout in S. aureus and Listeria monocytogenes does not change the phenotype of these bacteria, partially because of the absence of RNase E [37]. The first structure of Hfq from the Gram-positive bacterium Bacillus subtilis in complex with (AG)3A RNA was determined in 2011 [60]. It turned out that binding of A-rich RNAs to the surface of Hfq in Gram-positive bacteria differs from that in Gram-negative bacteria. Bacillus subtilis Hfq does not bind AU5G RNA. It has less affinity to A18 RNA as compared to Hfq proteins from Gram-negative bacteria, but it specifically binds AG-repeat-containing RNA. Instead of three binding pockets (A-R-E), only two are present, which were referred to as the A-L motif. The adenine base binds to the protein surface at the A-pocket, which is analogous to the R-pocket in Gram-negative bacteria. Binding occurs by means of stacking interactions of adenine with two phenylalanines of the protein, Phe24 and Phe29. Binding is further stabilized by hydrogen bonds between adenine and the Ser60 and Thr61 protein residues. The guanine of the (AG)3A RNA in complex with Hfq forms hydrogen bonds with Arg32, which is a strictly conserved amino acid residue among Hfq proteins in Gram-positive bacteria [60].
SPECIFICATION OF THE INTERACTION MODEL OF Hfq PROTEIN WITH
MESSENGER AND REGULATORY RNAs
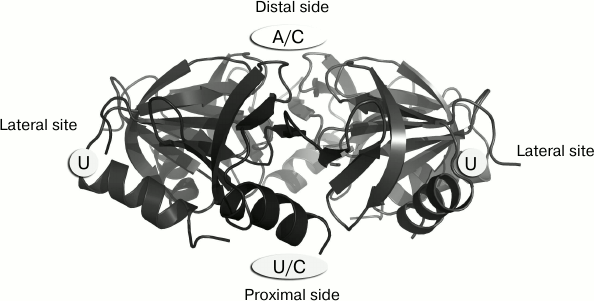
A complete model of Hfq–RNA interaction was missing for a long time due to the impossibility to obtain stable complexes of the protein with full length regulatory and messenger RNAs. To build models of interaction of Hfq protein with regulatory RNAs [61-63], data from the structures of Hfq complexed with short RNA fragments or with single nucleotides was used. It turned out that since Hfq protein binds single-stranded RNAs, it is capable of binding single nucleotides in the RNA-binding sites [64]. Complexes of the protein with UTP, CTP, and ATP that were obtained revealed structural details of the interaction of uridine in the Hfq protein lateral site whose existence was shown biochemically [65], and demonstrating the ability of Hfq to bind cytidines in both general RNA-interaction sites (Fig. 1). It is worth mentioning that, despite all efforts, complexes of the protein with guanine were not obtained. This is in agreement with biochemical data showing low affinity of Hfq to guanine-containing RNA sequences [64].
Fig. 1. Nucleotide specificity and arrangement of RNA-binding sites on the surface of Hfq hexamers.
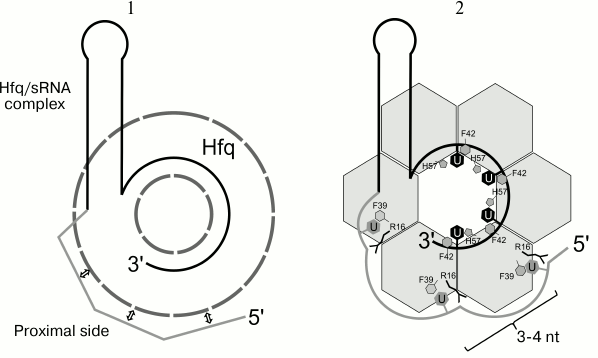
Many regulatory RNAs that interact with Hfq have polyuridylic sequences at both the 3′- and 5′-ends. It is assumed that Hfq protein first recognizes a U-rich sequence at the 3′-end of sRNA via the proximal site [66], then the 5′-end of sRNA binds to the repeated lateral binding sites of the protein [62] (Fig. 2). The uracils contacting the lateral binding site may be responsible for mRNA interaction [67]. The outcome of our work provided the initial model. We suggest that not less than four-five successive uridines should be present at the sRNA 3′-end. They could be partially replaced by cytosines. A sequence containing uridines in every 4th or 5th position should be present at the 5′-end for interaction with the Hfq lateral sites [64]. The data obtained were used later to build a model of interaction of Hfq with rpoS sRNA [63], and then were confirmed by the structure of the E. coli Hfq in complex with RydC sRNA from Salmonella sp. [49] (Fig. 3).
Fig. 2. Proposed model for arrangement of small regulatory RNAs on the Hfq protein surface. 1) Initial model based on mutagenesis data [62]. 2) Model for sRNA binding specified based on our structural data regarding UTP position on the Hfq surface. For the protein binding, the sRNA should have at the 3′-end four or five successive uridines, which could be partially replaced by cytidines. At the 5′-end, uridine should be present in every 3-4 nt.
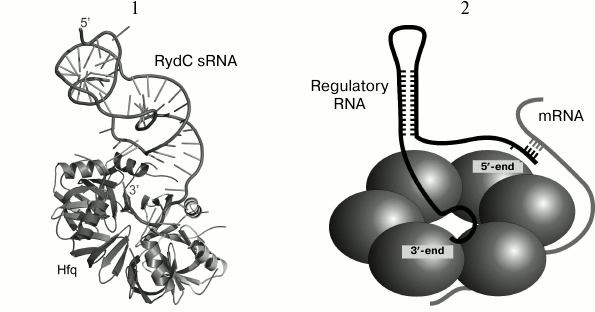
Fig. 3. 1) Structure of RydC sRNA–Hfq complex (HDB ID 4V2S). The Hfq hexamer is shown as a cartoon model, RydC sRNA is shown as a ribbon with indicated base directions (sticks). The 5′- and 3′-ends of sRNA are denoted. The 3′-end of RydC sRNA forms a network of contacts with amino acid residues of the proximal protein surface at the central part of the hexamer. Conserved uridines U23/24 and U46/47 of the sRNA interact with the lateral protein region. The 5′-end of RydC RNA forms additional contacts with the lateral part of the hexamer of the symmetrically bound Hfq protein molecule (not shown). 2) Proposed model for interaction of RydC sRNA and cfa mRNA from Salmonella sp. on the Hfq protein surface (a simplified scheme from [49]). The 3′-end of RydC sRNA (black) interacts with the protein central pore on the Hfq proximal surface. The cfa mRNA (gray) is associated with the distal Hfq surface and forms a duplex with the 5′-end of RydC (concurrently bound to Hfq) at the lateral protein surface. It has been suggested that the C-terminal part of Hfq directed towards a contact between the two RNAs and stabilizes their interaction.
An important role of small regulatory RNAs in regulation of biosynthesis in bacterial cells is now beyond doubt. An interesting feature of these processes is the participation of the bacterial protein Hfq that acts as an RNA chaperone and promotes interaction of sRNAs with mRNA targets. The unique feature of this hexameric protein is the presence of three RNA-interacting sites on its surface: one uridine-specific site, a second – adenine-specific site, and an additional site that is required for realization of interaction of two RNA molecules. The regulation of protein synthesis with participation of sRNAs and Hfq protein in different bacteria is extensively studied, and we still expect many interesting discoveries.
This work was supported by the Russian Science Foundation (project No. 14-14-00496).
REFERENCES
1.Dahm, R. (2005) Friedrich Miescher and the
discovery of DNA, Dev. Biol., 278, 274-288.
2.Portin, P. (2014) The birth and development of the
DNA theory of inheritance: sixty years since the discovery of the
structure of DNA, J. Genet., 93, 293-302.
3.Sanger, F., Nicklen, S., and Coulson, A. R. (1977)
DNA sequencing with chain-terminating inhibitors, Proc. Natl. Acad.
Sci. USA, 74, 5463-5467.
4.Ikekawa, F., and Ikekawa, S. (2001) Fruits of human
genome project and private venture, and their impact on life science,
Yakugaku Zasshi, 121, 845-873.
5.Hattori, M. (2005) Finishing the euchromatic
sequence of the human genome, Tanpakushitsu Kakusan Koso,
50, 162-168.
6.Ohno, S. (1972) So much “junk” DNA in
our genome, Brookhaven Symp. Biol., 23,
366-370.
7.Palazzo, A. F., and Gregory, T. R. (2014) The case
for junk DNA, PLoS Genet., 10, e1004351.
8.Tomizawa, J., and Som, T. (1984) Control of ColE1
plasmid replication: enhancement of binding of RNA I to the primer
transcript by the Rom protein, Cell, 38, 871-878.
9.Simons, R. W., and Kleckner, N. (1983)
Translational control of IS10 transposition, Cell, 34,
683-691.
10.Lee, R. C., Feinbaum, R. L., and Ambro, V. (1993)
The C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14, Cell, 75,
843-854.
11.Babski, J., Maier, L.-K., Heyer, R., Jaschinski,
K., Prasse, D., Jager, D., Randau, L., Schmitz, R. A., Marchfelder, A.,
and Soppa, J. (2014) Small regulatory RNAs in Archaea, RNA
Biol., 11, 484-493.
12.Prasse, D., Ehlers, C., Backofen, R., and
Schmitz, R. A. (2013) Regulatory RNAs in archaea: first target
identification in Methanoarchaea, Biochem. Soc. Trans.,
41, 344-349.
13.Jager, D., Sharma, C. M., Thomsen, J., Ehlers,
C., Vogel, J., and Schmitz, R. A. (2009) Deep sequencing analysis of
the Methanosarcina mazei Gö1 transcriptome in response to
nitrogen availability, Proc. Natl. Acad. Sci. USA,
106, 21878-21882.
14.Livny, J., and Waldor, M. K. (2007)
Identification of small RNAs in diverse bacterial species, Curr.
Opin. Microbiol., 10, 96-101.
15.Altuvia, S. (2007) Identification of bacterial
small non-coding RNAs: experimental approaches, Curr. Opin.
Microbiol., 10, 257-261.
16.Gottesman, S., and Storz, G. (2011) Bacterial
small RNA regulators: versatile roles and rapidly evolving variations,
Cold Spring Harb. Perspect. Biol., 3, 1-16.
17.Marzi, S., and Romby, P. (2012) RNA mimicry, a
decoy for regulatory proteins, Mol. Microbiol., 83,
1-6.
18.Gottesman, S., and Storz, G. (2015) RNA
reflections: converging on Hfq, RNA, 21, 511-512.
19.Zhang, A., Altuvia, S., Tiwari, A., Argaman, L.,
Hengge-Aronis, R., and Storz, G. (1998) The OxyS regulatory RNA
represses rpoS translation and binds the Hfq (HF-I) protein, EMBO
J., 17, 6061-6068.
20.Massу, E., and Gottesman, S. (2002) A small
RNA regulates the expression of genes involved in iron metabolism in
Escherichia coli, Proc. Natl. Acad. Sci. USA, 99,
4620-4625.
21.Massу, E., Escorcia, F. E., and Gottesman,
S. (2003) Coupled degradation of a small regulatory RNA and its mRNA
targets in Escherichia coli, Genes Dev., 17,
2374-2383.
22.Vanderpool, C. K., and Gottesman, S. (2004)
Involvement of a novel transcriptional activator and small RNA in
posttranscriptional regulation of the glucose phosphoenolpyruvate
phosphotransferase system, Mol. Microbiol., 54,
1076-1089.
23.Morita, T., Maki, K., and Aiba, H. (2005) RNase
E-based ribonucleoprotein complexes: mechanical basis of mRNA
destabilization mediated by bacterial noncoding RNAs, Genes
Dev., 19, 2176-2186.
24.Morita, T., Mochizuki, Y., and Aiba, H. (2006)
Translational repression is sufficient for gene silencing by bacterial
small noncoding RNAs in the absence of mRNA destruction, Proc. Natl.
Acad. Sci. USA, 103, 4858-4863.
25.Aiba, H. (2007) Mechanism of RNA silencing by
Hfq-binding small RNAs, Curr. Opin. Microbiol., 10,
134-139.
26.De Lay, N., Schu, D. J., and Gottesman, S. (2013)
Bacterial small RNA-based negative regulation: Hfq and its accomplices,
J. Biol. Chem., 288, 7996-8003.
27.Moller, T., Franch, T., Hojrup, P., Keene, D. R.,
Bachinger, H. P., Brennan, R. G., and Valentin-Hansen, P. (2002) Hfq: a
bacterial Sm-like protein that mediates RNA–RNA interaction,
Mol. Cell, 9, 23-30.
28.Gottesman, S. (2004) The small RNA regulators of
Escherichia coli: roles and mechanisms, Annu. Rev.
Microbiol., 58, 303-328.
29.Beisel, C. L., Updegrove, T. B., Janson, B. J.,
and Storz, G. (2012) Multiple factors dictate target selection by
Hfq-binding small RNAs, EMBO J., 31, 1961-1974.
30.Henderson, C. A., Vincent, H. A., Casamento, A.,
Stone, C. M., Phillips, J. O., Cary, P. D., Sobott, F., Gowers, D. M.,
Taylor, J. E., and Callaghan, A. J. (2013) Hfq binding changes the
structure of Escherichia coli small noncoding RNAs OxyS and
RprA, which are involved in the riboregulation of rpoS, RNA,
19, 1089-1104.
31.Lease, R. A., Cusick, M. E., and Belfort, M.
(1998) Riboregulation in Escherichia coli: DsrA RNA acts by
RNA–RNA interactions at multiple loci, Proc. Natl. Acad. Sci.
USA, 95, 12456-12461.
32.Majdalani, N., Cunning, C., Sledjeski, D.,
Elliott, T., and Gottesman, S. (1998) DsrA RNA regulates translation of
RpoS message by an anti-antisense mechanism, independent of its action
as an antisilencer of transcription, Proc. Natl. Acad. Sci. USA,
95, 12462-12467.
33.Kandror, O., DeLeon, A., and Goldberg, A. L.
(2002) Trehalose synthesis is induced upon exposure of Escherichia
coli to cold and is essential for viability at low temperatures,
Proc. Natl. Acad. Sci. USA, 99, 9727-9732.
34.Soper, T. J., and Woodson, S. A. (2008) The rpoS
mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA,
RNA, 14, 1907-1917.
35.McCullen, C. A., Benhammou, J. N., Majdalani, N.,
and Gottesman, S. (2010) Mechanism of positive regulation by DsrA and
RprA small noncoding RNAs: pairing increases translation and protects
rpoS mRNA from degradation, J. Bacteriol., 192,
5559-5571.
36.Mandin, P., and Gottesman, S. (2010) Integrating
anaerobic/aerobic sensing and the general stress response through the
ArcZ small RNA, EMBO J., 29, 3094-3107.
37.Nielsen, J. S., Lei, L. K., Ebersbach, T., Olsen,
A. S., Klitgaard, J. K., Valentin-Hansen, P., and Kallipolitis, B. H.
(2010) Defining a role for Hfq in Gram-positive bacteria: evidence for
Hfq-dependent antisense regulation in Listeria monocytogenes,
Nucleic Acids Res., 38, 907-919.
38.Geissmann, T. A., and Touati, D. (2004) Hfq, a
new chaperoning role: binding to messenger RNA determines access for
small RNA regulator, EMBO J., 23, 396-405.
39.Kawamoto, H., Koide, Y., Morita, T., and Aiba, H.
(2006) Base-pairing requirement for RNA silencing by a bacterial small
RNA and acceleration of duplex formation by Hfq, Mol.
Microbiol., 61, 1013-1022.
40.Rasmussen, A. A., Eriksen, M., Gilany, K.,
Udesen, C., Franch, T., Petersen, C., and Valentin-Hansen, P. (2005)
Regulation of ompA mRNA stability: the role of a small regulatory RNA
in growth phase-dependent control, Mol. Microbiol., 58,
1421-1429.
41.Sledjeski, D. D., Gupta, A., and Gottesman, S.
(1996) The small RNA, DsrA, is essential for the low temperature
expression of RpoS during exponential growth in Escherichia
coli, EMBO J., 15, 3993-4000.
42.Chen, S., Zhang, A., Blyn, L. B., and Storz, G.
(2004) MicC, a second small-RNA regulator of Omp protein expression in
Escherichia coli, J. Bacteriol., 186,
6689-6697.
43.Storz, G., Opdyke, J. A., and Zhang, A. (2004)
Controlling mRNA stability and translation with small, noncoding RNAs,
Curr. Opin. Microbiol., 7, 140-144.
44.Vassilieva, I. M., and Garber, M. B. (2002) The
regulatory role of the Hfq protein in bacterial cells, Mol.
Biol. (Moscow), 36, 785-791.
45.Masse, E., Escorcia, F. E., and Gottesman, S.
(2003) Coupled degradation of a small regulatory RNA and its mRNA
targets in Escherichia coli, Genes Dev., 17,
2374-2383.
46.Moll, I., Afonyushkin, T., Vytvytska, O., and
Kaberdin, V. R. (2003) Coincident Hfq binding and RNase E cleavage
sites on mRNA and small regulatory RNAs, RNA, 9,
1308-1314.
47.Vytvytska, O., Moll, I., Kaberdin, V. R., Von
Gabain, A., and Blasi, U. (2000) Hfq (HF1) stimulates ompA mRNA decay
by interfering with ribosome binding, Genes Dev., 14,
1109-1118.
48.Valentin-Hansen, P., and Eriksen, M. (2004)
MicroReview. The bacterial Sm-like protein Hfq: a key player in RNA
transactions, Mol. Microbiol., 51, 1525-1533.
49.Dimastrogiovanni, D., Frohlich, K. S., Bandyra,
K. J., Bruce, H. A., Hohensee, S., Vogel, J., and Luisi, B. F. (2014)
Recognition of the small regulatory RNA RydC by the bacterial Hfq
protein, Elife, 3, e05375.
50.Kovach, A. R., Hoff, K. E., Canty, J. T., Orans,
J., and Brennan, R. G. (2014) Recognition of U-rich RNA by Hfq from the
Gram-positive pathogen Listeria monocytogenes, RNA,
20, 1548-1559.
51.Murina, V. N., and Nikulin, A. D. (2011) RNA
binding Sm-Like proteins of Bacteria and Archaea. Similarity and
difference in structure and function, Biochemistry (Moscow),
76, 1434-1449.
52.Schumacher, M. A., Pearson, R. F., Moller, T.,
Valentin-Hansen, P., and Brennan, R. G. (2002) Structures of the
pleiotropic translational regulator Hfq and an Hfq–RNA complex: a
bacterial Sm-like protein, EMBO J., 21, 3546-3556.
53.Sauer, E., and Weichenrieder, O. (2011)
Structural basis for RNA 3′end recognition by Hfq, Proc.
Natl. Acad. Sci. USA, 108, 13065-13070.
54.Wang, W., Wang, L., Zou, Y., Zhang, J., Gong, Q.,
Wu, J., and Shi, Y. (2011) Cooperation of Escherichia coli Hfq
hexamers in DsrA binding, Genes Dev., 25, 2106-2117.
55.Link, T. M., Valentin-Hansen, P., and Brennan, R.
G. (2009) Structure of Escherichia coli Hfq bound to
polyriboadenylate RNA, Proc. Natl. Acad. Sci. USA,
106, 19292-19297.
56.Hammerle, H., Beich-Frandsen, M., Vecerek, B.,
Rajkowitsch, L., Carugo, O., Djinovic-Carugo, K., and Blasi, U. (2012)
Structural and biochemical studies on ATP binding and hydrolysis by the
Escherichia coli RNA chaperone Hfq, PLoS One,
7, e50892.
57.Murina, V. N., Melnik, B. S., Filimonov, V. V.,
Uhlein, M., Weiss, M. S., Muller, U., and Nikulin, A. D. (2014) Effect
of conserved intersubunit amino acid substitutions on Hfq protein
structure and stability, Biochemistry (Moscow), 79,
469-477.
58.Arluison, V., Mutyam, S. K., Mura, C., Marco, S.,
and Sukhodolets, M. V. (2007) Sm-like protein Hfq: location of the
ATP-binding site and the effect of ATP on Hfq–RNA complexes,
Protein Sci., 16, 1830-1841.
59.Wang, W., Wang, L., Wu, J., Gong, Q., and Shi, Y.
(2013) Hfq-bridged ternary complex is important for translation
activation of rpoS by DsrA, Nucleic Acids Res., 41,
5938-5948.
60.Someya, T., Baba, S., Fujimoto, M., Kawai, G.,
and Kumasaka, T. (2012) Crystal structure of Hfq from Bacillus
subtilis in complex with SELEX-derived RNA aptamer: insight into
RNA-binding properties of bacterial Hfq, Nucleic Acids Res.,
40, 1856-1867.
61.Robinson, K. E., Orans, J., Kovach, A. R., Link,
T. M., and Brennan, R. G. (2013) Mapping Hfq–RNA interaction
surfaces using tryptophan fluorescence quenching, Nucleic Acids
Res., 42, 1-14.
62.Sauer, E. (2013) Structure and RNA-binding
properties of the bacterial LSm protein Hfq, RNA Biol.,
10, 610-618.
63.Peng, Y., Curtis, J. E., Fang, X., and Woodson,
S. A. (2014) Structural model of an mRNA in complex with the bacterial
chaperone Hfq, Proc. Natl. Acad. Sci. USA, 111,
17134-17139.
64.Murina, V., Lekontseva, N., and Nikulin, A.
(2013) Hfq binds ribonucleotides in three different RNA-binding sites,
Acta Crystallogr. D Biol. Crystallogr., 69,
1504-1513.
65.Panja, S., Schu, D. J., and Woodson, S. A. (2013)
Conserved arginines on the rim of Hfq catalyze base pair formation and
exchange, Nucleic Acids Res., 41, 7536-7546.
66.Wilusz, C. J., and Wilusz, J. (2013) Lsm proteins
and Hfq: life at the 3′-end, RNA Biol., 10,
592-601.
67.Sauer, E., Schmidt, S., and Weichenrieder, O.
(2012) Small RNA binding to the lateral surface of Hfq hexamers and
structural rearrangements upon mRNA target recognition, Proc. Natl.
Acad. Sci. USA, 109, 9396-93401.